

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0108.9
(475 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
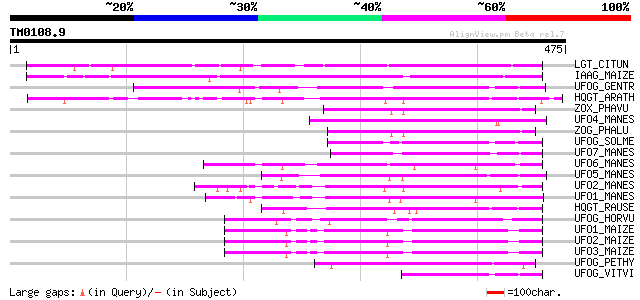
Score E
Sequences producing significant alignments: (bits) Value
LGT_CITUN (Q9MB73) Limonoid UDP-glucosyltransferase (EC 2.4.1.21... 225 2e-58
IAAG_MAIZE (Q41819) Indole-3-acetate beta-glucosyltransferase (E... 216 8e-56
UFOG_GENTR (Q96493) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 147 4e-35
HQGT_ARATH (Q9M156) Probable hydroquinone glucosyltransferase (E... 143 8e-34
ZOX_PHAVU (P56725) Zeatin O-xylosyltransferase (EC 2.4.2.40) (Ze... 131 4e-30
UFO4_MANES (Q40286) Flavonol 3-O-glucosyltransferase 4 (EC 2.4.1... 130 1e-29
ZOG_PHALU (Q9ZSK5) Zeatin O-glucosyltransferase (EC 2.4.1.203) (... 129 2e-29
UFOG_SOLME (Q43641) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 129 2e-29
UFO7_MANES (Q40289) Flavonol 3-O-glucosyltransferase 7 (EC 2.4.1... 128 4e-29
UFO6_MANES (Q40288) Flavonol 3-O-glucosyltransferase 6 (EC 2.4.1... 122 3e-27
UFO5_MANES (Q40287) Flavonol 3-O-glucosyltransferase 5 (EC 2.4.1... 120 6e-27
UFO2_MANES (Q40285) Flavonol 3-O-glucosyltransferase 2 (EC 2.4.1... 120 6e-27
UFO1_MANES (Q40284) Flavonol 3-O-glucosyltransferase 1 (EC 2.4.1... 119 1e-26
HQGT_RAUSE (Q9AR73) Hydroquinone glucosyltransferase (EC 2.4.1.2... 117 8e-26
UFOG_HORVU (P14726) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 108 2e-23
UFO1_MAIZE (P16166) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 108 3e-23
UFO2_MAIZE (P16165) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 103 7e-22
UFO3_MAIZE (P16167) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 102 2e-21
UFOG_PETHY (Q43716) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 98 5e-20
UFOG_VITVI (P51094) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 83 2e-15
>LGT_CITUN (Q9MB73) Limonoid UDP-glucosyltransferase (EC 2.4.1.210)
(Limonoid glucosyltransferase) (Limonoid GTase) (LGTase)
Length = 511
Score = 225 bits (573), Expect = 2e-58
Identities = 156/457 (34%), Positives = 239/457 (52%), Gaps = 35/457 (7%)
Query: 15 LIVSYPGQGQINPTLQFSKRLIAMGAHVTIVTTLHMLRRM------TTKTTIPGLSYTAF 68
L+VS+PG G +NP L+ + L + G +T+ T ++M T + T G + F
Sbjct: 10 LLVSFPGHGHVNPLLRLGRLLASKGFFLTLTTPESFGKQMRKAGNFTYEPTPVGDGFIRF 69
Query: 69 SDGFDDGFIVDVATHFNL--YTPELKRRGSEFITNLILFAENEGHPFTCLIHTLFVPWAP 126
+ F+DG+ D +L Y +L+ G + I +I + E P +CLI+ F+PW
Sbjct: 70 -EFFEDGWDEDDPRREDLDQYMAQLELIGKQVIPKIIKKSAEEYRPVSCLINNPFIPWVS 128
Query: 127 QVARELNLITAKLWIEPTTVMSILYNYFLGGDYIAKKNTVESSCSIEFSGLPFSLSPHDM 186
VA L L +A LW++ + Y+YF G + + E ++ +P L +M
Sbjct: 129 DVAESLGLPSAMLWVQSCACFAAYYHYFHG--LVPFPSEKEPEIDVQLPCMPL-LKHDEM 185
Query: 187 PSYLISKGSY------VLPLFKEDFKELRG*ANTTILVNTFEALEPEAPRAVDKLNMI-P 239
PS+L Y +L ++ K IL++TF LE E + K+ I P
Sbjct: 186 PSFLHPSTPYPFLRRAILGQYENLGKPF------CILLDTFYELEKEIIDYMAKICPIKP 239
Query: 240 IGLLIPSAFLDRKDFAHDTSFGGDIIQAPSNDYIEWLDSKPPSSVVYVSFGSYIVLSKTQ 299
+G L K+ T D P ++ I+WLD KPPSSVVY+SFG+ + L + Q
Sbjct: 240 VGPLF-------KNPKAPTLTVRDDCMKP-DECIDWLDKKPPSSVVYISFGTVVYLKQEQ 291
Query: 300 MDEIARALLDCRHPFLWVIREKEKFQDGEKEKEEELGNIEELEEVGKIAKWCSQVEVLSH 359
++EI ALL+ FLWV++ + G K + G +E++ + GK+ +W Q +VL+H
Sbjct: 292 VEEIGYALLNSGISFLWVMKPPPE-DSGVKIVDLPDGFLEKVGDKGKVVQWSPQEKVLAH 350
Query: 360 RSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKLIEDVWKIGVRVDLHVNEDGI 419
