

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
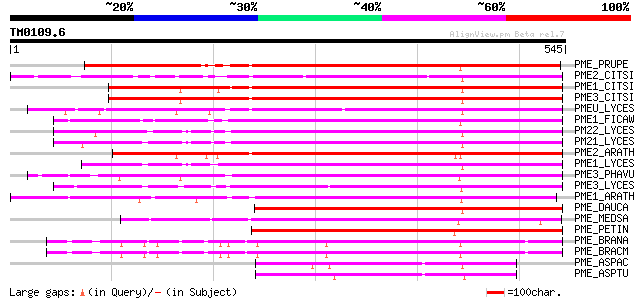
Query= TM0109.6
(545 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
PME_PRUPE (Q43062) Pectinesterase PPE8B precursor (EC 3.1.1.11) ... 474 e-133
PME2_CITSI (O04887) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 424 e-118
PME1_CITSI (O04886) Pectinesterase 1 precursor (EC 3.1.1.11) (Pe... 403 e-112
PME3_CITSI (P83948) Pectinesterase 3 precursor (EC 3.1.1.11) (Pe... 400 e-111
PMEU_LYCES (Q43143) Pectinesterase U1 precursor (EC 3.1.1.11) (P... 397 e-110
PME1_FICAW (P83947) Pectinesterase precursor (EC 3.1.1.11) (Pect... 396 e-110
PM22_LYCES (Q96575) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 394 e-109
PM21_LYCES (P09607) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 394 e-109
PME2_ARATH (Q42534) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 390 e-108
PME1_LYCES (P14280) Pectinesterase 1 precursor (EC 3.1.1.11) (Pe... 385 e-106
PME3_PHAVU (Q43111) Pectinesterase 3 precursor (EC 3.1.1.11) (Pe... 381 e-105
PME3_LYCES (Q96576) Pectinesterase 3 precursor (EC 3.1.1.11) (Pe... 362 2e-99
PME1_ARATH (Q43867) Pectinesterase 1 precursor (EC 3.1.1.11) (Pe... 344 4e-94
PME_DAUCA (P83218) Pectinesterase (EC 3.1.1.11) (Pectin methyles... 338 2e-92
PME_MEDSA (Q42920) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 300 5e-81
PME_PETIN (Q43043) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 295 2e-79
PME_BRANA (P41510) Probable pectinesterase precursor (EC 3.1.1.1... 254 3e-67
PME_BRACM (Q42608) Pectinesterase (EC 3.1.1.11) (Pectin methyles... 248 3e-65
PME_ASPAC (Q12535) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 125 2e-28
PME_ASPTU (P17872) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 118 4e-26
>PME_PRUPE (Q43062) Pectinesterase PPE8B precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 522
Score = 474 bits (1219), Expect = e-133
Identities = 237/479 (49%), Positives = 322/479 (66%), Gaps = 28/479 (5%)
Query: 74 SAALRTTLNEAQGAIDNLTKFSTFSVSNREQIAIGDCKELVDFSVSELAWSLGEMRRIRA 133
+ +L+ T++ Q L++F+ R AI DC +L+DFS EL WSL + +
Sbjct: 57 AGSLKDTIDAVQQVASILSQFANAFGDFRLANAISDCLDLLDFSADELNWSLSASQNQKG 116
Query: 134 GDTSV-QYEGNLEAWLSAALSNQDTCLEGFEGTDRRLESYISGSLTQVTQLISNVLSLYT 192
+ S + +L WLSAAL NQDTC GFEGT+ ++ IS L QVT L+ +L T
Sbjct: 117 KNNSTGKLSSDLRTWLSAALVNQDTCSNGFEGTNSIVQGLISAGLGQVTSLVQELL---T 173
Query: 193 QLHALPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKPHRSRADAVVALDGSGHY 252
Q+H P +N + + P W+ D++LL++ DA+VA DG+G++
Sbjct: 174 QVH------PNSNQQGPN------GQIPSWVKTKDRKLLQADG--VSVDAIVAQDGTGNF 219
Query: 253 RTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTIITSNRNFMQ 312
+T+AV AAP +S RRYVIY+K+G Y+ENV++K+K N+M++GDG+ TII+ NR+F+
Sbjct: 220 TNVTDAVLAAPDYSMRRYVIYIKRGTYKENVEIKKKKWNLMMIGDGMDATIISGNRSFVD 279
Query: 313 GWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLY 372
GWTTFR+AT AVSG+GFIARD++F NTAGP HQAVALR DSD S FYRC+I G+QDTLY
Sbjct: 280 GWTTFRSATFAVSGRGFIARDITFENTAGPEKHQAVALRSDSDLSVFYRCNIRGYQDTLY 339
Query: 373 AHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGF 432
H++RQFYR+C+I GT+DFIFG+ V QNC+I + LP QK ++TAQGRK P++ TG
Sbjct: 340 THTMRQFYRDCKISGTVDFIFGDATVVFQNCQILAKKGLPNQKNSITAQGRKDPNEPTGI 399
Query: 433 TIQDSFILA----------TQPTYLGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDFAL 482
+IQ I A + PTYLGRPWK YSRTV + +++S +++P GWLEW GDFAL
Sbjct: 400 SIQFCNITADSDLEAASVNSTPTYLGRPWKLYSRTVIMQSFLSNVIRPEGWLEWNGDFAL 459
Query: 483 TTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLPGTGVKFTA 541
+L+YGEY NYGPGA L RVKWPGY + ++ A +TV +FI G+ WLP TGVK+TA
Sbjct: 460 NSLFYGEYMNYGPGAGLGSRVKWPGYQVFNESTQAKNYTVAQFIEGNLWLPSTGVKYTA 518
>PME2_CITSI (O04887) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 510
Score = 424 bits (1090), Expect = e-118
Identities = 249/553 (45%), Positives = 326/553 (58%), Gaps = 53/553 (9%)
Query: 1 MALVNALFILMMLLILPSSSKSFSDEIPSEIQTQDMQALIAQACMDIENQNSCLQNIHNE 60
MAL + + ++L L +S +S P E+++ + Q C Q + + +I +
Sbjct: 1 MALRILITVSLVLFSLSHTSFGYS---PEEVKSWCGKTPNPQPCEYFLTQKTDVTSIKQD 57
Query: 61 LTKIGPPSPTSVLSAALRTTLNEAQGAIDNLTKFSTFSVSNREQIAIGDCKELVDFSVSE 120
T +L+ L A A + + RE+ A DC+EL + +V +
Sbjct: 58 ---------TDFYKISLQLALERATTAQSRTYTLGSKCRNEREKAAWEDCRELYELTVLK 108
Query: 121 LAWSLGEMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCLEGFEGTDRRLESYISGSLTQ- 179
L + G T V + + WLS AL+N +TC E D + Y+ L+
Sbjct: 109 L----NQTSNSSPGCTKV----DKQTWLSTALTNLETCRASLE--DLGVPEYVLPLLSNN 158
Query: 180 VTQLISNVLSLYTQLHALPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKPHRSR 239
VT+LISN LSL + +P+ P FP W+ GD++LL++ P R
Sbjct: 159 VTKLISNTLSL----NKVPYNEP-----------SYKDGFPTWVKPGDRKLLQTTP---R 200
Query: 240 ADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGI 299
A+ VVA DGSG+ +TI EAV AA RYVIY+K G Y EN+++K K NIM VGDGI
Sbjct: 201 ANIVVAQDGSGNVKTIQEAVAAASRAGGSRYVIYIKAGTYNENIEVKLK--NIMFVGDGI 258
