

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
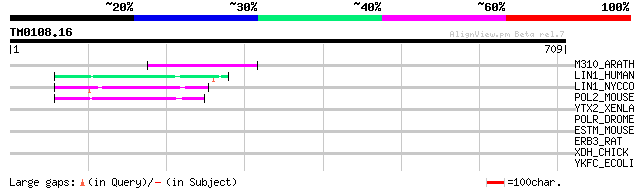
Query= TM0108.16
(709 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310... 67 1e-10
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 57 2e-07
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 51 1e-05
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 49 5e-05
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 39 0.047
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 35 0.68
ESTM_MOUSE (Q63880) Liver carboxylesterase 31 precursor (EC 3.1.... 34 1.5
ERB3_RAT (Q62799) Receptor tyrosine-protein kinase erbB-3 precur... 32 4.4
XDH_CHICK (P47990) Xanthine dehydrogenase/oxidase [Includes: Xan... 32 7.6
YKFC_ECOLI (Q47688) Hypothetical protein ykfC 31 9.9
>M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310
(ORF154)
Length = 154
Score = 67.4 bits (163), Expect = 1e-10
Identities = 37/142 (26%), Positives = 67/142 (47%), Gaps = 2/142 (1%)
Query: 177 AMPTYTMQVFWLPRSIIHHIDRAMRSFIWSKGSGQRGWNLVNWSTVVRDKSH-GGLGMKD 235
A+P Y M F L + + + AM F WS +R + V W + + K GGLG +D
Sbjct: 2 ALPVYAMSCFRLSKLLCKKLTSAMTEFWWSSCENKRKISWVAWQKLCKSKEDDGGLGFRD 61
Query: 236 MSDHNTALLGKAVWLLLKNSNKLWVQVMQHKYLRDHTILNAPHRASSSAVWKGILRARDR 295
+ N ALL K + ++ + L ++++ +Y +++ S W+ I+ R+
Sbjct: 62 LGWFNQALLAKQSFRIIHQPHTLLSRLLRSRYFPHSSMMECSVGTRPSYAWRSIIHGREL 121
Query: 296 LRGGFKFRLGTG-NSSIWYNDW 316
L G +G G ++ +W + W
Sbjct: 122 LSRGLLRTIGDGIHTKVWLDRW 143
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 56.6 bits (135), Expect = 2e-07
Identities = 52/232 (22%), Positives = 92/232 (39%), Gaps = 18/232 (7%)
Query: 58 LFADDVLLFCQASTTQVQLVADTLRDFFASSGLKVNIDKSKAISSKGVHPSIRDDIRGIA 117
LFADD++++ + Q + + +F SG K+N+ KS+A + I
Sbjct: 698 LFADDMIVYLENPIVSAQNLLKLISNFSKVSGYKINVQKSQAFLYTN-NRQTESQIMSEL 756
Query: 118 PIPLVNDLGKYLGFPLSGGRVSRGRFNF--LLENINRKMAAWKTNLLNLAGRACLAKSVI 175
P + + KYLG L+ + N+ LL I WK + GR + K I
Sbjct: 757 PFTIASKRIKYLGIQLTRDVKDLFKENYKPLLNEIKEDTNKWKNIPCSWVGRINIVKMAI 816
Query: 176 AAMPTYTMQV--FWLPRSIIHHIDRAMRSFIWSKGSGQRGWNLVNWSTVVRDKSHGGLGM 233
Y LP + +++ FIW++ + ST+ + GG+ +
Sbjct: 817 LPKVIYRFNAIPIKLPMTFFTELEKTTLKFIWNQKRAH-----IAKSTLSQKNKAGGITL 871
Query: 234 KDMSDHNTALLGKAVWLLLKNSN-KLW-----VQVMQHKYLRDHTILNAPHR 279
D + A + K W +N + W ++M H Y ++ I + P +
Sbjct: 872 PDFKLYYKATVTKTAWYWYQNRDIDQWNRTEPSEIMPHIY--NYLIFDKPEK 921
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 50.8 bits (120), Expect = 1e-05
Identities = 44/204 (21%), Positives = 87/204 (42%), Gaps = 16/204 (7%)