S+ CF+THCGWNSTMESL S VP++ FPQW DQ T+A + DV+K G+R+ E+ I
Sbjct: 351 PSVACFVTHCGWNSTMESLASGVPVITFPQWGDQVTDAMYLCDVFKTGLRLCRGEAENRI 410
Query: 420 VEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGLAKEA 456
+ +E+ + L + G K L NA KWK A+EA
Sbjct: 411 ISRDEVEKCL-LEATAGPKAVALEENALKWKKEAEEA 446
>IAAG_MAIZE (Q41819) Indole-3-acetate beta-glucosyltransferase (EC
2.4.1.121) (IAA-Glu synthetase) ((Uridine
5'-diphosphate-glucose:indol-3-ylacetyl)-beta-D-glucosyl
transferase)
Length = 471
Score = 216 bits (551), Expect = 8e-56
Identities = 143/451 (31%), Positives = 211/451 (46%), Gaps = 19/451 (4%)
Query: 15 LIVSYPGQGQINPTLQFSKRLIAMGAHVTIVTTLHMLRRMTTKTTIPGLSYTAFSDGFDD 74
L+V +PGQG +NP +QF+KRL + G T+VTT ++R P + A SDG D+
Sbjct: 6 LVVPFPGQGHMNPMVQFAKRLASKGVATTLVTT-RFIQRTADVDAHPAM-VEAISDGHDE 63
Query: 75 GFIVDVATHFNLYTPELKRRGSEFITNLILFAENEGHPFTCLIHTLFVPWAPQVARELNL 134
G A Y + S + +L+ + FTC+++ + W VAR + L
Sbjct: 64 GGFASAAGVAE-YLEKQAAAASASLASLVEARASSADAFTCVVYDSYEDWVLPVARRMGL 122
Query: 135 ITAKLWIEPTTVMSILYNYFLGGDYIAKKNTVESS--------CSIEFSGLPFSLSPHDM 186
+ V ++ Y++ G + + S S F GLP + ++
Sbjct: 123 PAVPFSTQSCAVSAVYYHFSQGRLAVPPGAAADGSDGGAGAAALSEAFLGLP-EMERSEL 181
Query: 187 PSYLISKGSYVLPLFKEDFKELRG*ANTTILVNTFEALEPEAPRAVDK-LNMIPIGLLIP 245
PS++ G Y + + + +L N+FE LE E + K L IG +P
Sbjct: 182 PSFVFDHGPYPTIAMQAIKQFAHAGKDDWVLFNSFEELETEVLAGLTKYLKARAIGPCVP 241
Query: 246 SAFLDRKDFAHDTSFGGDIIQAPSNDYIEWLDSKPPSSVVYVSFGSYIVLSKTQMDEIAR 305
R A+ G + P + +WLD+KP SV YVSFGS L Q +E+AR
Sbjct: 242 LPTAGRTAGANGRITYGANLVKPEDACTKWLDTKPDRSVAYVSFGSLASLGNAQKEELAR 301
Query: 306 ALLDCRHPFLWVIREKEKFQDGEKEKEEELGNIEELEEVGKIAKWCSQVEVLSHRSMGCF 365
LL PFLWV+R ++ Q E + WC Q++VL+H ++GCF
Sbjct: 302 GLLAAGKPFLWVVRASDEHQVPRYLLAEATAT-----GAAMVVPWCPQLDVLAHPAVGCF 356
Query: 366 LTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKLIEDVWKIGVRVDLHVNEDGIVEGEEI 425
+THCGWNST+E+L VPMVA WTDQ TNA+ +E W GVR + G E+
Sbjct: 357 VTHCGWNSTLEALSFGVPMVAMALWTDQPTNARNVELAWGAGVRARRDAGAGVFLRG-EV 415
Query: 426 RRGLTVVMKNGEKGEELRRNAKKWKGLAKEA 456
R + VM GE R+ A +W+ A+ A
Sbjct: 416 ERCVRAVMDGGEAASAARKAAGEWRDRARAA 446
>UFOG_GENTR (Q96493) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
Length = 453
Score = 147 bits (372), Expect = 4e-35
Identities = 106/359 (29%), Positives = 180/359 (49%), Gaps = 38/359 (10%)
Query: 107 ENEGHPFTCLIHTLFVPWAPQVARELNLITAKLWIEPTTVMSI-LYNYFLGGDYIAKKNT 165
E+ G +CL+ F+ +A + ++ + +W + + + +Y + +
Sbjct: 105 EDTGVNISCLLTDAFLWFAADFSEKIGVPWIPVWTAASCSLCLHVYTDEIRSRFAEFDIA 164
Query: 166 VESSCSIEFSGLPFSLSPHDMPSYLISKGS---YVLPLFKEDFKELRG*ANTTILVNTFE 222
++ +I+F ++S D+P LI + S + L L K + T + VN+FE
Sbjct: 165 EKAEKTIDFIPGLSAISFSDLPEELIMEDSQSIFALTLHNMGLKLHKA---TAVAVNSFE 221
Query: 223 ALEPEAP---RAVDKLNMIPIGLLIPSAFLDRKDFAHDTSFGGDIIQAPSNDYIEWLDSK 279
++P R+ ++LN++ IG L + I N+ ++WL ++
Sbjct: 222 EIDPIITNHLRSTNQLNILNIGPLQTLS---------------SSIPPEDNECLKWLQTQ 266
Query: 280 PPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEKFQDGEKEKEEELGNIE 339
SSVVY+SFG+ I +M +A L + PFLW +R+ E K I+
Sbjct: 267 KESSVVYLSFGTVINPPPNEMAALASTLESRKIPFLWSLRD-------EARKHLPENFID 319
Query: 340 ELEEVGKIAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKL 399
GKI W Q+ VL + ++G F+THCGWNST+ES+ RVP++ P + DQ NA++
Sbjct: 320 RTSTFGKIVSWAPQLHVLENPAIGVFVTHCGWNSTLESIFCRVPVIGRPFFGDQKVNARM 379
Query: 400 IEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGLAKEAGK 458