Query: 300 GQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAF 359
G+TIIT +++ G TTF++ATVAV G FIARD++ RNTAGP NHQAVALR SD S F
Sbjct: 259 GKTIITGSKSVGGGATTFKSATVAVVGDNFIARDITIRNTAGPNNHQAVALRSGSDLSVF 318
Query: 360 YRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVT 419
YRCS EG+QDTLY HS RQFYREC+IYGT+DFIFGN A VLQNC I+ R P P + T+T
Sbjct: 319 YRCSFEGYQDTLYVHSQRQFYRECDIYGTVDFIFGNAAVVLQNCNIFARKP-PNRTNTLT 377
Query: 420 AQGRKSPHQSTGFTIQDSFILATQ---------PTYLGRPWKQYSRTVYINTYMSGMVQP 470
AQGR P+QSTG I + + A T+LGRPWKQYSRTVYI T++ ++ P
Sbjct: 378 AQGRTDPNQSTGIIIHNCRVTAASDLKPVQSSVKTFLGRPWKQYSRTVYIKTFLDSLINP 437
Query: 471 RGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDS 530
GW+EW GDFAL TL+Y EY N GPG+S A RVKW GYH++ + S FTV FI G+S
Sbjct: 438 AGWMEWSGDFALNTLYYAEYMNTGPGSSTANRVKWRGYHVLTSPSQVSQFTVGNFIAGNS 497
Query: 531 WLPGTGVKFTAGL 543
WLP T V FT+GL
Sbjct: 498 WLPATNVPFTSGL 510
>PME1_CITSI (O04886) Pectinesterase 1 precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 584
Score = 403 bits (1036), Expect = e-112
Identities = 212/459 (46%), Positives = 295/459 (64%), Gaps = 16/459 (3%)
Query: 98 SVSNREQIAIGDCKELVDFSVSELAWSLGEMRRIRAGDTSVQYEGNLEAWLSAALSNQDT 157
+++ RE++A+ DC E +D ++ EL ++ ++ + Q+ +L+ +SAA++NQ T
Sbjct: 129 NLTKREKVALHDCLETIDETLDELHKAVEDLEEYPNKKSLSQHADDLKTLMSAAMTNQGT 188
Query: 158 CLEGFEGTD--RRLESYISGSLTQVTQLISNVLSLYTQLHALPFRPPR--NNTHKTSESL 213
CL+GF D + + +S V ++ SN L++ + R NN T E+
Sbjct: 189 CLDGFSHDDANKHVRDALSDGQVHVEKMCSNALAMIKNMTDTDMMIMRTSNNRKLTEETS 248
Query: 214 DLDSEFPEWMTEGDQELLRSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIY 273
+D +P W++ GD+ LL+S +AVVA DGSG+++T+ AV AAP +RY+I
Sbjct: 249 TVDG-WPAWLSPGDRRLLQSSS--VTPNAVVAADGSGNFKTVAAAVAAAPQGGTKRYIIR 305
Query: 274 VKKGLYRENVDMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARD 333
+K G+YRENV++ +K NIM +GDG +TIIT +RN + G TTF++AT AV G+GF+ARD
Sbjct: 306 IKAGVYRENVEVTKKHKNIMFIGDGRTRTIITGSRNVVDGSTTFKSATAAVVGEGFLARD 365
Query: 334 MSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIF 393
++F+NTAGP HQAVALRV +D SAFY C + +QDTLY HS RQF+ C I GT+DFIF
Sbjct: 366 ITFQNTAGPSKHQAVALRVGADLSAFYNCDMLAYQDTLYVHSNRQFFVNCLIAGTVDFIF 425
Query: 394 GNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSFILATQ---------P 444
GN AAVLQNC I+ R P QK VTAQGR P+Q+TG IQ S I AT P
Sbjct: 426 GNAAAVLQNCDIHARKPNSGQKNMVTAQGRTDPNQNTGIVIQKSRIGATSDLKPVQGSFP 485
Query: 445 TYLGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVK 504
TYLGRPWK+YSRTV + + ++ ++ P GW EW G+FAL TL+YGE++N G GA +GRVK
Sbjct: 486 TYLGRPWKEYSRTVIMQSSITDLIHPAGWHEWDGNFALNTLFYGEHQNSGAGAGTSGRVK 545
Query: 505 WPGYHIMKDAATASYFTVQRFIHGDSWLPGTGVKFTAGL 543
W G+ ++ A A FT FI G SWL TG F+ GL
Sbjct: 546 WKGFRVITSATEAQAFTPGSFIAGSSWLGSTGFPFSLGL 584
>PME3_CITSI (P83948) Pectinesterase 3 precursor (EC 3.1.1.11)
(Pectin methylesterase 3) (PE 3)
Length = 584
Score = 400 bits (1029), Expect = e-111
Identities = 209/458 (45%), Positives = 292/458 (63%), Gaps = 14/458 (3%)
Query: 98 SVSNREQIAIGDCKELVDFSVSELAWSLGEMRRIRAGDTSVQYEGNLEAWLSAALSNQDT 157
+++ RE++A+ DC E +D ++ EL ++ ++ + Q+ +L+ +SAA++NQ T
Sbjct: 129 NLTKREKVALHDCLETIDETLDELHKAVEDLEEYPNKKSLSQHADDLKTLMSAAMTNQGT 188
Query: 158 CLEGFEGTD--RRLESYISGSLTQVTQLISNVLSLYTQLHALPFRPPR-NNTHKTSESLD 214
CL+GF D + + +S V ++ SN L++ + R +N K E
Sbjct: 189 CLDGFSHDDANKHVRDALSDGQVHVEKMCSNALAMIKNMTDTDMMIMRTSNNRKLIEETS 248
Query: 215 LDSEFPEWMTEGDQELLRSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYV 274
+P W++ GD+ LL+S + VVA DGSG+++T+ +V AAP +RY+I +
Sbjct: 249 TVDGWPAWLSTGDRRLLQSSS--VTPNVVVAADGSGNFKTVAASVAAAPQGGTKRYIIRI 306
Query: 275 KKGLYRENVDMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDM 334
K G+YRENV++ +K NIM +GDG +TIIT +RN + G TTF++ATVAV G+GF+ARD+
Sbjct: 307 KAGVYRENVEVTKKHKNIMFIGDGRTRTIITGSRNVVDGSTTFKSATVAVVGEGFLARDI 366
Query: 335 SFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFG 394
+F+NTAGP HQAVALRV +D SAFY C + +QDTLY HS RQF+ C I GT+DFIFG
Sbjct: 367 TFQNTAGPSKHQAVALRVGADLSAFYNCDMLAYQDTLYVHSNRQFFVNCLIAGTVDFIFG 426
Query: 395 NGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSFILATQ---------PT 445
N AAVLQNC I+ R P QK VTAQGR P+Q+TG IQ S I AT PT
Sbjct: 427 NAAAVLQNCDIHARKPNSGQKNMVTAQGRADPNQNTGIVIQKSRIGATSDLKPVQGSFPT 486
Query: 446 YLGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKW 505
YLGRPWK+YSRTV + + ++ ++ P GW EW G+FAL TL+YGE++N G GA +GRVKW
Sbjct: 487 YLGRPWKEYSRTVIMQSSITDVIHPAGWHEWDGNFALNTLFYGEHQNAGAGAGTSGRVKW 546
Query: 506 PGYHIMKDAATASYFTVQRFIHGDSWLPGTGVKFTAGL 543
G+ ++ A A FT FI G SWL TG F+ GL
Sbjct: 547 KGFRVITSATEAQAFTPGSFIAGSSWLGSTGFPFSLGL 584
>PMEU_LYCES (Q43143) Pectinesterase U1 precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 583
Score = 397 bits (1020), Expect = e-110
Identities = 227/550 (41%), Positives = 329/550 (59%), Gaps = 34/550 (6%)
Query: 18 SSSKSFSDEIPSEIQTQDMQALIAQACMDIENQNSC---LQNIHNELTKIGPPSPTSVLS 74