Query: 58 LFADDVLLFCQASTTQVQLVADTLRDFFASSGLKVNIDKSKAI---SSKGVHPSIRDDIR 114
LFADD++++ + + + + ++++ SG K+N KS A ++ +++D I
Sbjct: 698 LFADDMIVYLENTRDSTTKLLEVIKEYSNVSGYKINTHKSVAFIYTNNNQAEKTVKDSI- 756
Query: 115 GIAPIPLVNDLGKYLGFPLSGG--RVSRGRFNFLLENINRKMAAWKTNLLNLAGRACLAK 172
P +V KYLG L+ + + + L + I + WK + GR + K
Sbjct: 757 ---PFTVVPKKMKYLGVYLTKDVKDLYKENYETLRKEIAEDVNKWKNIPCSWLGRINIVK 813
Query: 173 SVIAAMPTYTMQVFWL--PRSIIHHIDRAMRSFIWSKGSGQRGWNLVNWSTVVRDKSHGG 230
I Y + P S +++ + FIW++ Q L++ GG
Sbjct: 814 MSILPKAIYNFNAIPIKAPLSYFKDLEKIILHFIWNQKKPQIAKTLLS-----NKNKAGG 868
Query: 231 LGMKDMSDHNTALLGKAVWLLLKN 254
+ + D+ + +++ K W KN
Sbjct: 869 ITLPDLRLYYKSIVIKTAWYWHKN 892
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 48.9 bits (115), Expect = 5e-05
Identities = 43/197 (21%), Positives = 81/197 (40%), Gaps = 12/197 (6%)
Query: 58 LFADDVLLFCQASTTQVQLVADTLRDFFASSGLKVNIDKSKAISSKGVHPSIRDDIRGIA 117
L ADD++++ + + + + F G K+N +KS A + +IR
Sbjct: 725 LLADDMIVYISDPKNSTRELLNLINSFGEVVGYKINSNKSMAFLYT-KNKQAEKEIRETT 783
Query: 118 PIPLVNDLGKYLGFPLSG--GRVSRGRFNFLLENINRKMAAWKTNLLNLAGRACLAKSVI 175
P +V + KYLG L+ + F L + I + WK + GR + K I
Sbjct: 784 PFSIVTNNIKYLGVTLTKEVKDLYDKNFKSLKKEIKEDLRRWKDLPCSWIGRINIVKMAI 843
Query: 176 AAMPTYTMQV--FWLPRSIIHHIDRAMRSFIWSKGSGQRGWNLVNWSTVVRDK-SHGGLG 232
Y +P + ++ A+ F+W+ + ++++DK + GG+
Sbjct: 844 LPKAIYRFNAIPIKIPTQFFNELEGAICKFVWNNKKPRIA------KSLLKDKRTSGGIT 897
Query: 233 MKDMSDHNTALLGKAVW 249
M D+ + A++ K W
Sbjct: 898 MPDLKLYYRAIVIKTAW 914
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 38.9 bits (89), Expect = 0.047
Identities = 37/155 (23%), Positives = 66/155 (41%), Gaps = 16/155 (10%)
Query: 59 FADDVLLFCQASTTQVQLVADTLRDFFASSGLKVNIDKSKAISSKGVH-----PSIRDDI 113
+ADDV+L Q ++ + + A+S ++N KS + + P+ RD
Sbjct: 694 YADDVILVAQ-DLVDLERAQECQEVYAAASSARINWSKSSGLLEGSLKVDFLPPAFRD-- 750
Query: 114 RGIAPIPLVNDLGKYLGFPLSGGRVSRGR-FNFLLENINRKMAAWK--TNLLNLAGRACL 170
I + + KYLG LS + F L E + ++ WK +L++ GRA +
Sbjct: 751 -----ISWESKIIKYLGVYLSAEEYPVSQNFIELEECVLTRLGKWKGFAKVLSMRGRALV 805
Query: 171 AKSVIAAMPTYTMQVFWLPRSIIHHIDRAMRSFIW 205
++A+ Y + + I I R + F+W
Sbjct: 806 INQLVASQIWYRLICLSPTQEFIAKIQRRLLDFLW 840
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 35.0 bits (79), Expect = 0.68
Identities = 18/46 (39%), Positives = 28/46 (60%), Gaps = 1/46 (2%)
Query: 59 FADDVLLFCQASTTQVQLVADTLRDFFASSGLKVNIDKSKAISSKG 104
FADD++LF + + +Q++ D DF + GLK+N DK + KG
Sbjct: 579 FADDLVLFAE-TRMGLQVLLDKTLDFLSIVGLKLNADKCFTVGIKG 623