+EDVWKIGV V + G+ +E R L +V+ + +KG+E+R+N + K AK+A K
Sbjct: 380 VEDVWKIGVGV-----KGGVFTEDETTRVLELVLFS-DKGKEMRQNVGRLKEKAKDAVK 432
>HQGT_ARATH (Q9M156) Probable hydroquinone glucosyltransferase (EC
2.4.1.218) (Arbutin synthase)
Length = 480
Score = 143 bits (361), Expect = 8e-34
Identities = 147/505 (29%), Positives = 227/505 (44%), Gaps = 95/505 (18%)
Query: 16 IVSYPGQGQINPTLQFSKRLIAM-GAHVTIV------------TTLHMLRRMTTKTTIPG 62
I+ PG G + P ++F+KRL+ + G VT V T L L + +P
Sbjct: 11 IIPSPGMGHLIPLVEFAKRLVHLHGLTVTFVIAGEGPPSKAQRTVLDSLPSSISSVFLPP 70
Query: 63 LSYTAFSDGFD-DGFIVDVATHFNLYTPELKRRGSEFITNLILFAENEGHPFTCLIHTLF 121
+ T S + I T N PEL++ F+ G T L+ LF
Sbjct: 71 VDLTDLSSSTRIESRISLTVTRSN---PELRKVFDSFVEG--------GRLPTALVVDLF 119
Query: 122 VPWAPQVARELNLITAKLWIEPTTVMSILYNYFLGGDYIAKKNTVESSCSIEFSGLPFSL 181
A VA E ++ + V+S +FL ++ K ++ + S EF L
Sbjct: 120 GTDAFDVAVEFHVPPYIFYPTTANVLS----FFL---HLPK---LDETVSCEFRELT--- 166
Query: 182 SPHDMPSYLISKGSYVLPLF---KED-----------FKELRG*ANTTILVNTFEALEPE 227
P +P + G L K+D +KE G ILVNTF LEP
Sbjct: 167 EPLMLPGCVPVAGKDFLDPAQDRKDDAYKWLLHNTKRYKEAEG-----ILVNTFFELEPN 221
Query: 228 APRAV-----DKLNMIPIGLLIPSAFLDRKDFAHDTSFGGDIIQAPSNDYIEWLDSKPPS 282
A +A+ DK + P+G L+ + K Q ++ ++WLD++P
Sbjct: 222 AIKALQEPGLDKPPVYPVGPLVNIGKQEAK-------------QTEESECLKWLDNQPLG 268
Query: 283 SVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIRE-----KEKFQDGEKEKEEEL-- 335
SV+YVSFGS L+ Q++E+A L D FLWVIR + D + +
Sbjct: 269 SVLYVSFGSGGTLTCEQLNELALGLADSEQRFLWVIRSPSGIANSSYFDSHSQTDPLTFL 328
Query: 336 --GNIEELEEVGKIAK-WCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTD 392
G +E ++ G + W Q +VL+H S G FLTHCGWNST+ES+VS +P++A+P + +
Sbjct: 329 PPGFLERTKKRGFVIPFWAPQAQVLAHPSTGGFLTHCGWNSTLESVVSGIPLIAWPLYAE 388
Query: 393 QNTNAKLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGL 452
Q NA L+ + + +R +DG+V EE+ R + +M+ GE+G+ +R K+ K
Sbjct: 389 QKMNAVLLSEDIRAALRP--RAGDDGLVRREEVARVVKGLME-GEEGKGVRNKMKELKEA 445
Query: 453 A----KEAGKERKVVL*IQIYVLSW 473
A K+ G K + + L W
Sbjct: 446 ACRVLKDDGTSTKA---LSLVALKW 467
>ZOX_PHAVU (P56725) Zeatin O-xylosyltransferase (EC 2.4.2.40)
(Zeatin O-beta-D-xylosyltransferase)
Length = 454
Score = 131 bits (329), Expect = 4e-30
Identities = 74/187 (39%), Positives = 108/187 (57%), Gaps = 6/187 (3%)
Query: 269 SNDYIEWLDSKPPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEKFQ--D 326
S+ +EWLD + PSSV+YVSFG+ L Q+ E+A L + F+WV+R+ +K D
Sbjct: 243 SHPCMEWLDKQEPSSVIYVSFGTTTALRDEQIQELATGLEQSKQKFIWVLRDADKGDIFD 302
Query: 327 GEKEKEEEL--GNIEELEEVGKIAK-WCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVP 383
G + K EL G E +E +G + + W Q+E+LSH S G F++HCGWNS +ESL VP
Sbjct: 303 GSEAKRYELPEGFEERVEGMGLVVRDWAPQMEILSHSSTGGFMSHCGWNSCLESLTRGVP 362
Query: 384 MVAFPQWTDQNTNAKLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMKNGEKGEELR 443
M + +DQ NA L+ DV K+G+ V +V I + +M+ E G+E+R
Sbjct: 363 MATWAMHSDQPRNAVLVTDVLKVGLIVKDWEQRKSLVSASVIENAVRRLMETKE-GDEIR 421
Query: 444 RNAKKWK 450
+ A K K
Sbjct: 422 KRAVKLK 428
>UFO4_MANES (Q40286) Flavonol 3-O-glucosyltransferase 4 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
4) (Fragment)
Length = 241
Score = 130 bits (326), Expect = 1e-29
Identities = 77/212 (36%), Positives = 116/212 (54%), Gaps = 9/212 (4%)
Query: 257 DTSFGGDIIQAPSNDYIEWLDSKPPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLW 316
D + GD + + ++WLD P SV+Y GS L+ Q+ E+ L PF+W