S SK+ D + A++ AC + + C + N+ + K+ S V+
Sbjct: 44 SHSKNSDDHADIMAISSSAHAIVKSACSNTLHPELCYSAIVNVSDFSKKV--TSQKDVIE 101
Query: 75 AALRTTLNEAQG---AIDNLTKFSTFSVSNREQIAIGDCKELVDFSVSELAWSLGEMRRI 131
+L T+ + A+ L K + ++ RE++A+ DC E +D ++ EL ++ ++
Sbjct: 102 LSLNITVKAVRRNYYAVKELIK-TRKGLTPREKVALHDCLETMDETLDELHTAVEDLELY 160
Query: 132 RAGDTSVQYEGNLEAWLSAALSNQDTCLEGF--EGTDRRLESYISGSLTQVTQLISNVLS 189
+ ++ +L+ +S+A++NQ+TCL+GF + D+++ + V ++ SN L+
Sbjct: 161 PNKKSLKEHVEDLKTLISSAITNQETCLDGFSHDEADKKVRKVLLKGQKHVEKMCSNALA 220
Query: 190 LYTQL------HALPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKPHRSRADAV 243
+ + + + P NN ++ E+PEW++ GD+ LL+S D V
Sbjct: 221 MICNMTDTDIANEMKLSAPANNRKLVEDN----GEWPEWLSAGDRRLLQSST--VTPDVV 274
Query: 244 VALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTI 303
VA DGSG Y+T++EAV AP S++RYVI +K G+YRENVD+ +K TNIM +GDG TI
Sbjct: 275 VAADGSGDYKTVSEAVRKAPEKSSKRYVIRIKAGVYRENVDVPKKKTNIMFMGDGKSNTI 334
Query: 304 ITSNRNFMQGWTTFRTATVA-VSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRC 362
IT++RN G TTF +ATV V+GK +ARD++F+NTAG HQAVAL V SD SAFYRC
Sbjct: 335 ITASRNVQDGSTTFHSATVVRVAGK-VLARDITFQNTAGASKHQAVALCVGSDLSAFYRC 393
Query: 363 SIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQG 422
+ +QDTLY HS RQF+ +C + GT+DFIFGNGAAV Q+C I+ R P QK VTAQG
Sbjct: 394 DMLAYQDTLYVHSNRQFFVQCLVAGTVDFIFGNGAAVFQDCDIHARRPGSGQKNMVTAQG 453
Query: 423 RKSPHQSTGFTIQDSFILATQ---------PTYLGRPWKQYSRTVYINTYMSGMVQPRGW 473
R P+Q+TG IQ I AT PTYLGRPWK+YSRTV + + ++ ++QP GW
Sbjct: 454 RTDPNQNTGIVIQKCRIGATSDLRPVQKSFPTYLGRPWKEYSRTVIMQSSITDVIQPAGW 513
Query: 474 LEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLP 533
EW G+FAL TL+YGEY N G GA +GRVKW G+ ++ + A +T RFI G SWL
Sbjct: 514 HEWNGNFALDTLFYGEYANTGAGAPTSGRVKWKGHKVITSSTEAQAYTPGRFIAGGSWLS 573
Query: 534 GTGVKFTAGL 543
TG F+ GL
Sbjct: 574 STGFPFSLGL 583
>PME1_FICAW (P83947) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 545
Score = 396 bits (1017), Expect = e-110
Identities = 217/510 (42%), Positives = 304/510 (59%), Gaps = 24/510 (4%)
Query: 44 CMDIENQNSCLQNIHNELTKIGPPSPTSVLSAALRTTLNEAQGAIDNLTKFSTFSVSNRE 103
C N+ SCL I +E+T + ++L + L T Q A + S + +E
Sbjct: 48 CDQSVNKESCLAMI-SEVTGLNMADHRNLLKSFLEKTTPRIQKAFETANDASRRINNPQE 106
Query: 104 QIAIGDCKELVDFSVSELAWSLGEMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCLEGFE 163
+ A+ DC EL+D S + S+ + T+ +E +L WLS L+N TCL+G E
Sbjct: 107 RTALLDCAELMDLSKERVVDSISIL--FHQNLTTRSHE-DLHVWLSGVLTNHVTCLDGLE 163
Query: 164 -GTDRRLESYISGSLTQVTQLISNVLSLYTQLHALPFRPPRNNTHKTSESLDLDSEFPEW 222
G+ +++ + L ++ L+++ L P ++N + + FP W
Sbjct: 164 EGSTDYIKTLMESHLNELILRARTSLAIFVTLF-----PAKSNVIEP-----VTGNFPTW 213
Query: 223 MTEGDQELLRSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYREN 282
+T GD+ LL++ D VVA DGSG Y T+ EAV A P +S +R ++ V+ G+Y EN
Sbjct: 214 VTAGDRRLLQTLGKDIEPDIVVAKDGSGDYETLNEAVAAIPDNSKKRVIVLVRTGIYEEN 273
Query: 283 VDMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGP 342
VD + N+MLVG+G+ TIIT +RN + G TTF +ATVA G GFIA+D+ F+NTAGP
Sbjct: 274 VDFGYQKKNVMLVGEGMDYTIITGSRNVVDGSTTFDSATVAAVGDGFIAQDICFQNTAGP 333
Query: 343 VNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQN 402
+QAVALR+ +D++ RC I+ +QDTLY H+ RQFYR+ I GT+DFIFGN A V QN
Sbjct: 334 EKYQAVALRIGADETVINRCRIDAYQDTLYPHNYRQFYRDRNITGTVDFIFGNAAVVFQN 393
Query: 403 CKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSFILA---------TQPTYLGRPWKQ 453
C + R + Q+ T+TAQGR P+Q+TG +IQ+ I A T +YLGRPWK+
Sbjct: 394 CNLIPRKQMKGQENTITAQGRTDPNQNTGTSIQNCEIFASADLEPVEDTFKSYLGRPWKE 453
Query: 454 YSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKD 513
YSRTV + +Y+S ++ P GWLEW DFAL TL+YGEYRN GPG+ + RVKWPGYH++
Sbjct: 454 YSRTVVMESYISDVIDPAGWLEWDRDFALKTLFYGEYRNGGPGSGTSERVKWPGYHVITS 513
Query: 514 AATASYFTVQRFIHGDSWLPGTGVKFTAGL 543
A FTV I G SWL TGV +TAGL
Sbjct: 514 PEVAEQFTVAELIQGGSWLGSTGVDYTAGL 543
>PM22_LYCES (Q96575) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase 2) (PE 2)
Length = 550
Score = 394 bits (1012), Expect = e-109
Identities = 217/512 (42%), Positives = 305/512 (59%), Gaps = 29/512 (5%)
Query: 44 CMDIENQNSCLQNIHNELTKIGPPSPTSVLSAALRTTLN---EAQGAIDNLTKFSTFSVS 100
C ++ CL + + ++ + + LS ++ +N + AI ++K
Sbjct: 54 CKTAQDSQLCLSYVSDLMSNEIVTTDSDGLSILMKFLVNYVHQMNNAIPVVSKMKNQIND 113
Query: 101 NREQIAIGDCKELVDFSVSELAWSLGEMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCLE 160
R++ A+ DC EL+D SV ++ S+ + + + N ++WLS L+N TCL+
Sbjct: 114 IRQEGALTDCLELLDQSVDLVSDSIAAIDKRTHSE-----HANAQSWLSGVLTNHVTCLD 168
Query: 161 GFEGTDRRLESYISGSLTQVTQLISNVLSLYTQLHALPFRPPRNNTHKTSESLDLDSEFP 220
+ + + I+G T + +LIS L ++ P ++ + + P
Sbjct: 169 ELDSFTKAM---ING--TNLDELISRAKVALAMLASVT--TPNDDVLRPGLG-----KMP 216
Query: 221 EWMTEGDQELLRSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYR 280
W++ D++L+ S A+AVVA DG+G YRT+ EAV AAP S RYVIYVK+G+Y+
Sbjct: 217 SWVSSRDRKLMESSGKDIGANAVVAKDGTGKYRTLAEAVAAAPDKSKTRYVIYVKRGIYK 276
Query: 281 ENVDMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTA 340
