

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
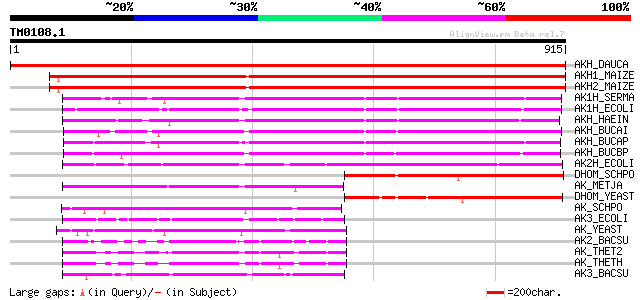
Query= TM0108.1
(915 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AKH_DAUCA (P37142) Bifunctional aspartokinase/homoserine dehydro... 1387 0.0
AKH1_MAIZE (P49079) Bifunctional aspartokinase/homoserine dehydr... 1315 0.0
AKH2_MAIZE (P49080) Bifunctional aspartokinase/homoserine dehydr... 1270 0.0
AK1H_SERMA (P27725) Bifunctional aspartokinase/homoserine dehydr... 553 e-156
AK1H_ECOLI (P00561) Bifunctional aspartokinase/homoserine dehydr... 552 e-156
AKH_HAEIN (P44505) Bifunctional aspartokinase/homoserine dehydro... 520 e-147
AKH_BUCAI (P57290) Bifunctional aspartokinase/homoserine dehydro... 513 e-145
AKH_BUCAP (Q8K9U9) Bifunctional aspartokinase/homoserine dehydro... 485 e-136
AKH_BUCBP (Q89AR4) Bifunctional aspartokinase/homoserine dehydro... 445 e-124
AK2H_ECOLI (P00562) Bifunctional aspartokinase/homoserine dehydr... 365 e-100
DHOM_SCHPO (O94671) Probable homoserine dehydrogenase (EC 1.1.1.... 291 4e-78
AK_METJA (Q57991) Probable aspartokinase (EC 2.7.2.4) (Aspartate... 265 3e-70
DHOM_YEAST (P31116) Homoserine dehydrogenase (EC 1.1.1.3) (HDH) 263 2e-69
AK_SCHPO (O60163) Probable aspartokinase (EC 2.7.2.4) (Aspartate... 170 1e-41
AK3_ECOLI (P08660) Lysine-sensitive aspartokinase III (EC 2.7.2.... 169 4e-41
AK_YEAST (P10869) Aspartokinase (EC 2.7.2.4) (Aspartate kinase) 163 2e-39
AK2_BACSU (P08495) Aspartokinase 2 (EC 2.7.2.4) (Aspartokinase I... 147 1e-34
AK_THET2 (P61488) Aspartokinase (EC 2.7.2.4) (Aspartate kinase) ... 140 2e-32
AK_THETH (P61489) Aspartokinase (EC 2.7.2.4) (Aspartate kinase) ... 139 3e-32
AK3_BACSU (P94417) Probable aspartokinase (EC 2.7.2.4) (Aspartat... 133 2e-30
>AKH_DAUCA (P37142) Bifunctional aspartokinase/homoserine
dehydrogenase, chloroplast precursor (AK-HD) (AK-HSDH)
[Includes: Aspartokinase (EC 2.7.2.4); Homoserine
dehydrogenase (EC 1.1.1.3)] (Fragment)
Length = 921
Score = 1387 bits (3589), Expect = 0.0
Identities = 709/920 (77%), Positives = 794/920 (86%), Gaps = 5/920 (0%)
Query: 1 MASLSASSLYHFSTISPSNNTPDITKISQCQCL-PFLPSHRSHSLRKALSLLPRGNQSPS 59
++S + S Y + S TP K L P H+S SL K L RG + S
Sbjct: 2 LSSAISPSSYAAIAAAYSARTPIFNKKKTAAVLSPLSLFHQSPSLSKTGIFLHRGRKESS 61
Query: 60 TK--ISASLTDVSPSV--LVEEQQLQKGETWSVHKFGGTCMGSSQRIKNVGDIVLEDDSE 115
+K I+AS+T PS+ VE+ L +G WS+HKFGGTC+GSS+RI+NV +IV+EDDSE
Sbjct: 62 SKFYIAASVTTAVPSLDDSVEKVHLPRGAMWSIHKFGGTCVGSSERIRNVAEIVVEDDSE 121
Query: 116 RKLVVVSAMSKVTDMMYELINKAQSRDESYVSSLDAVLEKHSLTAHDLLDGDGLATFLSQ 175
RKLVVVSAMSKVTDMMY+LI KAQSRD+SY S+LDAV+EKH LTA DLLD D LA FL++
Sbjct: 122 RKLVVVSAMSKVTDMMYDLIYKAQSRDDSYESALDAVMEKHKLTAFDLLDEDDLARFLTR 181
Query: 176 LHHDISNLKAMLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDV 235
L HD+ LKAMLRAIYIAGHATESF+DFVVGHGELWSAQ+LS VIRKNG DC WMDTRDV
Sbjct: 182 LQHDVITLKAMLRAIYIAGHATESFSDFVVGHGELWSAQLLSFVIRKNGGDCNWMDTRDV 241
Query: 236 LIVNPTSSNQVDPDYLESEKRLETWFSLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSA 295
L+VNP SNQVDPDYLESEKRLE WFS N C+ I+ATGFIASTPQ IPTTLKRDGSDFSA
Sbjct: 242 LVVNPAGSNQVDPDYLESEKRLEKWFSSNQCQTIVATGFIASTPQNIPTTLKRDGSDFSA 301
Query: 296 AIMGALFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTII 355
AIMGAL RA QVTIWTDV+GVYSADPRKVSEAV+LKTLSYQEAWEMSYFGANVLHPRTI
Sbjct: 302 AIMGALLRAGQVTIWTDVNGVYSADPRKVSEAVVLKTLSYQEAWEMSYFGANVLHPRTIN 361
Query: 356 PVMQYGIPILIRNIFNLSAPGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTG 415
PVM+Y IPI+IRNIFNLSAPGT IC SV + ED + +++ VKGFATIDNLALINVEGTG
Sbjct: 362 PVMRYDIPIVIRNIFNLSAPGTMICRESVGETEDGLKLESHVKGFATIDNLALINVEGTG 421
Query: 416 MAGVPGTASAIFGAVKDVGANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDN 475
MAGVPGTA+AIFGAVKDVGANVIMISQASSEHSICFAVPE EVKAVA+AL++RFR ALD
Sbjct: 422 MAGVPGTATAIFGAVKDVGANVIMISQASSEHSICFAVPESEVKAVAKALEARFRQALDA 481
Query: 476 GRLSQVAIIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLK 535
GRLSQVA PNCSILA VGQKMASTPGVSATLFNALAKANINVRAIAQGC+EYNITVVL
Sbjct: 482 GRLSQVANNPNCSILATVGQKMASTPGVSATLFNALAKANINVRAIAQGCTEYNITVVLS 541
Query: 536 REDCVKALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGI 595
REDCV+AL+AVHSRFYLSRTTIA+GI+GPGLIG+TLLDQLRDQA+ILKE IDLRVMGI
Sbjct: 542 REDCVRALKAVHSRFYLSRTTIAVGIVGPGLIGATLLDQLRDQAAILKENSKIDLRVMGI 601
Query: 596 LGSKSMLLSDWGIDLARWRELRDESGEVANLEKFVQHVHGNHFIPNTAIVDCTADSVIAG 655
GS++MLLS+ GIDL+RWRE++ E G+ A LEKFVQHV GNHFIP+T IVDCTADS +A
Sbjct: 602 TGSRTMLLSETGIDLSRWREVQKEKGQTAGLEKFVQHVRGNHFIPSTVIVDCTADSEVAS 661
Query: 656 HYYDWLRKGIHVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGL 715
HY+DWL +GIHV+TPNKKANSGPLDQYLKLRALQR+SYTHYFYEATV AGLPI++TL+GL
Sbjct: 662 HYHDWLCRGIHVITPNKKANSGPLDQYLKLRALQRRSYTHYFYEATVVAGLPIITTLQGL 721
Query: 716 LETGDRILQIEGIFSGTLSYIFNNFKDGRAFSEVVQEAKEAGYTEPDPRDDLSGTDVARK 775