Sbjct: 7 DKAERGDKASVDNTELLKWLDLWEPGSVIYACLGSISGLTSWQLAELGLGLESTNQPFIW 66
Query: 317 VIREKEKFQDGEKE-KEEELGNIEELEEVGKIAKWCSQVEVLSHRSMGCFLTHCGWNSTM 375
VIRE EK + EK EE + E I W QV +LSH ++G F THCGWNST+
Sbjct: 67 VIREGEKSEGLEKWILEEGYEERKRKREDFWIRGWSPQVLILSHPAIGAFFTHCGWNSTL 126
Query: 376 ESLVSRVPMVAFPQWTDQNTNAKLIEDVWKIGVRVDLHVN-----ED---GIVEGEEIRR 427
E + + VP+VA P + +Q N KL+ +V IGV V + ED +++ E++++
Sbjct: 127 EGISAGVPIVACPLFAEQFYNEKLVVEVLGIGVSVGVEAAVTWGLEDKCGAVMKKEQVKK 186
Query: 428 GLTVVMKNGEKGEELRRNAKKWKGLAKEAGKE 459
+ +VM G++GEE RR A++ +AK +E
Sbjct: 187 AIEIVMDKGKEGEERRRRAREIGEMAKRTIEE 218
>ZOG_PHALU (Q9ZSK5) Zeatin O-glucosyltransferase (EC 2.4.1.203)
(Trans-zeatin O-beta-D-glucosyltransferase)
Length = 459
Score = 129 bits (323), Expect = 2e-29
Identities = 69/183 (37%), Positives = 108/183 (58%), Gaps = 6/183 (3%)
Query: 273 IEWLDSKPPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEKFQ--DGEKE 330
+EWLD + PSSV+Y+SFG+ L Q+ +IA L + F+WV+RE +K G +
Sbjct: 252 MEWLDKQEPSSVIYISFGTTTALRDEQIQQIATGLEQSKQKFIWVLREADKGDIFAGSEA 311
Query: 331 KEEEL--GNIEELEEVGKIAK-WCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAF 387
K EL G E +E +G + + W Q+E+LSH S G F++HCGWNS +ES+ VP+ +
Sbjct: 312 KRYELPKGFEERVEGMGLVVRDWAPQLEILSHSSTGGFMSHCGWNSCLESITMGVPIATW 371
Query: 388 PQWTDQNTNAKLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAK 447
P +DQ NA L+ +V K+G+ V + +V + G+ +M+ E G+E+R+ A
Sbjct: 372 PMHSDQPRNAVLVTEVLKVGLVVKDWAQRNSLVSASVVENGVRRLMETKE-GDEMRQRAV 430
Query: 448 KWK 450
+ K
Sbjct: 431 RLK 433
>UFOG_SOLME (Q43641) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
Length = 433
Score = 129 bits (323), Expect = 2e-29
Identities = 72/184 (39%), Positives = 105/184 (56%), Gaps = 13/184 (7%)
Query: 273 IEWLDSKPPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEKFQDGEKEKE 332
I+WLD + SVVY+SFG+ L ++ IA AL + PF+W +R +G K
Sbjct: 249 IQWLDKQKEKSVVYLSFGTVTTLPPNEIGSIAEALETKKTPFIWSLRN-----NGVKNLP 303
Query: 333 EELGNIEELEEVGKIAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTD 392
+ G +E +E GKI W Q+E+L+H+S+G F+THCGWNS +E + VPM+ P + D
Sbjct: 304 K--GFLERTKEFGKIVSWAPQLEILAHKSVGVFVTHCGWNSILEGISFGVPMICRPFFGD 361
Query: 393 QNTNAKLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGL 452
Q N++++E VW+IG+++ E GI I L N EKG+ LR N + K
Sbjct: 362 QKLNSRMVESVWEIGLQI-----EGGIFTKSGIISALDTFF-NEEKGKILRENVEGLKEK 415
Query: 453 AKEA 456
A EA
Sbjct: 416 ALEA 419
>UFO7_MANES (Q40289) Flavonol 3-O-glucosyltransferase 7 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
7) (Fragment)
Length = 287
Score = 128 bits (321), Expect = 4e-29
Identities = 67/182 (36%), Positives = 103/182 (55%), Gaps = 13/182 (7%)
Query: 275 WLDSKPPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEKFQDGEKEKEEE 334
WLD + P+SV Y+SFGS ++ +A AL + PFLW +++ K
Sbjct: 93 WLDKQKPASVAYISFGSVATPPPHELVALAEALEASKVPFLWSLKDHSKVHLPN------ 146
Query: 335 LGNIEELEEVGKIAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQN 394
G ++ + G + W QVE+L H ++G F+THCGWNS +ES+V VPM+ P + DQ
Sbjct: 147 -GFLDRTKSHGIVLSWAPQVEILEHAALGVFVTHCGWNSILESIVGGVPMICRPFFGDQR 205
Query: 395 TNAKLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGLAK 454
N +++EDVW+IG+ +D G++ GL ++ G KG+++R N K+ K LAK
Sbjct: 206 LNGRMVEDVWEIGLLMD-----GGVLTKNGAIDGLNQILLQG-KGKKMRENIKRLKELAK 259
Query: 455 EA 456
A
Sbjct: 260 GA 261
>UFO6_MANES (Q40288) Flavonol 3-O-glucosyltransferase 6 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