ENV++ + +M+VGDG+ TIIT N N + G TTF +AT+A GKGFI +D+ +NTA
Sbjct: 277 ENVEVSSRKMKLMIVGDGMHATIITGNLNVVDGSTTFHSATLAAVGKGFILQDICIQNTA 336
Query: 341 GPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVL 400
GP HQAVALRV +D+S RC I+ +QDTLYAHS RQFYR+ + GTIDFIFGN A V
Sbjct: 337 GPAKHQAVALRVGADKSVINRCRIDAYQDTLYAHSQRQFYRDSYVTGTIDFIFGNAAVVF 396
Query: 401 QNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSFILATQ---------PTYLGRPW 451
Q CK+ R P Q+ VTAQGR P+Q+TG +IQ I+A+ PTYLGRPW
Sbjct: 397 QKCKLVARKPGKYQQNMVTAQGRTDPNQATGTSIQFCNIIASSDLEPVLKEFPTYLGRPW 456
Query: 452 KQYSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIM 511
K+YSRTV + +Y+ G++ P GW EW GDFAL TL+YGE+ N GPGA + RVKWPGYH +
Sbjct: 457 KKYSRTVVMESYLGGLINPAGWAEWDGDFALKTLYYGEFMNNGPGAGTSKRVKWPGYHCI 516
Query: 512 KDAATASYFTVQRFIHGDSWLPGTGVKFTAGL 543
D A A FTV + I G SWL TGV + GL
Sbjct: 517 TDPAEAMPFTVAKLIQGGSWLRSTGVAYVDGL 548
>PM21_LYCES (P09607) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase 2) (PE 2)
Length = 550
Score = 394 bits (1012), Expect = e-109
Identities = 218/512 (42%), Positives = 303/512 (58%), Gaps = 29/512 (5%)
Query: 44 CMDIENQNSCLQNIHNELTKIGPPSPT---SVLSAALRTTLNEAQGAIDNLTKFSTFSVS 100
C ++ CL + + ++ S + S+L L ++++ AI + K
Sbjct: 54 CKTAQDSQLCLSYVSDLISNEIVTSDSDGLSILKKFLVYSVHQMNNAIPVVRKIKNQIND 113
Query: 101 NREQIAIGDCKELVDFSVSELAWSLGEMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCLE 160
REQ A+ DC EL+D SV + S+ + + S N ++WLS L+N TCL+
Sbjct: 114 IREQGALTDCLELLDLSVDLVCDSIAAIDK-----RSRSEHANAQSWLSGVLTNHVTCLD 168
Query: 161 GFEGTDRRLESYISGSLTQVTQLISNVLSLYTQLHALPFRPPRNNTHKTSESLDLDSEFP 220
+ + + I+G T + +LIS L ++ P + + + P
Sbjct: 169 ELDSFTKAM---ING--TNLDELISRAKVALAMLASVT--TPNDEVLRPGLG-----KMP 216
Query: 221 EWMTEGDQELLRSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYR 280
W++ D++L+ S A+AVVA DG+G YRT+ EAV AAP S RYVIYVK+G Y+
Sbjct: 217 SWVSSRDRKLMESSGKDIGANAVVAKDGTGKYRTLAEAVAAAPDKSKTRYVIYVKRGTYK 276
Query: 281 ENVDMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTA 340
ENV++ + N+M++GDG+ TIIT + N + G TTF +AT+A GKGFI +D+ +NTA
Sbjct: 277 ENVEVSSRKMNLMIIGDGMYATIITGSLNVVDGSTTFHSATLAAVGKGFILQDICIQNTA 336
Query: 341 GPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVL 400
GP HQAVALRV +D+S RC I+ +QDTLYAHS RQFYR+ + GTIDFIFGN A V
Sbjct: 337 GPAKHQAVALRVGADKSVINRCRIDAYQDTLYAHSQRQFYRDSYVTGTIDFIFGNAAVVF 396
Query: 401 QNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSFILATQ---------PTYLGRPW 451
Q C++ R P Q+ VTAQGR P+Q+TG +IQ I+A+ PTYLGRPW
Sbjct: 397 QKCQLVARKPGKYQQNMVTAQGRTDPNQATGTSIQFCDIIASPDLKPVVKEFPTYLGRPW 456
Query: 452 KQYSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIM 511
K+YSRTV + +Y+ G++ P GW EW GDFAL TL+YGE+ N GPGA + RVKWPGYH++
Sbjct: 457 KKYSRTVVMESYLGGLIDPSGWAEWHGDFALKTLYYGEFMNNGPGAGTSKRVKWPGYHVI 516
Query: 512 KDAATASYFTVQRFIHGDSWLPGTGVKFTAGL 543
D A A FTV + I G SWL T V + GL
Sbjct: 517 TDPAEAMSFTVAKLIQGGSWLRSTDVAYVDGL 548
>PME2_ARATH (Q42534) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase 2) (PE 2)
Length = 587
Score = 390 bits (1003), Expect = e-108
Identities = 216/465 (46%), Positives = 293/465 (62%), Gaps = 25/465 (5%)
Query: 102 REQIAIGDCKELVDFSVSELAWSLGEMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCLEG 161
RE A+ DC E +D ++ EL ++ ++ + + ++ +L+ +S+A++NQ TCL+G
Sbjct: 125 REVTALHDCLETIDETLDELHVAVEDLHQYPKQKSLRKHADDLKTLISSAITNQGTCLDG 184
Query: 162 F--EGTDRRLESYISGSLTQVTQLISNVLSLY-----TQLHALPFRPP-----RNNTHKT 209
F + DR++ + V + SN L++ T + R NN K
Sbjct: 185 FSYDDADRKVRKALLKGQVHVEHMCSNALAMIKNMTETDIANFELRDKSSTFTNNNNRKL 244
Query: 210 SESL-DLDSE-FPEWMTEGDQELLRSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHSN 267
E DLDS+ +P+W++ GD+ LL+ +ADA VA DGSG + T+ AV AAP SN
Sbjct: 245 KEVTGDLDSDGWPKWLSVGDRRLLQGST--IKADATVADDGSGDFTTVAAAVAAAPEKSN 302
Query: 268 RRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGK 327
+R+VI++K G+YRENV++ +K TNIM +GDG G+TIIT +RN + G TTF +ATVA G+
Sbjct: 303 KRFVIHIKAGVYRENVEVTKKKTNIMFLGDGRGKTIITGSRNVVDGSTTFHSATVAAVGE 362
Query: 328 GFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYG 387
F+ARD++F+NTAGP HQAVALRV SD SAFY+C + +QDTLY HS RQF+ +C I G
Sbjct: 363 RFLARDITFQNTAGPSKHQAVALRVGSDFSAFYQCDMFAYQDTLYVHSNRQFFVKCHITG 422
Query: 388 TIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQD------SFILA 441
T+DFIFGN AAVLQ+C I R P QK VTAQGR P+Q+TG IQ+ S +LA
Sbjct: 423 TVDFIFGNAAAVLQDCDINARRPNSGQKNMVTAQGRSDPNQNTGIVIQNCRIGGTSDLLA 482
Query: 442 ---TQPTYLGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGAS 498
T PTYLGRPWK+YSRTV + + +S +++P GW EW G FAL TL Y EY N G GA
Sbjct: 483 VKGTFPTYLGRPWKEYSRTVIMQSDISDVIRPEGWHEWSGSFALDTLTYREYLNRGGGAG 542
Query: 499 LAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLPGTGVKFTAGL 543
A RVKW GY ++ A FT +FI G WL TG F+ L
Sbjct: 543 TANRVKWKGYKVITSDTEAQPFTAGQFIGGGGWLASTGFPFSLSL 587
>PME1_LYCES (P14280) Pectinesterase 1 precursor (EC 3.1.1.11)