LETGD+IL+IEGIFSGTLSYIFNNFK FSEVV EAK AGYTEPDPRDDL+GTDVARK
Sbjct: 722 LETGDKILRIEGIFSGTLSYIFNNFKSTTPFSEVVSEAKAAGYTEPDPRDDLAGTDVARK 781
Query: 776 VIILARESGLKLELSNIPVESLVPEPLQACASAQEFMQRLPEFDQMFAKKLEEAENAGEV 835
VIILAR SGLKLELS+IPV+SLVPEPL+ ASA+EF+ +LP+FD +K E+AENAGEV
Sbjct: 782 VIILARGSGLKLELSDIPVQSLVPEPLRGIASAEEFLLQLPQFDSDMTRKREDAENAGEV 841
Query: 836 LRYVGVVDVTSKKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQV 895
LRYVGVVD ++KGVVEL+RYKK+HPFAQLSGSDNI AFTT RY QP I+RGPGAGA+V
Sbjct: 842 LRYVGVVDAVNQKGVVELKRYKKEHPFAQLSGSDNINAFTTERYNKQPPIIRGPGAGAEV 901
Query: 896 TAGGIFSDILRLASYLGAPS 915
TAGG+FSDILRLASYLGAPS
Sbjct: 902 TAGGVFSDILRLASYLGAPS 921
>AKH1_MAIZE (P49079) Bifunctional aspartokinase/homoserine
dehydrogenase 1, chloroplast precursor (AK-HD 1)
(AK-HSDH 1) [Includes: Aspartokinase (EC 2.7.2.4);
Homoserine dehydrogenase (EC 1.1.1.3)]
Length = 920
Score = 1315 bits (3404), Expect = 0.0
Identities = 660/854 (77%), Positives = 744/854 (86%), Gaps = 7/854 (0%)
Query: 66 LTDVSPSVLVEEQQ----LQKGETWSVHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVV 121
L + +V VE+ + L KG+ WSVHKFGGTCMG+S+RI NV DIVL D SERKLVVV
Sbjct: 70 LAPTAGAVSVEQAEAIADLPKGDMWSVHKFGGTCMGTSERIHNVADIVLRDPSERKLVVV 129
Query: 122 SAMSKVTDMMYELINKAQSRDESYVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDIS 181
SAMSKVTDMMY L+NKAQSRD+SY++ LD V +KH TA DLL G+ LA FLSQLH DIS
Sbjct: 130 SAMSKVTDMMYNLVNKAQSRDDSYIAVLDEVFDKHMTTAKDLLAGEDLARFLSQLHADIS 189
Query: 182 NLKAMLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPT 241
NLKAMLRAIYIAGHATESF+DFVVGHGELWSAQMLS I+K+G C WMDTR+VL+VNP+
Sbjct: 190 NLKAMLRAIYIAGHATESFSDFVVGHGELWSAQMLSYAIQKSGTPCSWMDTREVLVVNPS 249
Query: 242 SSNQVDPDYLESEKRLETWFSLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGAL 301
+NQVDPDYLESEKRLE WFS P + IIATGFIASTP+ IPTTLKRDGSDFSAAI+G+L
Sbjct: 250 GANQVDPDYLESEKRLEKWFSRCPAETIIATGFIASTPENIPTTLKRDGSDFSAAIIGSL 309
Query: 302 FRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYG 361
+ARQVTIWTDVDGV+SADPRKVSEAVIL TLSYQEAWEMSYFGANVLHPRTIIPVM+Y
Sbjct: 310 VKARQVTIWTDVDGVFSADPRKVSEAVILSTLSYQEAWEMSYFGANVLHPRTIIPVMKYN 369
Query: 362 IPILIRNIFNLSAPGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPG 421
IPI+IRNIFN SAPGT IC N+N D+ + VK FATID LAL+NVEGTGMAGVPG
Sbjct: 370 IPIVIRNIFNTSAPGTMICQQPANENGDL---EACVKAFATIDKLALVNVEGTGMAGVPG 426
Query: 422 TASAIFGAVKDVGANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQV 481
TA+AIFGAVKDVGANVIMISQASSEHS+CFAVPEKEV V+ AL +RFR AL GRLS+V
Sbjct: 427 TANAIFGAVKDVGANVIMISQASSEHSVCFAVPEKEVALVSAALHARFREALAAGRLSKV 486
Query: 482 AIIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVK 541
+I NCSILA VG +MASTPGVSATLF+ALAKANINVRAIAQGCSEYNIT+VLK+EDCV+
Sbjct: 487 EVIHNCSILATVGLRMASTPGVSATLFDALAKANINVRAIAQGCSEYNITIVLKQEDCVR 546
Query: 542 ALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGILGSKSM 601
ALRA HSRF+LS+TT+A+GIIGPGLIG TLL+QL+DQA++LKE NIDLRVMGI GS++M
Sbjct: 547 ALRAAHSRFFLSKTTLAVGIIGPGLIGRTLLNQLKDQAAVLKENMNIDLRVMGIAGSRTM 606
Query: 602 LLSDWGIDLARWRELRDESGEVANLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWL 661
LLSD G+DL +W+E E ANL+KFV H+ NHF PN +VDCTAD+ +A HYYDWL
Sbjct: 607 LLSDIGVDLTQWKEKLQTEAEPANLDKFVHHLSENHFFPNRVLVDCTADTSVASHYYDWL 666
Query: 662 RKGIHVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDR 721
+KGIHV+TPNKKANSGPLD+YLKLR LQR SYTHYFYEATVGAGLPI+STLRGLLETGD+
Sbjct: 667 KKGIHVITPNKKANSGPLDRYLKLRTLQRASYTHYFYEATVGAGLPIISTLRGLLETGDK 726
Query: 722 ILQIEGIFSGTLSYIFNNFKDGRAFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVIILAR 781
IL+IEGIFSGTLSYIFNNF+ R FS+VV EAK+AGYTEPDPRDDLSGTDVARKVIILAR
Sbjct: 727 ILRIEGIFSGTLSYIFNNFEGARTFSDVVAEAKKAGYTEPDPRDDLSGTDVARKVIILAR 786
Query: 782 ESGLKLELSNIPVESLVPEPLQACASAQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGV 841
ESGL LELS+IPV SLVPE L++C SA E+MQ+LP FD+ +A++ + AE AGEVLRYVGV
Sbjct: 787 ESGLGLELSDIPVRSLVPEALKSCTSADEYMQKLPSFDEDWARERKNAEAAGEVLRYVGV 846
Query: 842 VDVTSKKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIF 901
VDV SKKG VELR YK+DHPFAQLSGSDNIIAFTT RYKDQPLIVRGPGAGA+VTAGG+F
Sbjct: 847 VDVVSKKGQVELRAYKRDHPFAQLSGSDNIIAFTTSRYKDQPLIVRGPGAGAEVTAGGVF 906
Query: 902 SDILRLASYLGAPS 915
DILRL+SYLGAPS
Sbjct: 907 CDILRLSSYLGAPS 920
>AKH2_MAIZE (P49080) Bifunctional aspartokinase/homoserine
dehydrogenase 2, chloroplast precursor (AK-HD 2)
(AK-HSDH 2) [Includes: Aspartokinase (EC 2.7.2.4);
Homoserine dehydrogenase (EC 1.1.1.3)]
Length = 917
Score = 1270 bits (3287), Expect = 0.0
Identities = 632/854 (74%), Positives = 730/854 (85%), Gaps = 7/854 (0%)
Query: 66 LTDVSPSVLVEEQQ----LQKGETWSVHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVV 121
L + ++ VE+ + L KG+ WSVHKFGGTCMG+ +RI+ V +IVL D SERKL++V
Sbjct: 67 LRPAAAAISVEQDEVNTYLPKGDMWSVHKFGGTCMGTPKRIQCVANIVLGDSSERKLIIV 126