6) (Fragment)
Length = 394
Score = 122 bits (305), Expect = 3e-27
Identities = 90/304 (29%), Positives = 145/304 (47%), Gaps = 33/304 (10%)
Query: 167 ESSCSIEFSGLPFSLSPHDMP-SYLISKGSYVLPLFKEDFKELRG*ANTTILVNTFEALE 225
+S + L SL +P S L+ Y +E +G I+VNTF LE
Sbjct: 87 DSDAELSVPSLANSLPARVLPASMLVKDRFYAFIRIIRGLREAKG-----IMVNTFMELE 141
Query: 226 PEAPRAV--DKLNMIPIGLLIPSAFLDRKDFAHDTSFGGDIIQAPSNDYIEWLDSKPPSS 283
A ++ D+ + PI + P L ++ + + ++ IEWLD +PPSS
Sbjct: 142 SHALNSLKDDQSKIPPIYPVGPILKLSNQE---------NDVGPEGSEIIEWLDDQPPSS 192
Query: 284 VVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEKFQDGEKEKEEELGNIEELEE 343
VV++ FGS Q EIA AL RH FLW +R G+ E + N++E+
Sbjct: 193 VVFLCFGSMGGFDMDQAKEIACALEQSRHRFLWSLRRPPP--KGKIETSTDYENLQEILP 250
Query: 344 VG---------KIAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQN 394
VG K+ W QV +L H ++G F++HCGWNS +ES+ VP+ +P + +Q
Sbjct: 251 VGFSERTAGMGKVVGWAPQVAILEHPAIGGFVSHCGWNSILESIWFSVPIATWPLYAEQQ 310
Query: 395 TNA--KLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGL 452
NA + E + +++D + I+ ++I RG+ VM E E+R+ K+
Sbjct: 311 FNAFTMVTELGLAVEIKMDYKKESEIILSADDIERGIKCVM---EHHSEIRKRVKEMSDK 367
Query: 453 AKEA 456
+++A
Sbjct: 368 SRKA 371
>UFO5_MANES (Q40287) Flavonol 3-O-glucosyltransferase 5 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
5)
Length = 487
Score = 120 bits (302), Expect = 6e-27
Identities = 87/266 (32%), Positives = 137/266 (50%), Gaps = 41/266 (15%)
Query: 216 ILVNTFEALEPEAPRA---------VDKLNMIPIGLLIPSAFLDRKDFAHDTSFGGDIIQ 266
IL+NT+EALEP A V K+ + PIG L A
Sbjct: 212 ILMNTWEALEPTTFGALRDVKFLGRVAKVPVFPIGPLRRQAG-----------------P 254
Query: 267 APSN-DYIEWLDSKPPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEK-- 323
SN + ++WLD +P SVVYVSFGS LS QM E+A L + F+WV+R+
Sbjct: 255 CGSNCELLDWLDQQPKESVVYVSFGSGGTLSLEQMIELAWGLERSQQRFIWVVRQPTVKT 314
Query: 324 -----FQDGEKEKEEE----LGNIEELEEVGKIA-KWCSQVEVLSHRSMGCFLTHCGWNS 373
F G+ + G + ++ VG + +W Q+ ++SH S+G FL+HCGWNS
Sbjct: 315 GDAAFFTQGDGADDMSGYFPEGFLTRIQNVGLVVPQWSPQIHIMSHPSVGVFLSHCGWNS 374
Query: 374 TMESLVSRVPMVAFPQWTDQNTNAKLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVM 433
+ES+ + VP++A+P + +Q NA L+ + + VR ++ +V+ EEI R + +M
Sbjct: 375 VLESITAGVPIIAWPIYAEQRMNATLLTEELGVAVR-PKNLPAKEVVKREEIERMIRRIM 433
Query: 434 KNGEKGEELRRNAKKWKGLAKEAGKE 459
+ E+G E+R+ ++ K ++A E
Sbjct: 434 VD-EEGSEIRKRVRELKDSGEKALNE 458
>UFO2_MANES (Q40285) Flavonol 3-O-glucosyltransferase 2 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
2) (Fragment)
Length = 346
Score = 120 bits (302), Expect = 6e-27
Identities = 97/318 (30%), Positives = 162/318 (50%), Gaps = 44/318 (13%)
Query: 159 YIAKKNTVESSCSIEFSG------LPFSLSPHD---MPSYLISKGSY--VLPLFKEDFKE 207
Y+ K + E+ IEF +P ++P +PS +++K + +L + K+ F++
Sbjct: 32 YVQKIHDEENFNPIEFKDSDTELIVPSLVNPFPTRILPSSILNKERFGQLLAIAKK-FRQ 90
Query: 208 LRG*ANTTILVNTFEALEPEAPRAVDKLNMIPIGLLIPSAFLDRKDFAHDTSFGGDIIQA 267
+G I+VNTF LE RA++ + P+ + P LD K +T
Sbjct: 91 AKG-----IIVNTFLELES---RAIESFKVPPLYHVGP--ILDVKSDGRNTH-------- 132
Query: 268 PSNDYIEWLDSKPPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIR-----EKE 322
+ ++WLD +P SVV++ FGS S+ Q+ EIA AL + H FLW IR +K
Sbjct: 133 --PEIMQWLDDQPEGSVVFLCFGSMGSFSEDQLKEIAYALENSGHRFLWSIRRPPPPDKI 190
Query: 323 KFQDGEKEKEEEL--GNIEELEEVGKIAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVS 380