(Pectin methylesterase 1) (PE 1)
Length = 546
Score = 385 bits (990), Expect = e-106
Identities = 211/482 (43%), Positives = 288/482 (58%), Gaps = 26/482 (5%)
Query: 71 SVLSAALRTTLNEAQGAIDNLTKFSTFSVSNREQIAIGDCKELVDFSVSELAWSLGEMRR 130
S+L L +++ AI + K R+ A+ DC EL+D SV + S+ + +
Sbjct: 80 SILMKFLVNYVHQMNNAIPVVRKMKNQINDIRQHGALTDCLELLDQSVDFASDSIAAIDK 139
Query: 131 IRAGDTSVQYEGNLEAWLSAALSNQDTCLEGFEGTDRRLESYISGSLTQVTQLISNVLSL 190
S N ++WLS L+N TCL+ + + + I+G T + +LIS
Sbjct: 140 -----RSRSEHANAQSWLSGVLTNHVTCLDELDSFTKAM---ING--TNLEELISRAKVA 189
Query: 191 YTQLHALPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKPHRSRADAVVALDGSG 250
L +L T + + + P W++ D++L+ S A+AVVA DG+G
Sbjct: 190 LAMLASL-------TTQDEDVFMTVLGKMPSWVSSMDRKLMESSGKDIIANAVVAQDGTG 242
Query: 251 HYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTIITSNRNF 310
Y+T+ EAV AAP S RYVIYVK+G Y+ENV++ N+M+VGDG+ T IT + N
Sbjct: 243 DYQTLAEAVAAAPDKSKTRYVIYVKRGTYKENVEVASNKMNLMIVGDGMYATTITGSLNV 302
Query: 311 MQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQDT 370
+ G TTFR+AT+A G+GFI +D+ +NTAGP QAVALRV +D S RC I+ +QDT
Sbjct: 303 VDGSTTFRSATLAAVGQGFILQDICIQNTAGPAKDQAVALRVGADMSVINRCRIDAYQDT 362
Query: 371 LYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQST 430
LYAHS RQFYR+ + GT+DFIFGN A V Q C++ R P Q+ VTAQGR P+Q+T
Sbjct: 363 LYAHSQRQFYRDSYVTGTVDFIFGNAAVVFQKCQLVARKPGKYQQNMVTAQGRTDPNQAT 422
Query: 431 GFTIQDSFILATQ---------PTYLGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDFA 481
G +IQ I+A+ PTYLGRPWK+YSRTV + +Y+ G++ P GW EW GDFA
Sbjct: 423 GTSIQFCNIIASSDLEPVLKEFPTYLGRPWKEYSRTVVMESYLGGLINPAGWAEWDGDFA 482
Query: 482 LTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLPGTGVKFTA 541
L TL+YGE+ N GPGA + RVKWPGYH++ D A A FTV + I G SWL TGV +
Sbjct: 483 LKTLYYGEFMNNGPGAGTSKRVKWPGYHVITDPAKAMPFTVAKLIQGGSWLRSTGVAYVD 542
Query: 542 GL 543
GL
Sbjct: 543 GL 544
>PME3_PHAVU (Q43111) Pectinesterase 3 precursor (EC 3.1.1.11)
(Pectin methylesterase 3) (PE 3)
Length = 581
Score = 381 bits (979), Expect = e-105
Identities = 226/542 (41%), Positives = 302/542 (55%), Gaps = 33/542 (6%)
Query: 18 SSSKSFSDEIPSEIQTQDMQALIAQACMDIENQ-NSCLQNIHNELTKIGPPSPTSVLSAA 76
S S SD +P QT+ A +A D +SC +I + L + P + +
Sbjct: 57 SESSPSSDSVP---QTELSPAASLKAVCDTTRYPSSCFSSI-SSLPESNTTDPELLFKLS 112
Query: 77 LRTTLNEAQGAIDNLTKFSTFSVSNREQIA-----IGDCKELVDFSVSELAWSLGEMRRI 131
LR AID L+ F + +N EQ A I C + ++ L S+ + +
Sbjct: 113 LRV-------AIDELSSFPSKLRANAEQDARLQKAIDVCSSVFGDALDRLNDSISALGTV 165
Query: 132 RAGDTSVQYEGNLEAWLSAALSNQDTCLEGFEGTD----RRLESYISGSLTQVTQLISNV 187
S N+E WLSAAL++QDTCL+ + R I ++ T+ SN
Sbjct: 166 AGRIASSASVSNVETWLSAALTDQDTCLDAVGELNSTAARGALQEIETAMRNSTEFASNS 225
Query: 188 LSLYTQLHALPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKPHRSRADAVVALD 247
L++ T++ L R H+ FPEW+ ++ LL K + S DAVVA D
Sbjct: 226 LAIVTKILGLLSRFETPIHHRRLLG------FPEWLGAAERRLLEEKNNDSTPDAVVAKD 279
Query: 248 GSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTIITSN 307
GSG ++TI EA+ S R+ +YVK+G Y EN+D+ + N+M+ GDG +T + +
Sbjct: 280 GSGQFKTIGEALKLVKKKSEERFSVYVKEGRYVENIDLDKNTWNVMIYGDGKDKTFVVGS 339
Query: 308 RNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGF 367
RNFM G TF TAT AV GKGFIA+D+ F N AG HQAVALR SD+S F+RCS +GF
Sbjct: 340 RNFMDGTPTFETATFAVKGKGFIAKDIGFVNNAGASKHQAVALRSGSDRSVFFRCSFDGF 399
Query: 368 QDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPH 427
QDTLYAHS RQFYR+C+I GTIDFIFGN A V Q+CKI R PLP Q T+TAQG+K P+
Sbjct: 400 QDTLYAHSNRQFYRDCDITGTIDFIFGNAAVVFQSCKIMPRQPLPNQFNTITAQGKKDPN 459
Query: 428 QSTGFTIQDSFIL-----ATQPTYLGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDF-A 481
Q+TG IQ S I T PTYLGRPWK +S TV + + + ++ P GW+ W +
Sbjct: 460 QNTGIIIQKSTITPFGNNLTAPTYLGRPWKDFSTTVIMQSDIGALLNPVGWMSWVPNVEP 519
Query: 482 LTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLPGTGVKFTA 541
TT++Y EY+N GPGA ++ RVKW GY A FTVQ FI G WLP V+F +
Sbjct: 520 PTTIFYAEYQNSGPGADVSQRVKWAGYKPTITDRNAEEFTVQSFIQGPEWLPNAAVQFDS 579
Query: 542 GL 543
L
Sbjct: 580 TL 581
>PME3_LYCES (Q96576) Pectinesterase 3 precursor (EC 3.1.1.11)
(Pectin methylesterase 3) (PE 3)
Length = 544
Score = 362 bits (928), Expect = 2e-99
Identities = 209/509 (41%), Positives = 289/509 (56%), Gaps = 29/509 (5%)
Query: 44 CMDIENQNSCLQNIHNELTKIGPPSPTSVLSAALRTTLNEAQGAIDNLTKFSTFSVSNRE 103
C ++ CL + +T +VL L +++ AI + K R+
Sbjct: 54 CKTAQDSQLCLSYVSEIVTT--ESDGVTVLKKFLVKYVHQMNNAIPVVRKIKNQINDIRQ 111
Query: 104 QIAIGDCKELVDFSVSELAWSLGEMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCLEGFE 163
Q A+ DC EL+D SV ++ S+ + + S N ++WLS L+N TCL+
Sbjct: 112 QGALTDCLELLDQSVDLVSDSIAAIDK-----RSRSEHANAQSWLSGVLTNHVTCLDELT 166
Query: 164 GTDRRLESYISGSLTQVTQLISNVLSLYTQLHALPFRPPRNNTHKTSESLDLDSEFPEWM 223
S + + T + +LI+ L ++ N + L + P W+
Sbjct: 167 SF-----SLSTKNGTVLDELITRAKVALAMLASVT----TPNDEVLRQGL---GKMPYWV 214
Query: 224 TEGDQELLRSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENV 283