Query: 122 SAMSKVTDMMYELINKAQSRDESYVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDIS 181
SAMSKVTDMMY L+ KAQSRD+SY +L V EKH A DLLDG+ LA FLSQLH D+S
Sbjct: 127 SAMSKVTDMMYNLVQKAQSRDDSYAIALAEVFEKHMTAAKDLLDGEDLARFLSQLHSDVS 186
Query: 182 NLKAMLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPT 241
NL+AMLRAIYIAGHATESF+DFVVGHGELWSAQMLS I+K+GA C WMDTR+VL+V P+
Sbjct: 187 NLRAMLRAIYIAGHATESFSDFVVGHGELWSAQMLSYAIKKSGAPCSWMDTREVLVVTPS 246
Query: 242 SSNQVDPDYLESEKRLETWFSLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGAL 301
NQVDPDYLE EKRL+ WFS P ++I+ATGFIAST IPTTLKRDGSDFSAAI+G+L
Sbjct: 247 GCNQVDPDYLECEKRLQKWFSRQPAEIIVATGFIASTAGNIPTTLKRDGSDFSAAIVGSL 306
Query: 302 FRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYG 361
RARQVTIWTDVDGV+SADPRKVSEAVIL TLSYQEAWEMSYFGANVLHPRTIIPVM+
Sbjct: 307 VRARQVTIWTDVDGVFSADPRKVSEAVILSTLSYQEAWEMSYFGANVLHPRTIIPVMKDN 366
Query: 362 IPILIRNIFNLSAPGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPG 421
IPI+IRN+FNLSAPGT IC N+N D+ VK FAT+DNLAL+NVEGTGMAGVPG
Sbjct: 367 IPIVIRNMFNLSAPGTMICKQPANENGDL---DACVKSFATVDNLALVNVEGTGMAGVPG 423
Query: 422 TASAIFGAVKDVGANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQV 481
TASAIF AVKDVGANVIMISQASSEHS+CFAVPEKEV V+ L RFR AL GRLS+V
Sbjct: 424 TASAIFSAVKDVGANVIMISQASSEHSVCFAVPEKEVAVVSAELHDRFREALAAGRLSKV 483
Query: 482 AIIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVK 541
+I CSILAAVG +MASTPGVSA LF+ALAKANINVRAIAQGCSEYNITVVLK++DCV+
Sbjct: 484 EVINGCSILAAVGLRMASTPGVSAILFDALAKANINVRAIAQGCSEYNITVVLKQQDCVR 543
Query: 542 ALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGILGSKSM 601
ALRA HSRF+LS+TT+A+GIIGPGLIG LL+QL++Q ++LKE NIDLRV+GI GS +M
Sbjct: 544 ALRAAHSRFFLSKTTLAVGIIGPGLIGGALLNQLKNQTAVLKENMNIDLRVIGITGSSTM 603
Query: 602 LLSDWGIDLARWRELRDESGEVANLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWL 661
LLSD GIDL +W++L + E A++ FV H+ NH PN +VDCTAD+ +A HYYDWL
Sbjct: 604 LLSDTGIDLTQWKQLLQKEAEPADIGSFVHHLSDNHVFPNKVLVDCTADTSVASHYYDWL 663
Query: 662 RKGIHVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDR 721
+KGIHV+TPNKKANSGPLDQYLKLR +QR SYTHYFYEATVGAGLPI+STLRGLLETGD+
Sbjct: 664 KKGIHVITPNKKANSGPLDQYLKLRTMQRASYTHYFYEATVGAGLPIISTLRGLLETGDK 723
Query: 722 ILQIEGIFSGTLSYIFNNFKDGRAFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVIILAR 781
IL+IEGIFSGTLSYIFNNF+ RAFS+VV EA+EAGYTEPDPRDDLSGTDVARKV++LAR
Sbjct: 724 ILRIEGIFSGTLSYIFNNFEGTRAFSDVVAEAREAGYTEPDPRDDLSGTDVARKVVVLAR 783
Query: 782 ESGLKLELSNIPVESLVPEPLQACASAQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGV 841
ESGL+LELS+IPV+SLVPE L +C+SA EFMQ+LP FD+ +A++ +AE AGEVLRYVG
Sbjct: 784 ESGLRLELSDIPVKSLVPETLASCSSADEFMQKLPSFDEDWARQRSDAEAAGEVLRYVGA 843
Query: 842 VDVTSKKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIF 901
+D ++ G VELRRY++DHPFAQLSGSDNIIAFTT RYK+QPLIVRGPGAGA+VTAGG+F
Sbjct: 844 LDAVNRSGQVELRRYRRDHPFAQLSGSDNIIAFTTSRYKEQPLIVRGPGAGAEVTAGGVF 903
Query: 902 SDILRLASYLGAPS 915
DILRLASYLGAPS
Sbjct: 904 CDILRLASYLGAPS 917
>AK1H_SERMA (P27725) Bifunctional aspartokinase/homoserine
dehydrogenase I (AKI-HDI) [Includes: Aspartokinase I (EC
2.7.2.4); Homoserine dehydrogenase I (EC 1.1.1.3)]
Length = 819
Score = 553 bits (1424), Expect = e-156
Identities = 325/839 (38%), Positives = 479/839 (56%), Gaps = 42/839 (5%)
Query: 88 VHKFGGTCMGSSQRIKNVGDIVLEDDSERKLV-VVSAMSKVTDMMYELINKAQSRDESYV 146
V KFGGT + +++R V DI+ + + ++ V+SA +K+T+ + +I+K + +
Sbjct: 3 VLKFGGTSVANAERFLRVADIMESNARQGQVATVLSAPAKITNHLVAMIDKTVAGQDILP 62
Query: 147 SSLDAVLEKHSLTAHDLLDGDGLATFLSQLHHD---------ISNLKAMLRAIYIAGHAT 197
+ DA DLL G LA L +D + LK +L + + G
Sbjct: 63 NMSDA-----ERIFADLLSG--LAQALPGFEYDRLKGVVDQEFAQLKQVLHGVSLLGQCP 115
Query: 198 ESFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQVDPDYLES---- 253
+S ++ GE S ++ V R G V ++NP YLES
Sbjct: 116 DSVNAAIICRGEKLSIAIMEGVFRAKGYP--------VTVINPVEKLLAQGHYLESTVDI 167
Query: 254 -EKRLETWFSLNPCK-VIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWT 311
E L + P +++ GF A + L R+GSD+SAA++ A RA IWT
Sbjct: 168 AESTLRIAAAAIPADHIVLMAGFTAGNDKGELVVLGRNGSDYSAAVLAACLRADCCEIWT 227
Query: 312 DVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFN 371
DVDGVY+ DPR V +A +LK++SYQEA E+SYFGA VLHPRTI P+ Q+ IP LI+N N
Sbjct: 228 DVDGVYTCDPRTVPDARLLKSMSYQEAMELSYFGAKVLHPRTITPIAQFQIPCLIKNTSN 287
Query: 372 LSAPGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVK 431
APGT I +D + VKG ++N+A+INV G GM G+ G A+ +F +
Sbjct: 288 PQAPGTLI-------GKDSTDADMPVKGITNLNNMAMINVSGPGMKGMVGMAARVFAVMS 340
Query: 432 DVGANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIPNCSILA 491
G +V++I+Q+SSE+SI F VP+ E++ AL+ F L +G L + ++ +I++
Sbjct: 341 RAGISVVLITQSSSEYSISFCVPQGELQRARRALEEEFYLELKDGVLDPLDVMERLAIIS 400
Query: 492 AVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFY 551
VG M + G+SA F+ALA+ANIN+ AIAQG SE +I+VV+ + +R H +