++ + L G +E VGK+ W QV VL+H ++G F++HCGWNS +ESL
Sbjct: 191 ASPTDYEDPRDVLPEGFLERTVAVGKVIGWAPQVAVLAHPAIGGFVSHCGWNSVLESLWF 250
Query: 381 RVPMVAFPQWTDQNTNAKLIEDVWKIGVRVDLHVNEDG--IVEGEEIRRGLTVVMKNGEK 438
VP+ +P + +Q NA + +GV +D+ ++ IV ++I R + +M+N
Sbjct: 251 GVPIATWPMYAEQQFNAFEMVVELGLGVEIDMGYRKESGIIVNSDKIERAIRKLMEN--- 307
Query: 439 GEELRRNAKKWKGLAKEA 456
+E R+ K+ + +K A
Sbjct: 308 SDEKRKKVKEMREKSKMA 325
>UFO1_MANES (Q40284) Flavonol 3-O-glucosyltransferase 1 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
1)
Length = 449
Score = 119 bits (299), Expect = 1e-26
Identities = 87/302 (28%), Positives = 152/302 (49%), Gaps = 33/302 (10%)
Query: 168 SSCSIEFSGLPFSLSPHDMPSYLISKGSYVLPLFKED--FKELRG*ANTTILVNTFEALE 225
S ++ GL S MP+ ++SK + PL + + E +G +++NTF LE
Sbjct: 154 SDGELQVPGLVNSFPSKAMPTAILSK-QWFPPLLENTRRYGEAKG-----VIINTFFELE 207
Query: 226 PEAPRAVDKLNMIPIGLLIPSAFLDRKDFAHDTSFGGDIIQAPSNDYIEWLDSKPPSSVV 285
A + + P+G + LD + +T+ + ++WLD +PPSSVV
Sbjct: 208 SHAIESFKDPPIYPVGPI-----LDVRSNGRNTN----------QEIMQWLDDQPPSSVV 252
Query: 286 YVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEK--FQDGEKEKEE-----ELGNI 338
++ FGS SK Q+ EIA AL D H FLW + + F + + E+ G +
Sbjct: 253 FLCFGSNGSFSKDQVKEIACALEDSGHRFLWSLADHRAPGFLESPSDYEDLQEVLPEGFL 312
Query: 339 EELEEVGKIAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNA- 397
E + K+ W QV VL+H + G ++H GWNS +ES+ VP+ +P + +Q NA
Sbjct: 313 ERTSGIEKVIGWAPQVAVLAHPATGGLVSHSGWNSILESIWFGVPVATWPMYAEQQFNAF 372
Query: 398 -KLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMK-NGEKGEELRRNAKKWKGLAKE 455
+IE + +++D + IV+ ++I RG+ +MK + ++ ++++ ++K +G E
Sbjct: 373 QMVIELGLAVEIKMDYRNDSGEIVKCDQIERGIRCLMKHDSDRRKKVKEMSEKSRGALME 432
Query: 456 AG 457
G
Sbjct: 433 GG 434
Score = 30.8 bits (68), Expect = 8.0
Identities = 20/71 (28%), Positives = 33/71 (46%), Gaps = 8/71 (11%)
Query: 140 WIEPTTVMSILYNYFLGGDYIAKKNTVESSCSIEFSGLPF--SLSPHDMPSYLISKGSYV 197
W++ S+++ F +K E +C++E SG F SL+ H P +L S Y
Sbjct: 242 WLDDQPPSSVVFLCFGSNGSFSKDQVKEIACALEDSGHRFLWSLADHRAPGFLESPSDY- 300
Query: 198 LPLFKEDFKEL 208
ED +E+
Sbjct: 301 -----EDLQEV 306
>HQGT_RAUSE (Q9AR73) Hydroquinone glucosyltransferase (EC 2.4.1.218)
(Arbutin synthase)
Length = 470
Score = 117 bits (292), Expect = 8e-26
Identities = 84/257 (32%), Positives = 131/257 (50%), Gaps = 34/257 (13%)
Query: 216 ILVNTFEALEPEAPRAVD-----KLNMIPIGLLIPSAFLDRKDFAHDTSFGGDIIQAPSN 270
I+VNTF LEP +A+ K + PIG LI + + D
Sbjct: 207 IMVNTFNDLEPGPLKALQEEDQGKPPVYPIGPLIRADSSSKVDDC--------------- 251
Query: 271 DYIEWLDSKPPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIRE-KEKFQDGE- 328
+ ++WLD +P SV+++SFGS +S Q E+A L FLWV+R +K +
Sbjct: 252 ECLKWLDDQPRGSVLFISFGSGGAVSHNQFIELALGLEMSEQRFLWVVRSPNDKIANATY 311
Query: 329 ---KEKEEELGNIEE--LEEV-GK---IAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLV 379
+ + + L + E LE G+ + W Q E+LSH S G FLTHCGWNS +ES+V
Sbjct: 312 FSIQNQNDALAYLPEGFLERTKGRCLLVPSWAPQTEILSHGSTGGFLTHCGWNSILESVV 371
Query: 380 SRVPMVAFPQWTDQNTNAKLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMKNGEKG 439
+ VP++A+P + +Q NA ++ + K+ +R E+G++ EI + +M+ GE+G
Sbjct: 372 NGVPLIAWPLYAEQKMNAVMLTEGLKVALRP--KAGENGLIGRVEIANAVKGLME-GEEG 428
Query: 440 EELRRNAKKWKGLAKEA 456
++ R K K A A
Sbjct: 429 KKFRSTMKDLKDAASRA 445
>UFOG_HORVU (P14726) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1)
Length = 455
Score = 108 bits (271), Expect = 2e-23