+ D++L+ S A+ VVA DG+G Y+T+ EAV AAP + RYVIYVK G+Y+ENV
Sbjct: 215 SSRDRKLMESSGKDIIANRVVAQDGTGDYQTLAEAVAAAPDKNKTRYVIYVKMGIYKENV 274
Query: 284 DMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPV 343
+ +K N+M+VGDG+ TIIT + N + G +TF + T+A G+GFI +D+ +NTAGP
Sbjct: 275 VVTKKKMNLMIVGDGMNATIITGSLNVVDG-STFPSNTLAAVGQGFILQDICIQNTAGPE 333
Query: 344 NHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNC 403
QAVALRV +D S RC I+ +QDTLYAHS RQFYR+ + GT+DFIFGN A V Q C
Sbjct: 334 KDQAVALRVGADMSVINRCRIDAYQDTLYAHSQRQFYRDSYVTGTVDFIFGNAAVVFQKC 393
Query: 404 KIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSFILAT---------QPTYLGRPWKQY 454
+I R P QK VTAQGR P+Q+TG +IQ I+A+ TYLGRPWK++
Sbjct: 394 QIVARKPNKRQKNMVTAQGRTDPNQATGTSIQFCDIIASPDLEPVMNEYKTYLGRPWKKH 453
Query: 455 SRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDA 514
SRTV + +Y+ G + P GW EW GDFAL TL+YGE+ N GPGA + RVKWPGYH++ D
Sbjct: 454 SRTVVMQSYLDGHIDPSGWFEWRGDFALKTLYYGEFMNNGPGAGTSKRVKWPGYHVITDP 513
Query: 515 ATASYFTVQRFIHGDSWLPGTGVKFTAGL 543
A FTV I G SWL T V + GL
Sbjct: 514 NEAMPFTVAELIQGGSWLNSTSVAYVEGL 542
>PME1_ARATH (Q43867) Pectinesterase 1 precursor (EC 3.1.1.11)
(Pectin methylesterase 1) (PE 1)
Length = 586
Score = 344 bits (882), Expect = 4e-94
Identities = 198/561 (35%), Positives = 302/561 (53%), Gaps = 37/561 (6%)
Query: 1 MALVNALFILMMLLILPSSSKSFSDEIPSEIQTQDMQALIAQACMDIENQNSCLQNIHNE 60
+++V + +++ ++ K+ ++ PS + C SC+ +I ++
Sbjct: 34 ISVVVLIAVIIAAVVATVVHKNKNESTPSPPPELTPSTSLKAICSVTRFPESCISSI-SK 92
Query: 61 LTKIGPPSPTSVLSAALRTTLNEAQGAIDNLTKFSTFSVSNREQIAIGDCKELVDFSVSE 120
L P ++ +L+ ++E D K S + R + A+ C +L++ ++
Sbjct: 93 LPSSNTTDPETLFKLSLKVIIDELDSISDLPEKLSKETEDERIKSALRVCGDLIEDALDR 152
Query: 121 LAWSLG----EMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCLEGFEGTDRRLESYISGS 176
L ++ E ++ + ++ +L+ WLSA +++ +TC + + + Y + +
Sbjct: 153 LNDTVSAIDDEEKKKTLSSSKIE---DLKTWLSATVTDHETCFDSLDELKQNKTEYANST 209
Query: 177 LTQVTQ---------------LISNVLSLYTQLHALPFRPPRNNTHKTSESLDLDSEFPE 221
+TQ + ++S +LS + L R R +H +S+D F +
Sbjct: 210 ITQNLKSAMSRSTEFTSNSLAIVSKILSALSDLGIPIHRRRRLMSHHHQQSVD----FEK 265
Query: 222 WMTEGDQELLRSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRE 281
W + KP D VA DG+G T+ EAV P S + +VIYVK G Y E
Sbjct: 266 WARRRLLQTAGLKP-----DVTVAGDGTGDVLTVNEAVAKVPKKSLKMFVIYVKSGTYVE 320
Query: 282 NVDMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAG 341
NV M + N+M+ GDG G+TII+ ++NF+ G T+ TAT A+ GKGFI +D+ NTAG
Sbjct: 321 NVVMDKSKWNVMIYGDGKGKTIISGSKNFVDGTPTYETATFAIQGKGFIMKDIGIINTAG 380
Query: 342 PVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQ 401
HQAVA R SD S +Y+CS +GFQDTLY HS RQFYR+C++ GTIDFIFG+ A V Q
Sbjct: 381 AAKHQAVAFRSGSDFSVYYQCSFDGFQDTLYPHSNRQFYRDCDVTGTIDFIFGSAAVVFQ 440
Query: 402 NCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSFILAT----QPTYLGRPWKQYSRT 457
CKI R PL Q T+TAQG+K P+QS+G +IQ I A PTYLGRPWK++S T
Sbjct: 441 GCKIMPRQPLSNQFNTITAQGKKDPNQSSGMSIQRCTISANGNVIAPTYLGRPWKEFSTT 500
Query: 458 VYINTYMSGMVQPRGWLEWF-GDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAAT 516
V + T + +V+P GW+ W G ++ YGEY+N GPG+ + RVKW GY + A
Sbjct: 501 VIMETVIGAVVRPSGWMSWVSGVDPPASIVYGEYKNTGPGSDVTQRVKWAGYKPVMSDAE 560
Query: 517 ASYFTVQRFIHGDSWLPGTGV 537
A+ FTV +HG W+P TGV
Sbjct: 561 AAKFTVATLLHGADWIPATGV 581
>PME_DAUCA (P83218) Pectinesterase (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 319
Score = 338 bits (868), Expect = 2e-92
Identities = 171/312 (54%), Positives = 213/312 (67%), Gaps = 9/312 (2%)
Query: 241 DAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIG 300
+ VVA DGSG Y+T++EAV AAP S RYVI +K G+YRENVD+ +K NIM +GDG
Sbjct: 8 NVVVAADGSGDYKTVSEAVAAAPEDSKTRYVIRIKAGVYRENVDVPKKKKNIMFLGDGRT 67
Query: 301 QTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFY 360
TIIT+++N G TTF +ATVA G GF+ARD++F+NTAG HQAVALRV SD SAFY
Sbjct: 68 STIITASKNVQDGSTTFNSATVAAVGAGFLARDITFQNTAGAAKHQAVALRVGSDLSAFY 127
Query: 361 RCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTA 420
RC I +QD+LY HS RQF+ C I GT+DFIFGN A VLQ+C I+ R P QK VTA
Sbjct: 128 RCDILAYQDSLYVHSNRQFFINCFIAGTVDFIFGNAAVVLQDCDIHARRPGSGQKNMVTA 187
Query: 421 QGRKSPHQSTGFTIQDSFILATQ---------PTYLGRPWKQYSRTVYINTYMSGMVQPR 471
QGR P+Q+TG IQ S I AT PTYLGRPWK+YSRTV + + ++ ++ P
Sbjct: 188 QGRTDPNQNTGIVIQKSRIGATSDLQPVQSSFPTYLGRPWKEYSRTVVMQSSITNVINPA 247
Query: 472 GWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSW 531
GW W G+FAL TL+YGEY+N G GA+ +GRV W G+ ++ + A FT FI G SW
Sbjct: 248 GWFPWDGNFALDTLYYGEYQNTGAGAATSGRVTWKGFKVITSSTEAQGFTPGSFIAGGSW 307
Query: 532 LPGTGVKFTAGL 543
L T F+ GL
Sbjct: 308 LKATTFPFSLGL 319
>PME_MEDSA (Q42920) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE) (P65)
Length = 447
Score = 300 bits (769), Expect = 5e-81
Identities = 161/446 (36%), Positives = 247/446 (55%), Gaps = 17/446 (3%)
Query: 110 CKELVDFSVSELAWSLGEMRRIRAGDTSVQYEGNLEAWLSAALSNQDTCLEGFEGTDRRL 169
C E++D++V + S+G + + S +Y +L+ WL+ LS+Q TCL+GF T +