Sbjct: 401 VVGDGMRTLRGISARFFSALARANINIVAIAQGSSERSISVVVSNDSATTGVRVSHQMLF 460
Query: 552 LSRTTIAMGIIGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGILGSKSMLLSDWGIDLA 611
+ I + +IG G +G L++Q+ Q LK++ +IDLRV GI S+ ML + GI L
Sbjct: 461 NTDQVIEVFVIGVGGVGGALIEQIYRQQPWLKQK-HIDLRVCGIANSRVMLTNVHGIALD 519
Query: 612 RWRELRDESGEVANLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWLRKGIHVVTPN 671
WR+ + E NL + ++ V H + N IVDCT+ +A Y D+L G HVVTPN
Sbjct: 520 SWRDALAGAQEPFNLGRLIRLVKEYHLL-NPVIVDCTSSQAVADQYVDFLADGFHVVTPN 578
Query: 672 KKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDRILQIEGIFSG 731
KKAN+ ++ Y +LRA S+ + Y+ VGAGLP++ L+ LL GD +++ GI SG
Sbjct: 579 KKANTSSMNYYQQLRAAAAGSHRKFLYDTNVGAGLPVIENLQNLLNAGDELVRFSGILSG 638
Query: 732 TLSYIFNNFKDGRAFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELSN 791
+LS+IF +G + S +A+ GYTEPDPRDDLSG DVARK++ILARE+G KLELS+
Sbjct: 639 SLSFIFGKLDEGLSLSAATLQARANGYTEPDPRDDLSGMDVARKLLILAREAGYKLELSD 698
Query: 792 IPVESLVPEPLQACASAQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGVVDVTSKKGVV 851
I VE ++P A F+ RLPE D+ FA+ + A G+VLRYVG++D K V
Sbjct: 699 IEVEPVLPPSFDASGDVDTFLARLPELDKEFARNVANAAEQGKVLRYVGLIDEGRCK--V 756
Query: 852 ELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIFSDILRLASY 910
+ + P ++ +N +AF +R Y+ PL++RG GAG VTA G+F+D+LR S+
Sbjct: 757 RIEAVDGNDPLYKVKNGENALAFYSRYYQPLPLVLRGYGAGNDVTAAGVFADLLRTLSW 815
>AK1H_ECOLI (P00561) Bifunctional aspartokinase/homoserine
dehydrogenase I (AKI-HDI) [Includes: Aspartokinase I (EC
2.7.2.4); Homoserine dehydrogenase I (EC 1.1.1.3)]
Length = 820
Score = 552 bits (1422), Expect = e-156
Identities = 323/831 (38%), Positives = 491/831 (58%), Gaps = 25/831 (3%)
Query: 88 VHKFGGTCMGSSQRIKNVGDIVLEDDSERKLV--VVSAMSKVTDMMYELINKAQSRDESY 145
V KFGGT + +++R V DI LE ++ + V V+SA +K+T+ + +I K S ++
Sbjct: 3 VLKFGGTSVANAERFLRVADI-LESNARQGQVATVLSAPAKITNHLVAMIEKTISGQDAL 61
Query: 146 --VSSLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDISNLKAMLRAIYIAGHATESFTDF 203
+S + + + G LA + + + + +K +L I + G +S
Sbjct: 62 PNISDAERIFAELLTGLAAAQPGFPLAQLKTFVDQEFAQIKHVLHGISLLGQCPDSINAA 121
Query: 204 VVGHGELWSAQMLSLVIRKNGADCKWMD-TRDVLIVNPTSSNQVDPDYLESEKRLETWFS 262
++ GE S +++ V+ G + +D +L V + VD ES +R+ S
Sbjct: 122 LICRGEKMSIAIMAGVLEARGHNVTVIDPVEKLLAVGHYLESTVD--IAESTRRIAA--S 177
Query: 263 LNPCK-VIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWTDVDGVYSADP 321
P +++ GF A + L R+GSD+SAA++ A RA IWTDVDGVY+ DP
Sbjct: 178 RIPADHMVLMAGFTAGNEKGELVVLGRNGSDYSAAVLAACLRADCCEIWTDVDGVYTCDP 237
Query: 322 RKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPGTKICH 381
R+V +A +LK++SYQEA E+SYFGA VLHPRTI P+ Q+ IP LI+N N APGT I
Sbjct: 238 RQVPDARLLKSMSYQEAMELSYFGAKVLHPRTITPIAQFQIPCLIKNTGNPQAPGTLI-- 295
Query: 382 PSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDVGANVIMIS 441
+ +ED + VK G + ++N+A+ +V G GM G+ G A+ +F A+ +V++I+
Sbjct: 296 -GASRDEDELPVK----GISNLNNMAMFSVSGPGMKGMVGMAARVFAAMSRARISVVLIT 350
Query: 442 QASSEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIPNCSILAAVGQKMASTP 501
Q+SSE+SI F VP+ + A+Q F L G L +A+ +I++ VG M +
Sbjct: 351 QSSSEYSISFCVPQSDCVRAERAMQEEFYLELKEGLLEPLAVTERLAIISVVGDGMRTLR 410
Query: 502 GVSATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFYLSRTTIAMGI 561
G+SA F ALA+ANIN+ AIAQG SE +I+VV+ +D +R H + + I + +
Sbjct: 411 GISAKFFAALARANINIVAIAQGSSERSISVVVNNDDATTGVRVTHQMLFNTDQVIEVFV 470
Query: 562 IGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGILGSKSMLLSDWGIDLARWRELRDESG 621
IG G +G LL+QL+ Q S LK + +IDLRV G+ SK++L + G++L W+E ++
Sbjct: 471 IGVGGVGGALLEQLKRQQSWLKNK-HIDLRVCGVANSKALLTNVHGLNLENWQEELAQAK 529
Query: 622 EVANLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWLRKGIHVVTPNKKANSGPLDQ 681
E NL + ++ V H + N IVDCT+ +A Y D+LR+G HVVTPNKKAN+ +D
Sbjct: 530 EPFNLGRLIRLVKEYHLL-NPVIVDCTSSQAVADQYADFLREGFHVVTPNKKANTSSMDY 588
Query: 682 YLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDRILQIEGIFSGTLSYIFNNFK 741
Y +LR +S + Y+ VGAGLP++ L+ LL GD +++ GI SG+LSYIF
Sbjct: 589 YHQLRYAAEKSRRKFLYDTNVGAGLPVIENLQNLLNAGDELMKFSGILSGSLSYIFGKLD 648
Query: 742 DGRAFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPVESLVPEP 801
+G +FSE A+E GYTEPDPRDDLSG DVARK++ILARE+G +LEL++I +E ++P
Sbjct: 649 EGMSFSEATTLAREMGYTEPDPRDDLSGMDVARKLLILARETGRELELADIEIEPVLPAE 708
Query: 802 LQACASAQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGVVDVTSKKGV--VELRRYKKD 859
A FM L + D +FA ++ +A + G+VLRYVG +D + GV V++ +
Sbjct: 709 FNAEGDVAAFMANLSQLDDLFAARVAKARDEGKVLRYVGNID---EDGVCRVKIAEVDGN 765
Query: 860 HPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIFSDILRLASY 910
P ++ +N +AF + Y+ PL++RG GAG VTA G+F+D+LR S+
Sbjct: 766 DPLFKVKNGENALAFYSHYYQPLPLVLRGYGAGNDVTAAGVFADLLRTLSW 816
>AKH_HAEIN (P44505) Bifunctional aspartokinase/homoserine