>ESTM_MOUSE (Q63880) Liver carboxylesterase 31 precursor (EC
3.1.1.1) (ES-Male) (Esterase-31)
Length = 554
Score = 33.9 bits (76), Expect = 1.5
Identities = 16/47 (34%), Positives = 26/47 (55%), Gaps = 3/47 (6%)
Query: 114 RGIAPIPLVNDLGKYLGF---PLSGGRVSRGRFNFLLENINRKMAAW 157
+G+ P P +N L +YL P +G ++ +GR F E + RK+ W
Sbjct: 495 KGLPPWPQLNQLEQYLEIGLEPRTGVKLKKGRLQFWTETLPRKIQEW 541
>ERB3_RAT (Q62799) Receptor tyrosine-protein kinase erbB-3 precursor
(EC 2.7.1.112) (c-erbB3)
Length = 1339
Score = 32.3 bits (72), Expect = 4.4
Identities = 15/42 (35%), Positives = 24/42 (56%), Gaps = 2/42 (4%)
Query: 405 CLHDALPVNSKRFHCHLANSEACSRCSYIREDGIHSLRDCTH 446
C + LP+ C+ + S+AC+RC++ R DG H + C H
Sbjct: 552 CHPECLPMEGTST-CNGSGSDACARCAHFR-DGPHCVNSCPH 591
>XDH_CHICK (P47990) Xanthine dehydrogenase/oxidase [Includes:
Xanthine dehydrogenase (EC 1.1.1.204) (XD); Xanthine
oxidase (EC 1.1.3.22) (XO) (Xanthine oxidoreductase)]
Length = 1358
Score = 31.6 bits (70), Expect = 7.6
Identities = 29/108 (26%), Positives = 47/108 (42%), Gaps = 5/108 (4%)
Query: 250 LLLKNSNKLWVQVMQHKYLRDHTILNAPHRASSSAVWKGILRARDRLRGGFKFRLGTGNS 309
L LK +L + K L+D L A S + G++ R L F F+
Sbjct: 491 LALKTCRELAGRDWNEKLLQDACRLLAGEMDLSPSAPGGMVEFRRTLTLSFFFKFYLTVL 550
Query: 310 SIWYNDWSGHGNLCEHLPFVHITDTQLHLRDLVVNNAWDFSKLFTSIP 357
D +G NLCE +P +I+ T+L +D + + ++LF +P
Sbjct: 551 QKLSKDQNGPNNLCEPVPPNYISATELFHKDPIAS-----TQLFQEVP 593
>YKFC_ECOLI (Q47688) Hypothetical protein ykfC
Length = 376
Score = 31.2 bits (69), Expect = 9.9
Identities = 26/95 (27%), Positives = 41/95 (42%), Gaps = 26/95 (27%)
Query: 52 PPVSHLLFADDVLLFCQASTTQVQLVADTLRDFFASS-GLKVNIDKSKAISSKGVHPSIR 110
P V++ +ADD +L + + QV+ + + R S L++N+DK+K
Sbjct: 270 PAVAYCRYADDFVLIVKGTKAQVEAIREECRGVLEGSLKLRLNMDKTK------------ 317
Query: 111 DDIRGIAPIPLVNDLGKYLGFPLSGGRVSRGRFNF 145
IP VND GF G R+ R R +
Sbjct: 318 --------IPHVND-----GFIFLGHRLIRKRSRY 339
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.324 0.137 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 89,237,427
Number of Sequences: 164201
Number of extensions: 3821387
Number of successful extensions: 9580
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 9573
Number of HSP's gapped (non-prelim): 12
length of query: 709
length of database: 59,974,054
effective HSP length: 117
effective length of query: 592
effective length of database: 40,762,537
effective search space: 24131421904
effective search space used: 24131421904
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0108.16