Sbjct: 4 CNEVLDYAVDGIHKSVGTLDQFDFHKLS-EYAFDLKVWLTGTLSHQQTCLDGFANTTTKA 62
Query: 170 ESYISGSLTQVTQLISNVLSLYTQLHALPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQE 229
++ L +L SN + + + + + + + S L D P W+ +G +
Sbjct: 63 GETMTKVLKTSMELSSNAIDMMDAVSRI-LKGFDTSQYSVSRRLLSDDGIPSWVNDGHRR 121
Query: 230 LLRSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKM 289
LL + +AVVA DGSG ++T+T+A+ P + +VI+VK G+Y+E V++ ++M
Sbjct: 122 LLAGG--NVQPNAVVAQDGSGQFKTLTDALKTVPPKNAVPFVIHVKAGVYKETVNVAKEM 179
Query: 290 TNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVA 349
+ ++GDG +T T + N+ G T+ TAT V+G F+A+D+ F NTAG HQAVA
Sbjct: 180 NYVTVIGDGPTKTKFTGSLNYADGINTYNTATFGVNGANFMAKDIGFENTAGTGKHQAVA 239
Query: 350 LRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRA 409
LRV +DQ+ FY C ++GFQDTLY S RQFYR+C I GTIDF+FG V QNCK+ R
Sbjct: 240 LRVTADQAIFYNCQMDGFQDTLYVQSQRQFYRDCSISGTIDFVFGERFGVFQNCKLVCRL 299
Query: 410 PLPLQKVTVTAQGRKSPHQSTGFTIQDSFI--------LATQPTYLGRPWKQYSRTVYIN 461
P Q+ VTA GR+ + ++ Q S + + +YLGRPWKQYS+ V ++
Sbjct: 300 PAKGQQCLVTAGGREKQNSASALVFQSSHFTGEPALTSVTPKVSYLGRPWKQYSKVVIMD 359
Query: 462 TYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASY-- 519
+ + + P G++ W G T Y EY N GPGA RVKW G ++ A Y
Sbjct: 360 STIDAIFVPEGYMPWMGSAFKETCTYYEYNNKGPGADTNLRVKWHGVKVLTSNVAAEYYP 419
Query: 520 ---FTVQRFIHGDSWLPGTGVKFTAG 542
F + D+W+ +GV ++ G
Sbjct: 420 GKFFEIVNATARDTWIVKSGVPYSLG 445
>PME_PETIN (Q43043) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 374
Score = 295 bits (756), Expect = 2e-79
Identities = 144/317 (45%), Positives = 202/317 (63%), Gaps = 11/317 (3%)
Query: 238 SRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGD 297
++ +A VALDGSG Y+TI EA++A P + ++I++K G+Y+E +D+ + MTN++L+G+
Sbjct: 55 AKPNATVALDGSGQYKTIKEALDAVPKKNTEPFIIFIKAGVYKEYIDIPKSMTNVVLIGE 114
Query: 298 GIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQS 357
G +T IT N++ G +TF T TV V+G F+A+++ F NTAGP QAVALRV +D++
Sbjct: 115 GPTKTKITGNKSVKDGPSTFHTTTVGVNGANFVAKNIGFENTAGPEKEQAVALRVSADKA 174
Query: 358 AFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVT 417
Y C I+G+QDTLY H+ RQFYR+C I GT+DFIFGNG AVLQNCK+ R P Q
Sbjct: 175 IIYNCQIDGYQDTLYVHTYRQFYRDCTITGTVDFIFGNGEAVLQNCKVIVRKPAQNQSCM 234
Query: 418 VTAQGRKSPHQSTGFTIQ--------DSFILA-TQPTYLGRPWKQYSRTVYINTYMSGMV 468
VTAQGR P Q +Q D F L+ TYLGRPWK+YSRT+ + +Y+ +
Sbjct: 235 VTAQGRTEPIQKGAIVLQNCEIKPDTDYFSLSPPSKTYLGRPWKEYSRTIIMQSYIDKFI 294
Query: 469 QPRGWLEW-FGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIH 527
+P GW W +F T +Y EY+N GPGA+L R+ W G+ A FT +I+
Sbjct: 295 EPEGWAPWNITNFGRDTSYYAEYQNRGPGAALDKRITWKGFQKGFTGEAAQKFTAGVYIN 354
Query: 528 GD-SWLPGTGVKFTAGL 543
D +WL V + AG+
Sbjct: 355 NDENWLQKANVPYEAGM 371
>PME_BRANA (P41510) Probable pectinesterase precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 584
Score = 254 bits (650), Expect = 3e-67
Identities = 179/562 (31%), Positives = 274/562 (47%), Gaps = 75/562 (13%)
Query: 37 QALIAQACMDIENQNSCLQNIHNELTKIGPPSPTSVLSAALRTTLNEAQGAIDNLTKFST 96
Q + C ++ SC + L + P+ ++ A + T D +TK +
Sbjct: 43 QKAVESLCASATDKGSCAKT----LDPVKSDDPSKLIKAFMLATK-------DAVTKSTN 91
Query: 97 FSVSNREQIAIG---DCKELVDFSVSELAWSLGEMRRI--RAGDTSVQYEGN----LEAW 147
F+ S E + K ++D+ L ++L ++ I G+ +Q G+ L+ W
Sbjct: 92 FTASTEEGMGKNINATSKAVLDYCKRVLMYALEDLETIVEEMGE-DLQQSGSKMDQLKQW 150
Query: 148 LSAALSNQDTCLEGFEGTDRRLESYISGSLTQVTQLISNVLSLYTQLHALPFRPPRNN-- 205
L+ + Q C++ E ++ L + + L SN + ++ HAL + N
Sbjct: 151 LTGVFNYQTDCIDDIEESE--LRKVMGEGIAHSKILSSNAIDIF---HALTTAMSQMNVK 205
Query: 206 -----------THKTSESL--DLDSE-FPEWMTEGDQELLRSKPHRSRADA--------- 242
T L DLD + P+W ++ D++L+ ++ R A A
Sbjct: 206 VDDMKKGNLGETPAPDRDLLEDLDQKGLPKWHSDKDRKLM-AQAGRPGAPADEGIGEGGG 264
Query: 243 ---------VVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIM 293
VVA DGSG ++TI+EAV A P + R +IY+K G+Y+E V + +K+ N+
Sbjct: 265 GGGKIKPTHVVAKDGSGQFKTISEAVKACPEKNPGRCIIYIKAGVYKEQVTIPKKVNNVF 324
Query: 294 LVGDGIGQTIITSNRN--FMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALR 351
+ GDG QTIIT +R+ G TT + TV V +GF+A+ + F+NTAGP+ HQAVA R
Sbjct: 325 MFGDGATQTIITFDRSVGLSPGTTTSLSGTVQVESEGFMAKWIGFQNTAGPLGHQAVAFR 384
Query: 352 VDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPL 411
V+ D++ + C +G+QDTLY ++ RQFYR + GT+DFIFG A V+QN I R
Sbjct: 385 VNGDRAVIFNCRFDGYQDTLYVNNGRQFYRNIVVSGTVDFIFGKSATVIQNSLILCRKGS 444
Query: 412 PLQKVTVTAQG-RKSPHQSTGFTIQDSFILA---------TQPTYLGRPWKQYSRTVYIN 461
P Q VTA G K G + + I+A T +YLGRPWK ++ T I
Sbjct: 445 PGQTNHVTADGNEKGKAVKIGIVLHNCRIMADKELEADRLTVKSYLGRPWKPFATTAVIG 504
Query: 462 TYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFT 521
T + ++QP GW EW G+ T Y E+ N GPGA+ A RV W + K AA FT
Sbjct: 505 TEIGDLIQPTGWNEWQGEKFHLTATYVEFNNRGPGANTAARVPWA--KMAKSAAEVERFT 562
Query: 522 VQRFIHGDSWLPGTGVKFTAGL 543
V ++ +W+ V GL
Sbjct: 563 VANWLTPANWIQEANVPVQLGL 584
>PME_BRACM (Q42608) Pectinesterase (EC 3.1.1.11) (Pectin