dehydrogenase (AK-HD) [Includes: Aspartokinase (EC
2.7.2.4); Homoserine dehydrogenase (EC 1.1.1.3)]
Length = 815
Score = 520 bits (1340), Expect = e-147
Identities = 310/833 (37%), Positives = 476/833 (56%), Gaps = 38/833 (4%)
Query: 88 VHKFGGTCMGSSQRIKNVGDIVLEDDSERKLV-VVSAMSKVTDMMYELINKA---QSRDE 143
V KFGGT + + +R ++ + E + V+SA +K+T+ + L KA QS D
Sbjct: 3 VLKFGGTSLANPERFSQAAKLIEQAHLEEQAAGVLSAPAKITNHLVALSEKAALNQSTDT 62
Query: 144 SYVSSLDAVLE--KHSLTAHDLLDGDGLATFLSQLHHDISNLKAMLRAIYIAGHATESFT 201
+ +++ T ++ D +G + + +K +L I AG ++
Sbjct: 63 HFNEAIEIFYNIINGLHTENNQFDLNGTKALIDA---EFVQIKGLLEEIRQAGKVEDAVK 119
Query: 202 DFVVGHGELWSAQMLSLVIRKNGADCKWMDTR--DVLIVNPTSSNQVDPDYLESEKRLET 259
+ GE S M+ W + R V IV+P YLES +E
Sbjct: 120 ATIDCRGEKLSIAMMKA----------WFEARGYSVHIVDPVKQLLAKGGYLESSVEIEE 169
Query: 260 WF------SLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWTDV 313
++ KV++ GF A + L R+GSD+SAA + A A IWTDV
Sbjct: 170 STKRVDAANIAKDKVVLMAGFTAGNEKGELVLLGRNGSDYSAACLAACLGASVCEIWTDV 229
Query: 314 DGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLS 373
DGVY+ DPR V +A +L TLSY+EA E+SYFGA V+HPRTI P++ IP +I+N N S
Sbjct: 230 DGVYTCDPRLVPDARLLPTLSYREAMELSYFGAKVIHPRTIGPLLPQNIPCVIKNTGNPS 289
Query: 374 APGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDV 433
APG+ I + ++ + VKG +DNLA+ NV G GM G+ G AS +F A+
Sbjct: 290 APGSII-------DGNVKSESLQVKGITNLDNLAMFNVSGPGMQGMVGMASRVFSAMSGA 342
Query: 434 GANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIPNCSILAAV 493
G +VI+I+Q+SSE+SI F VP K + L++ F + L+ +L + +I + SI++ V
Sbjct: 343 GISVILITQSSSEYSISFCVPVKSAEVAKTVLETEFANELNEHQLEPIEVIKDLSIISVV 402
Query: 494 GQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFYLS 553
G M G++A F+ALA+ANI++ AIAQG SE +I+ V+ + ++A++A H + +
Sbjct: 403 GDGMKQAKGIAARFFSALAQANISIVAIAQGSSERSISAVVPQNKAIEAVKATHQALFNN 462
Query: 554 RTTIAMGIIGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGILGSKSMLLSDWGIDLARW 613
+ + M ++G G +G L++Q++ Q L ++ N+++RV I S MLL + G++L W
Sbjct: 463 KKVVDMFLVGVGGVGGELIEQVKRQKEYLAKK-NVEIRVCAIANSNRMLLDENGLNLEDW 521
Query: 614 RELRDESGEVANLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWLRKGIHVVTPNKK 673
+ + + + ++ + + + +H + N VDCT+ +AG Y L++G HVVTPNKK
Sbjct: 522 KNDLENATQPSDFDVLLSFIKLHHVV-NPVFVDCTSAESVAGLYARALKEGFHVVTPNKK 580
Query: 674 ANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDRILQIEGIFSGTL 733
AN+ L Y +LR + S + YE VGAGLP++ L+ LL GD + EGI SG+L
Sbjct: 581 ANTRELVYYNELRQNAQASQHKFLYETNVGAGLPVIENLQNLLAAGDELEYFEGILSGSL 640
Query: 734 SYIFNNFKDGRAFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELSNIP 793
S+IF ++G + SEV A+E G+TEPDPRDDLSG DVARK++ILARE+G++LELS++
Sbjct: 641 SFIFGKLEEGLSLSEVTALAREKGFTEPDPRDDLSGQDVARKLLILAREAGIELELSDVE 700
Query: 794 VESLVPEPLQACASAQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGVVDVTSKKGVVEL 853
VE ++P+ SA EFM LP+ D+ F ++ A+ G+VLRYVG ++ K V +
Sbjct: 701 VEGVLPKGFSDGKSADEFMAMLPQLDEEFKTRVATAKAEGKVLRYVG--KISEGKCKVSI 758
Query: 854 RRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIFSDILR 906
++P ++ +N +AF TR Y+ PL++RG GAG VTA GIF+DILR
Sbjct: 759 VAVDLNNPLYKVKDGENALAFYTRYYQPIPLLLRGYGAGNAVTAAGIFADILR 811
>AKH_BUCAI (P57290) Bifunctional aspartokinase/homoserine
dehydrogenase (AK-HD) [Includes: Aspartokinase (EC
2.7.2.4); Homoserine dehydrogenase (EC 1.1.1.3)]
Length = 816
Score = 513 bits (1321), Expect = e-145
Identities = 292/839 (34%), Positives = 484/839 (56%), Gaps = 46/839 (5%)
Query: 90 KFGGTCMGSSQRIKNVGDIVLED-DSERKLVVVSAMSKVTDMMYELINKAQSRDES---- 144
KFGGT + ++++ +V I+ E+ +++ VV+SA +K+T+ + ++I ++
Sbjct: 5 KFGGTSLANAEKFLSVSSIIEENTQTDQIAVVLSAPAKITNYLVKIIENTIKNNQILETV 64
Query: 145 ------YVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDISNLKAMLRAIYIAGHATE 198
++ ++ L S H ++ + + + LK +++ I + +
Sbjct: 65 HLAENIFMQLINNFLNIQSNFPHKEIE--------KIIKKEFNELKNIIQGILLLKQCPD 116
Query: 199 SFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSS------NQVDP--DY 250
+ ++ GE+ S ++ +++ + V I+NP + N +D D
Sbjct: 117 NIRAIIISRGEILSVFIMKSILQSKNYN--------VTIINPVKNLVAIGDNYLDSTVDI 168
Query: 251 LESEKRLETWFSLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIW 310
ES+K ++ ++N +I+ GFIA K L R+GSD+SAA++ A A IW
Sbjct: 169 SESKKNIQN-MNINQSNIILMAGFIAGNKDKKLVVLGRNGSDYSAAVLAACLDANCCEIW 227
Query: 311 TDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIF 370
TDVDGV+++DPRKV A +LK++SYQEA E+SYFGA VLHPRTI P+ Q+ IP LI+N
Sbjct: 228 TDVDGVFTSDPRKVPNARLLKSISYQEAMELSYFGAKVLHPRTIEPIAQFKIPCLIKNTN 287
Query: 371 NLSAPGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAV 430
N+ + GT IC + ++ K+F+KG +D +A+ N+ G + V S IF +
Sbjct: 288 NVKSIGTLICEQNCSE-------KDFLKGVTHLDEIAMFNISGPHIKDVGSVISRIFTMM 340
Query: 431 KDVGANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIPNCSIL 490
+++I+Q+SSE+ I F V E ++ + F+ L +G L+ I N SIL