Identities = 81/278 (29%), Positives = 124/278 (44%), Gaps = 32/278 (11%)
Query: 185 DMPSYLISKG-SYVLPLFKEDFKELRG*ANTTILVNTFEALEPE---APRAVDKLNMIPI 240
D+P ++S +YV+ L + A T + +NTF L+P A A + N +P+
Sbjct: 183 DLPDGVVSGDFNYVISLLVHRQAQRLPKAATAVALNTFPGLDPPDLIAALAAELPNCLPL 242
Query: 241 GLLIPSAFLDRKDFAHDTSFGGDIIQAPSNDY--IEWLDSKPPSSVVYVSFGSYIVLSKT 298
G P L + DT+ +AP++ + + WLD +P SV YVSFG+
Sbjct: 243 G---PYHLLPGAEPTADTN------EAPADPHGCLAWLDRRPARSVAYVSFGTNATARPD 293
Query: 299 QMDEIARALLDCRHPFLWVIREKEKFQDGEKEKEEELGNIEELEEVGKIAKWCSQVEVLS 358
++ E+A L PFLW +R G +E G + W QV VL
Sbjct: 294 ELQELAAGLEASGAPFLWSLRGVVA--------AAPRGFLERAP--GLVVPWAPQVGVLR 343
Query: 359 HRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKLIEDVWKIGVRVDLHVNEDG 418
H ++G F+TH GW S ME + S VPM P + DQ NA+ + VW G D +
Sbjct: 344 HAAVGAFVTHAGWASVMEGVSSGVPMACRPFFGDQTMNARSVASVWGFGTAFDGPMTRGA 403
Query: 419 IVEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGLAKEA 456
+ + GE GE +R A++ + + +A
Sbjct: 404 VANA-------VATLLRGEDGERMRAKAQELQAMVGKA 434
>UFO1_MAIZE (P16166) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1) (Bz-McC allele)
Length = 471
Score = 108 bits (270), Expect = 3e-23
Identities = 85/279 (30%), Positives = 128/279 (45%), Gaps = 31/279 (11%)
Query: 185 DMPSYLISKG-SYVLPLFKEDFKELRG*ANTTILVNTFEALEP-EAPRAVDKL--NMIPI 240
D+P ++S +YV+ L + + + +NTF L+P + A+ ++ N +P
Sbjct: 196 DLPDGVVSGDFNYVINLLVHRMGQCLPRSAAAVALNTFPGLDPPDVTAALAEILPNCVPF 255
Query: 241 GLLIPSAFLDRKDFAHDTSFGGDIIQAPSNDYIEWLDSKPPSSVVYVSFGSYIVLSKTQM 300
G P L +D A DT+ D + + WL +P V YVSFG+ ++
Sbjct: 256 G---PYHLLLAEDDA-DTAAPAD-----PHGCLAWLGRQPARGVAYVSFGTVACPRPDEL 306
Query: 301 DEIARALLDCRHPFLWVIREKE--KFQDGEKEKEEELGNIEELEEVGKIAKWCSQVEVLS 358
E+A L D PFLW +RE G ++ G+ G + W QV VL
Sbjct: 307 RELAAGLEDSGAPFLWSLREDSWPHLPPGFLDRAAGTGS-------GLVVPWAPQVAVLR 359
Query: 359 HRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKLIEDVWKIGVRVDLHVNEDG 418
H S+G F+TH GW S +E L S VPM P + DQ NA+ + VW G + + G
Sbjct: 360 HPSVGAFVTHAGWASVLEGLSSGVPMACRPFFGDQRMNARSVAHVWGFGAAFEGAMTSAG 419
Query: 419 IVEG-EEIRRGLTVVMKNGEKGEELRRNAKKWKGLAKEA 456
+ EE+ R GE+G +R AK+ + L EA
Sbjct: 420 VATAVEELLR--------GEEGARMRARAKELQALVAEA 450
>UFO2_MAIZE (P16165) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1) (Bz-Mc2 allele)
Length = 471
Score = 103 bits (258), Expect = 7e-22
Identities = 83/279 (29%), Positives = 127/279 (44%), Gaps = 31/279 (11%)
Query: 185 DMPSYLISKG-SYVLPLFKEDFKELRG*ANTTILVNTFEALEP-EAPRAVDKL--NMIPI 240
D+P ++S +YV+ L + + + +NTF L+P + A+ ++ N +P
Sbjct: 196 DLPDGVVSGDFNYVISLLVHRMGQCLPRSAAAVALNTFPGLDPPDVTAALAEILPNCVPF 255
Query: 241 GLLIPSAFLDRKDFAHDTSFGGDIIQAPSNDYIEWLDSKPPSSVVYVSFGSYIVLSKTQM 300
G P L +D A DT+ D + + WL +P V YVSFG+ ++
Sbjct: 256 G---PYHLLLAEDDA-DTAAPAD-----PHGCLAWLGRQPARGVAYVSFGTVACPRPDEL 306
Query: 301 DEIARALLDCRHPFLWVIREKE--KFQDGEKEKEEELGNIEELEEVGKIAKWCSQVEVLS 358
E+A L PFLW +RE G ++ G+ G + W QV VL
Sbjct: 307 RELAAGLEASAAPFLWSLREDSWTLLPPGFLDRAAGTGS-------GLVVPWAPQVAVLR 359
Query: 359 HRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKLIEDVWKIGVRVDLHVNEDG 418
H S+G F+TH GW S +E + S VPM P + DQ NA+ + VW G + + G
Sbjct: 360 HPSVGAFVTHAGWASVLEGVSSGVPMACRPFFGDQRMNARSVAHVWGFGAAFEGAMTSAG 419
Query: 419 IVEG-EEIRRGLTVVMKNGEKGEELRRNAKKWKGLAKEA 456
+ EE+ R GE+G +R AK+ + L EA
Sbjct: 420 VAAAVEELLR--------GEEGAGMRARAKELQALVAEA 450
>UFO3_MAIZE (P16167) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1) (Bz-W22 allele)