methylesterase) (PE) (Fragment)
Length = 571
Score = 248 bits (633), Expect = 3e-65
Identities = 177/562 (31%), Positives = 274/562 (48%), Gaps = 75/562 (13%)
Query: 37 QALIAQACMDIENQNSCLQNIHNELTKIGPPSPTSVLSAALRTTLNEAQGAIDNLTKFST 96
Q + C ++ SC + L + P+ ++ A + T D +TK +
Sbjct: 30 QKAVESLCASATDKGSCAKT----LDPVKSDDPSKLIKAFMLATK-------DAVTKSTN 78
Query: 97 FSVSNREQIAIG---DCKELVDFSVSELAWSLGEMRRI--RAGDTSVQYEGN----LEAW 147
F+ S E + K ++D+ L ++L ++ I G+ +Q G+ L+ W
Sbjct: 79 FTASTEEGMGKNMNATSKAVLDYCKRVLMYALEDLETIVEEMGE-DLQQSGSKMDQLKQW 137
Query: 148 LSAALSNQDTCLEGFEGTDRRLESYISGSLTQVTQLISNVLSLYTQLHALPFRPPRNN-- 205
L+ + Q C++ E ++ L + + L SN + ++ HAL + N
Sbjct: 138 LTGVFNYQTDCIDDIEESE--LRKVMGEGIRHSKILSSNAIDIF---HALTTAMSQMNVK 192
Query: 206 -----------THKTSESL--DLDSE-FPEWMTEGDQELLRSKPHRSRADA--------- 242
T L DLD + P+W ++ D++L+ ++ R A A
Sbjct: 193 VDDMKKGNLGETPAPDRDLLEDLDQKGLPKWHSDKDRKLM-AQAGRPGAPADEGIGEGGG 251
Query: 243 ---------VVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIM 293
VVA DGSG ++TI+EAV A P + R +IY+K G+Y+E V + +K+ N+
Sbjct: 252 GGGKIKPTHVVAKDGSGQFKTISEAVKACPEKNPGRCIIYIKAGVYKEQVTIPKKVNNVF 311
Query: 294 LVGDGIGQTIITSNRN--FMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALR 351
+ GDG QTIIT +R+ G TT + TV V +GF+A+ + F+NTAGP+ +QAVA R
Sbjct: 312 MFGDGATQTIITFDRSVGLSPGTTTSLSGTVQVESEGFMAKWIGFQNTAGPLGNQAVAFR 371
Query: 352 VDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPL 411
V+ D++ + C +G+QDTLY ++ RQFYR + GT+DFI G A V+QN I R
Sbjct: 372 VNGDRAVIFNCRFDGYQDTLYVNNGRQFYRNIVVSGTVDFINGKSATVIQNSLILCRKGS 431
Query: 412 PLQKVTVTAQGR-KSPHQSTGFTIQDSFILA---------TQPTYLGRPWKQYSRTVYIN 461
P Q VTA G+ K G + + I+A T +YLGRPWK ++ T I
Sbjct: 432 PGQTNHVTADGKQKGKAVKIGIVLHNCRIMADKELEADRLTVKSYLGRPWKPFATTAVIG 491
Query: 462 TYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFT 521
T + ++QP GW EW G+ T Y E+ N GPGA+ A RV W + K AA FT
Sbjct: 492 TEIGDLIQPTGWNEWQGEKFHLTATYVEFNNRGPGANPAARVPWA--KMAKSAAEVERFT 549
Query: 522 VQRFIHGDSWLPGTGVKFTAGL 543
V ++ +W+ V GL
Sbjct: 550 VANWLTPANWIQEANVTVQLGL 571
>PME_ASPAC (Q12535) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 331
Score = 125 bits (315), Expect = 2e-28
Identities = 84/273 (30%), Positives = 127/273 (45%), Gaps = 19/273 (6%)
Query: 242 AVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVG----- 296
A+V G Y TI +A++A + + I++++G Y E V + +++ G
Sbjct: 27 AIVVAKSGGDYTTIGDAIDALSTSTTDTQTIFIEEGTYDEQVYLPAMTGKVIIYGQTENT 86
Query: 297 DGIGQTIITSNRNFMQ---GWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVD 353
D ++T G + TAT G +++ NT G HQA+AL
Sbjct: 87 DSYADNLVTITHAISYEDAGESDDLTATFRNKAVGSQVYNLNIANTCGQACHQALALSAW 146
Query: 354 SDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGA-AVLQNCKIYTRAPLP 412
+DQ +Y C+ G+QDTL A + Q Y I G +DFIFG A A QN I R
Sbjct: 147 ADQQGYYGCNFTGYQDTLLAQTGNQLYINSYIEGAVDFIFGQHARAWFQNVDI--RVVEG 204
Query: 413 LQKVTVTAQGRKSPHQSTGFTIQDSFILATQ-------PTYLGRPWKQYSRTVYINTYMS 465
++TA GR S ++ + I S + A + YLGRPW +Y+R V+ T M+
Sbjct: 205 PTSASITANGRSSETDTSYYVINKSTVAAKEGDDVAEGTYYLGRPWSEYARVVFQQTSMT 264
Query: 466 GMVQPRGWLEWFGDFALTT-LWYGEYRNYGPGA 497
++ GW EW T + +GEY N G G+
Sbjct: 265 NVINSLGWTEWSTSTPNTEYVTFGEYANTGAGS 297
>PME_ASPTU (P17872) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 331
Score = 118 bits (295), Expect = 4e-26
Identities = 83/272 (30%), Positives = 121/272 (43%), Gaps = 18/272 (6%)
Query: 242 AVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQ 301
A+V G Y TI+ AV+A + S I++++G Y E V + +++ G
Sbjct: 27 AIVVAKSGGDYDTISAAVDALSTTSTETQTIFIEEGSYDEQVYIPALSGKLIVYGQTEDT 86
Query: 302 TIITSNRNFMQGWTTF-------RTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDS 354
T TSN + TAT+ +G +++ NT G HQA+A+ +
Sbjct: 87 TTYTSNLVNITHAIALADVDNDDETATLRNYAEGSAIYNLNIANTCGQACHQALAVSAYA 146
Query: 355 DQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGA-AVLQNCKIYTRAPLPL 413
+ +Y C G+QDTL A + Q Y I G +DFIFG A A C I R
Sbjct: 147 SEQGYYACQFTGYQDTLLAETGYQVYAGTYIEGAVDFIFGQHARAWFHECDI--RVLEGP 204
Query: 414 QKVTVTAQGRKSPHQSTGFTIQDSFILATQPT-------YLGRPWKQYSRTVYINTYMSG 466
++TA GR S + + I S + A YLGRPW QY+R + T M+
Sbjct: 205 SSASITANGRSSESDDSYYVIHKSTVAAADGNDVSSGTYYLGRPWSQYARVCFQKTSMTD 264
Query: 467 MVQPRGWLEWFGDFALT-TLWYGEYRNYGPGA 497
++ GW EW T + + EY N G GA
Sbjct: 265 VINHLGWTEWSTSTPNTENVTFVEYGNTGTGA 296
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.133 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 62,062,879
Number of Sequences: 164201
Number of extensions: 2553100
Number of successful extensions: 5924
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 5819
Number of HSP's gapped (non-prelim): 45
length of query: 545
length of database: 59,974,054
effective HSP length: 115
effective length of query: 430
effective length of database: 41,090,939
effective search space: 17669103770
effective search space used: 17669103770
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0109.6