Sbjct: 341 SRGNIKILLITQSSSENKINFCVYEHDIYKILYLFNKEFQLELKDGLLNPFKIKKNLSIL 400
Query: 491 AAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRF 550
+ VG + +++ +F+ L INV AI+QG S+++I++V+K+E+ +KA++ VH+
Sbjct: 401 SIVGSNIYKKHNIASKIFSVLGALKINVIAISQGSSKHSISLVIKKENILKAVQHVHNTL 460
Query: 551 YLSRTTIAMGIIGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGILGSKSMLLSDWGIDL 610
+ ++ TI M +IG G IGSTLL+Q+ Q L ++ NI++++ I SK +LL D DL
Sbjct: 461 FFNKKTIHMFLIGIGGIGSTLLNQILKQKQFLDKK-NIEIKICAIANSKKILLLDNTNDL 519
Query: 611 ARWRELRDESGEVANLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWLRKGIHVVTP 670
+ W+ +S + NLE + NHF N+ IVDCT+ +++ Y +++ HVVT
Sbjct: 520 SNWKNDFKKSTQKFNLELLNNLIKNNHF-SNSVIVDCTSSQLLSEQYVNFIDNNFHVVTS 578
Query: 671 NKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDRILQIEGIFS 730
NKKAN+ + Y K+R Q+ + YE VGAGLP++ TL+ L ++GD ++ +GI S
Sbjct: 579 NKKANTSEWNYYKKIRKSVAQTGKKFLYETNVGAGLPVIETLQNLFKSGDNLICFKGILS 638
Query: 731 GTLSYIFNNFKDGRAFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELS 790
G+LS+IF ++G S+ +EAKE G+TEP+P DDLSG DVARK++ILARESG +EL
Sbjct: 639 GSLSFIFGRLEEGILLSQATREAKELGFTEPNPCDDLSGIDVARKLLILARESGYNIELK 698
Query: 791 NIPVESLVPEPLQACASAQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGVVDVTSKKGV 850
+I +E L+P + +F+ +L E D F+ K+++A N G VLR+V ++ ++
Sbjct: 699 DIKIEPLLPNNFKIYEDTDKFLLKLKELDVYFSNKIKQALNVGNVLRFVATIE-QKRQFF 757
Query: 851 VELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIFSDILRLAS 909
++L ++P ++ +N +AF T Y+ PL++RG GAG VTA G+FSD+LR S
Sbjct: 758 IKLEEVNINNPLYKVKNGENALAFYTNYYQPIPLVLRGYGAGNNVTASGVFSDLLRTLS 816
>AKH_BUCAP (Q8K9U9) Bifunctional aspartokinase/homoserine
dehydrogenase (AK-HD) [Includes: Aspartokinase (EC
2.7.2.4); Homoserine dehydrogenase (EC 1.1.1.3)]
Length = 814
Score = 485 bits (1249), Expect = e-136
Identities = 275/829 (33%), Positives = 471/829 (56%), Gaps = 32/829 (3%)
Query: 90 KFGGTCMGSSQRIKNVGDIVLEDDSERKL-VVVSAMSKVTDMMYELINKAQSRDESYVSS 148
KFGGT + ++++ V DI+ + + + ++ VV+SA +K+T+ + +I D+ +
Sbjct: 5 KFGGTSLANAKKFLCVADIIEKKNKKEQIAVVLSAPAKITNYLATIIEN--KIDDEVLKK 62
Query: 149 LDAVLEKHSLTAHDLLDGDGLATF---LSQLHHDISNLKAMLRAIYIAGHATESFTDFVV 205
++ D+ L + S + + + LK ++ I + E ++
Sbjct: 63 INLAKNIFIELIQDIKRIQPLFPYENTKSTIEIEFNKLKKIINGILLIKQCPEGIKPIII 122
Query: 206 GHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSS-----NQVDP--DYLESEKRLE 258
GE+ S ++ +++ +V I+NP ++ N +D D ES+KR++
Sbjct: 123 SRGEILSVDIMKNILQSRN--------HEVTILNPVTNLLSIGNYLDSTIDIKESKKRIK 174
Query: 259 TWFSLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWTDVDGVYS 318
+++ +I+ GFIA + L R+GSD+SAAI+ + A+ IWTDVDGV +
Sbjct: 175 K-INIDQKNIILMAGFIAGNKEGELVVLGRNGSDYSAAILASCLNAKCCEIWTDVDGVLT 233
Query: 319 ADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPGTK 378
ADPR VS +L +SYQEA E+SYFGA VLHPRTI P+ Q+ IP +I+N N + GT
Sbjct: 234 ADPRIVSNTYLLDYISYQEAMELSYFGAKVLHPRTIEPISQFQIPCVIKNTNNTESKGTW 293
Query: 379 ICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDVGANVI 438
I N+ D N +KG +DN+ + N+ G+ + T + IF + +I
Sbjct: 294 I--GKENNPSD-----NSLKGVTYLDNIIMFNISGSCLKDSGNTIARIFTILSRESMKII 346
Query: 439 MISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIPNCSILAAVGQKMA 498
+I Q+SSE+ I F EK++ + L+ F + G L+ I+ N +IL+ +G ++
Sbjct: 347 LIIQSSSENQINFCTFEKDIDYILLILKKEFTLEIKEGLLNDFNIVKNLTILSVIGSNIS 406
Query: 499 STPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFYLSRTTIA 558
+++ +F++L + INV AIA G S+++I++V+K+E+ ++ ++ +H+ + +T I
Sbjct: 407 EKNNIASKIFSSLGSSKINVLAIAHGSSKHSISIVIKKENLLQGIQNIHNTLFFKKTIIN 466
Query: 559 MGIIGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGILGSKSMLLSDWGIDLARWRELRD 618
+ +IG G +G LL Q+ Q L ++ NI ++ I SK +L I+L W E
Sbjct: 467 VFLIGIGGVGKALLKQILKQEKFLDQK-NIKIQFRMIANSKKLLFLKNSINLNNWEENFK 525
Query: 619 ESGEVANLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWLRKGIHVVTPNKKANSGP 678
+S E NL + + N N+ I+DCT+D +++ Y +++KG H++T NKKAN+
Sbjct: 526 KSKEKFNLT-ILNELLKNTCDSNSVIIDCTSDYILSKQYISFIKKGFHIITSNKKANTDS 584
Query: 679 LDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDRILQIEGIFSGTLSYIFN 738
L Y ++R + + YE VGAGLP+++TL+ L TGD ++ +GI SG+LS+IF
Sbjct: 585 LKYYSEIRTTALKENKKFLYETNVGAGLPVINTLQSLFSTGDCLISFKGILSGSLSFIFG 644
Query: 739 NFKDGRAFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPVESLV 798
++G SE +EAK+ G+TEP+P DDLSG DVARK+++LARE G +EL +I +E ++
Sbjct: 645 KLEEGVLLSEATKEAKKLGFTEPNPFDDLSGIDVARKLLVLAREIGYSIELKDISIEPIL 704
Query: 799 PEPLQACASAQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGVVDVTSKKGVVELRRYKK 858
PE + +++EF+ +L E D F++++ +A + G VLR++G ++ K V++
Sbjct: 705 PERFKKYQNSEEFLFKLKELDSFFSERVNKARDIGNVLRFIGSIEKNGKCS-VKIEEINS 763
Query: 859 DHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIFSDILRL 907
++P ++ +N + F T Y+ PL++RG GAG VTA G+FSD+LR+