Length = 471
Score = 102 bits (255), Expect = 2e-21
Identities = 83/279 (29%), Positives = 126/279 (44%), Gaps = 31/279 (11%)
Query: 185 DMPSYLISKG-SYVLPLFKEDFKELRG*ANTTILVNTFEALEP-EAPRAVDKL--NMIPI 240
D+P ++S +YV+ L + + + +NTF L+P + A+ ++ N +P
Sbjct: 196 DLPDGVVSGDFNYVINLLVHRMGQCLPRSAAAVALNTFPGLDPPDVTAALAEILPNCVPF 255
Query: 241 GLLIPSAFLDRKDFAHDTSFGGDIIQAPSNDYIEWLDSKPPSSVVYVSFGSYIVLSKTQM 300
G P L +D A DT+ D + + WL +P V YVSFG+ ++
Sbjct: 256 G---PYHLLLAEDDA-DTAAPAD-----PHGCLAWLGRQPARGVAYVSFGTVACPRPDEL 306
Query: 301 DEIARALLDCRHPFLWVIREKE--KFQDGEKEKEEELGNIEELEEVGKIAKWCSQVEVLS 358
E+A L PFLW +RE G ++ G+ G + W QV VL
Sbjct: 307 RELAAGLEASGAPFLWSLREDSWTLLPPGFLDRAAGTGS-------GLVVPWAPQVAVLR 359
Query: 359 HRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKLIEDVWKIGVRVDLHVNEDG 418
H S+G F+TH GW S +E + S VPM P + DQ NA+ + VW G + + G
Sbjct: 360 HPSVGAFVTHAGWASVLEGVSSGVPMACRPFFGDQRMNARSVAHVWGFGAAFEGAMTSAG 419
Query: 419 IVEG-EEIRRGLTVVMKNGEKGEELRRNAKKWKGLAKEA 456
+ EE+ R GE+G +R AK + L EA
Sbjct: 420 VAAAVEELLR--------GEEGARMRARAKVLQALVAEA 450
>UFOG_PETHY (Q43716) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Anthocyanin rhamnosyl transferase)
Length = 473
Score = 97.8 bits (242), Expect = 5e-20
Identities = 64/198 (32%), Positives = 103/198 (51%), Gaps = 10/198 (5%)
Query: 262 GDIIQAPSNDYIE-----WLDSKPPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLW 316
G ++ P + +E WL+ +V+Y SFGS L+ Q+ E+A L PF
Sbjct: 248 GPVVPDPPSGKLEEKWATWLNKFEGGTVIYCSFGSETFLTDDQVKELALGLEQTGLPFFL 307
Query: 317 VIREKEKFQ-DGEKEKEEELGNIEELEEVGKI-AKWCSQVEVLSHRSMGCFLTHCGWNST 374
V+ E + G +E +++ G I + W Q +L+H S+GC++ H G++S
Sbjct: 308 VLNFPANVDVSAELNRALPEGFLERVKDKGIIHSGWVQQQNILAHSSVGCYVCHAGFSSV 367
Query: 375 MESLVSRVPMVAFPQWTDQNTNAKLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMK 434
+E+LV+ +V PQ DQ NAKL+ + GV ++ +EDG E+I+ + VM
Sbjct: 368 IEALVNDCQVVMLPQKGDQILNAKLVSGDMEAGVEINRR-DEDGYFGKEDIKEAVEKVMV 426
Query: 435 NGEK--GEELRRNAKKWK 450
+ EK G+ +R N KKWK
Sbjct: 427 DVEKDPGKLIRENQKKWK 444
>UFOG_VITVI (P51094) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Fragment)
Length = 154
Score = 82.8 bits (203), Expect = 2e-15
Identities = 43/121 (35%), Positives = 65/121 (53%), Gaps = 6/121 (4%)
Query: 336 GNIEELEEVGKIAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNT 395
G +E+ G + W Q EVL+ R G F+THCGWNS ES+ VP++ P + DQ
Sbjct: 16 GFLEKTRGYGMVVPWAPQAEVLALRQFGAFVTHCGWNSLWESVAGGVPLICRPFFGDQRL 75
Query: 396 NAKLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGLAKE 455
N +++EDV +IGVR+ E G+ + ++ EKG++LR N + + A
Sbjct: 76 NGRMVEDVLEIGVRI-----EGGVFTKSGLMSCFDQILSQ-EKGKKLRENLRALRETADR 129
Query: 456 A 456
A
Sbjct: 130 A 130
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.139 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 57,675,383
Number of Sequences: 164201
Number of extensions: 2540810
Number of successful extensions: 6959
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 89
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 6812
Number of HSP's gapped (non-prelim): 118
length of query: 475
length of database: 59,974,054
effective HSP length: 114
effective length of query: 361
effective length of database: 41,255,140
effective search space: 14893105540
effective search space used: 14893105540
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0108.9