Sbjct: 764 NNPLYKVKNGENALTFYTNYYQPIPLVLRGYGAGNDVTASGVFSDLLRI 812
>AKH_BUCBP (Q89AR4) Bifunctional aspartokinase/homoserine
dehydrogenase (AK-HD) [Includes: Aspartokinase (EC
2.7.2.4); Homoserine dehydrogenase (EC 1.1.1.3)]
Length = 816
Score = 445 bits (1144), Expect = e-124
Identities = 261/825 (31%), Positives = 453/825 (54%), Gaps = 22/825 (2%)
Query: 90 KFGGTCMGSSQRIKNVGDIVLED-DSERKLVVVSAMSKVTDMMYELINKAQSRDESYVSS 148
KFGGT + +S+ +V I+ + ++E+ +V+SA T+++ IN Q+ + +
Sbjct: 5 KFGGTSLSNSELFFHVATIIENNLNNEQIAIVLSAPGNTTNLLEIAIN--QTINNKNIIP 62
Query: 149 LDAVLEKHSLTAHDLLDGDGLATFLSQLHHDISN----LKAMLRAIYIAGHATESFTDFV 204
+ +EK+ L + + ++ ++I N LK +L+ I + + +
Sbjct: 63 IVQKIEKNFLKLINDIYQVEQKLLYEKIKNNIENKLLELKNLLQGINLLRQCPDKIRAKI 122
Query: 205 VGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQVDPDYLESEKRLETWFSLN 264
+ GE S +++ ++ G + +D L+ + + S+ R+ + +
Sbjct: 123 ISSGEYLSISIMNSILISRGYNTTIIDPVKKLLTKEDTYLNATVNIKISKFRILS-MKIP 181
Query: 265 PCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWTDVDGVYSADPRKV 324
+I+ GF A Q TL R+GSD+SA I+ + IWTDV+GVY+ DP+ V
Sbjct: 182 KHHIILMPGFTAGNKQGELVTLGRNGSDYSATILSVCLNSTMCEIWTDVNGVYTCDPKLV 241
Query: 325 SEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPGTKICHPSV 384
S+A +L +LSY+EA E+SY GA +LHP TI P+ ++ IP I+N N S+ GTKI V
Sbjct: 242 SDAKLLTSLSYREAIELSYLGAKILHPNTIYPIQKFKIPCTIKNTHNPSSIGTKISCNHV 301
Query: 385 NDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDVGANVIMISQAS 444
+ KN + G ++N+ + ++ + IF + +I+ Q S
Sbjct: 302 KN-------KNLITGVTYLENVHMFSISCLYSKNIETIIPKIFSCMSLSKIWIILTIQTS 354
Query: 445 SEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIPNCSILAAVGQKMASTPGVS 504
S+++I F + + L L + L + + ++++ + + + ++
Sbjct: 355 SQNTISFCILKTMTNTALHVLHKALYLELKHKLLKPIKVEKKLTLISVISSDILNNTKIT 414
Query: 505 ATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFYLSRTTIAMGIIGP 564
+F+ L NIN AI++G S+ +I++V+K +D + +RA+H + T + +IG
Sbjct: 415 EKVFSILKHVNINTLAISKGASKNSISIVVKHDDGILGVRALHKGIFNKNCTAEIFLIGI 474
Query: 565 GLIGSTLLDQLRDQASILKEEFNIDLRVMGILGSKSMLLSDWGIDLARWRELRDESGEVA 624
G +G T L Q+ +Q + LK + NIDL++ GI SK+ +L+ GI+ W+ + S +
Sbjct: 475 GRVGQTFLKQIIEQKNWLKSK-NIDLKICGIANSKNFILNLDGINPKNWKRDLNLSKKSF 533
Query: 625 NLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWLRKGIHVVTPNKKANSGPLDQYLK 684
N + + + N+++ N IVDCT+D IA Y ++ G H+VTPNKKAN+ Y
Sbjct: 534 NFKHLLNSIQ-NYYLINPIIVDCTSDKNIANQYISFINCGFHIVTPNKKANTTNWKYYED 592
Query: 685 LRALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDRILQIEGIFSGTLSYIFNNFKDGR 744
+R ++ +FYE VGAGLP++ L+ LL TGD ++ +GI SG+LS+IF +D
Sbjct: 593 IRLAAQKEKKKFFYETNVGAGLPVIENLKNLLRTGDTLIHFKGILSGSLSFIFGKLEDNI 652
Query: 745 AFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPVESLVPEPLQA 804
+ SE ++A+ G+TEP+P+DDLSG DVARK++ILARE G KLEL +I +E L+P+
Sbjct: 653 SLSEATKQAQSLGFTEPNPKDDLSGIDVARKLLILAREVGYKLELKDIKIEPLLPKEFNN 712
Query: 805 CASAQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGVVDVTSKKGVVELRRYKKDH--PF 862
++ +F+ +L E DQ+F ++++A G+ LR+VG++ ++KG +++ + DH P
Sbjct: 713 ISNTTDFITKLKELDQIFCNRVKKARKLGKRLRFVGII---NQKGNCQVKIDEVDHNDPL 769
Query: 863 AQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIFSDILRL 907
+ +N +AF ++ Y+ PL++RG GAG VTA GIFSD+LR+
Sbjct: 770 YNIKNGENALAFYSKYYQPIPLVLRGYGAGNNVTASGIFSDVLRI 814
>AK2H_ECOLI (P00562) Bifunctional aspartokinase/homoserine
dehydrogenase II (AKII-HDII) [Includes: Aspartokinase II
(EC 2.7.2.4); Homoserine dehydrogenase II (EC 1.1.1.3)]
Length = 809
Score = 365 bits (936), Expect = e-100
Identities = 253/825 (30%), Positives = 395/825 (47%), Gaps = 29/825 (3%)
Query: 88 VHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVVSAMSKVTDMMYELINKAQSRDESYVS 147
+HKFGG+ + + V I+ E ++VVSA T+ + + +Q+ D
Sbjct: 13 LHKFGGSSLADVKCYLRVAGIMAEYSQPDDMMVVSAAGSTTNQLINWLKLSQT-DRLSAH 71
Query: 148 SLDAVLEKHSLTA-HDLLDGDGLATFLSQLHHDISNLKAMLRAIYIAGHATESFTDFVVG 206
+ L ++ LL + + +S D+ L A+L + ++ VVG
Sbjct: 72 QVQQTLRRYQCDLISGLLPAEEADSLISAFVSDLERLAALLDS-----GINDAVYAEVVG 126
Query: 207 HGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQVDPDYLESEKRLETWFSLNPC 266
HGE+WSA+++S V+ + G W+D R+ L + QVD S L+ +P
Sbjct: 127 HGEVWSARLMSAVLNQQGLPAAWLDAREFLRAERAAQPQVDEGL--SYPLLQQLLVQHPG 184
Query: 267 KVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWTDVDGVYSADPRKVSE 326
K ++ TGFI+ L R+GSD+SA +GAL +VTIW+DV GVYSADPRKV +
Sbjct: 185 KRLVVTGFISRNNAGETVLLGRNGSDYSATQIGALAGVSRVTIWSDVAGVYSADPRKVKD 244
Query: 327 AVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPGTKICHPSVND 386
A +L L EA E++ A VLH RT+ PV I + +R + T+I
Sbjct: 245 ACLLPLLRLDEASELARLAAPVLHARTLQPVSGSEIDLQLRCSYTPDQGSTRI------- 297
Query: 387 NEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDVGANVIMISQASSE 446
E ++ + + D++ LI + I +K + + +
Sbjct: 298 -ERVLASGTGARIVTSHDDVCLIEFQVPASQDFKLAHKEIDQILKRAQVRPLAVGVHNDR 356
Query: 447 HSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIPNCSILAAVGQKMASTPGVSAT 506
+ F +E S + + G ++ + +++A VG + P
Sbjct: 357 QLLQFCY-------TSEVADSALKILDEAGLPGELRLRQGLALVAMVGAGVTRNPLHCHR 409
Query: 507 LFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFYLSRTTIAMGIIGPGL 566
+ L + Q ++ VL+ ++ +H + + I + + G G
Sbjct: 410 FWQQLKGQPVEFTW--QSDDGISLVAVLRTGPTESLIQGLHQSVFRAEKRIGLVLFGKGN 467
Query: 567 IGSTLLDQLRDQASILKEEFNIDLRVMGILGSKSMLLSDWGIDLARWRELRDESGEVANL 626
IGS L+ + S L + + G++ S+ LLS G+D +R ++ +
Sbjct: 468 IGSRWLELFAREQSTLSARTGFEFVLAGVVDSRRSLLSYDGLDASRALAFFNDEAVEQDE 527
Query: 627 EKFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWLRKGIHVVTPNKKANSGPLDQYLKLR 686
E + H + ++D TA +A Y D+ G HV++ NK A + ++Y ++
Sbjct: 528 ESLFLWMRA-HPYDDLVVLDVTASQQLADQYLDFASHGFHVISANKLAGASDSNKYRQIH 586
Query: 687 ALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDRILQIEGIFSGTLSYIFNNFKDGRAF 746
++ H+ Y ATVGAGLPI T+R L+++GD IL I GIFSGTLS++F F F
Sbjct: 587 DAFEKTGRHWLYNATVGAGLPINHTVRDLIDSGDTILSISGIFSGTLSWLFLQFDGSVPF 646
Query: 747 SEVVQEAKEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPVESLVPEPLQACA 806
+E+V +A + G TEPDPRDDLSG DV RK++ILARE+G +E + VESLVP +
Sbjct: 647 TELVDQAWQQGLTEPDPRDDLSGKDVMRKLVILAREAGYNIEPDQVRVESLVPAHCEG-G 705
Query: 807 SAQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGVVDVTSKKGVVELRRYKKDHPFAQLS 866
S F + E ++ ++LE A G VLRYV D K V + ++DHP A L
Sbjct: 706 SIDHFFENGDELNEQMVQRLEAAREMGLVLRYVARFDANG-KARVGVEAVREDHPLASLL 764
Query: 867 GSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIFSDILRLASYL 911
DN+ A +R Y+D PL++RGPGAG VTAG I SDI RLA L
Sbjct: 765 PCDNVFAIESRWYRDNPLVIRGPGAGRDVTAGAIQSDINRLAQLL 809
>DHOM_SCHPO (O94671) Probable homoserine dehydrogenase (EC 1.1.1.3)
(HDH)
Length = 376
Score = 291 bits (746), Expect = 4e-78
Identities = 169/370 (45%), Positives = 234/370 (62%), Gaps = 12/370 (3%)
Query: 553 SRTTIAMGIIGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGILGSKSMLLSD--WGIDL 610
SRT + + I+G G IG LL+Q++ V+ I + +S I+L
Sbjct: 4 SRTNVNVAIVGTGNIGGELLNQIKGFNENASTNGTTSFNVVAISSMEGHYVSKDYQPINL 63
Query: 611 ARWRELRDESGEVANLEKFVQHVHGNHFIPNTAI-VDCTADSVIAGHYYDWLRKGIHVVT 669
+ W+ L ++ +L+ V + + P AI VD TA IA Y +L K I++ T
Sbjct: 64 SEWKNLTSQNQGAYSLDALVDFLAKS---PLPAILVDNTASEAIAQSYPKFLSKKINIAT 120
Query: 670 PNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDRILQIEGIF 729
PNKKA S D Y + ++S +EA+VGAGLPI+STL+ L+ TGD I++IEGIF
Sbjct: 121 PNKKAFSASNDVYQNIIKASKESGALLMHEASVGAGLPIISTLKELIATGDEIIKIEGIF 180
Query: 730 SGTLSYIFN----NFKDGRA-FSEVVQEAKEAGYTEPDPRDDLSGTDVARKVIILARESG 784
SGTLSYIFN N K G A FS++V+ AK+ GYTEPDPRDDL+G DVARKV IL+R +G
Sbjct: 181 SGTLSYIFNVWSPNGKKGTASFSDIVKIAKQNGYTEPDPRDDLNGMDVARKVTILSRIAG 240
Query: 785 LKLE-LSNIPVESLVPEPLQACASAQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGVVD 843
+ +E S+ PV+SL+PEPL++ +A+EF+ LP FD FA EEAE G+V+R+VG D
Sbjct: 241 VHVESASSFPVKSLIPEPLKSAVNAEEFLAGLPNFDSEFASMREEAEKEGKVVRFVGEAD 300
Query: 844 VTSKKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIFSD 903
V +K +V+L +Y HPFA L SDNII+FTT+RY +PL+V G GAGA VTA G+ D
Sbjct: 301 VANKTTLVKLEKYDASHPFANLQSSDNIISFTTKRYHTRPLVVIGAGAGAAVTAAGVLGD 360
Query: 904 ILRLASYLGA 913
++++ S + A
Sbjct: 361 MIKIMSQVRA 370
>AK_METJA (Q57991) Probable aspartokinase (EC 2.7.2.4) (Aspartate
kinase)
Length = 473
Score = 265 bits (678), Expect = 3e-70
Identities = 171/474 (36%), Positives = 262/474 (55%), Gaps = 19/474 (4%)
Query: 87 SVHKFGGTCMGSSQRIKNVGDIVLEDDSERK--LVVVSAMSKVTDMMYELINKAQS-RDE 143
+V KFGGT +GS +RI++V IV + E +VVVSAMS+VT+ + E+ +A RD
Sbjct: 3 TVMKFGGTSVGSGERIRHVAKIVTKRKKEDDDVVVVVSAMSEVTNALVEISQQALDVRDI 62
Query: 144 SYVSS-LDAVLEKHSLTAHDLLDGDGLATFLSQLHHD-ISNLKAMLRAIYIAGHATESFT 201
+ V + + EKH + + + + + ++ I L+ +L + G T
Sbjct: 63 AKVGDFIKFIREKHYKAIEEAIKSEEIKEEVKKIIDSRIEELEKVLIGVAYLGELTPKSR 122
Query: 202 DFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQVDPDYLESEKRLETWF 261
D+++ GE S+ +LS IR G ++ + I+ + LE ++RL
Sbjct: 123 DYILSFGERLSSPILSGAIRDLGEKSIALEGGEAGIITDNNFGSARVKRLEVKERLLPL- 181
Query: 262 SLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWTDVDGVYSADP 321
L + + TGFI +T + TTL R GSD+SAA++G A + IWTDV GVY+ DP
Sbjct: 182 -LKEGIIPVVTGFIGTTEEGYITTLGRGGSDYSAALIGYGLDADIIEIWTDVSGVYTTDP 240
Query: 322 RKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPGTKICH 381
R V A + LSY EA E++YFGA VLHPRTI P M+ GIPIL++N F + GT I
Sbjct: 241 RLVPTARRIPKLSYIEAMELAYFGAKVLHPRTIEPAMEKGIPILVKNTFEPESEGTLI-- 298
Query: 382 PSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDVGANVIMIS 441
DM + VK +TI N+ALIN+ G GM GV GTA+ IF A+ + NVI+IS
Sbjct: 299 -----TNDMEMSDSIVKAISTIKNVALINIFGAGMVGVSGTAARIFKALGEEEVNVILIS 353
Query: 442 QASSEHSICFAVPEKEVKAVAEALQSRF-----RHALDNGRLSQVAIIPNCSILAAVGQK 496
Q SSE +I V E++V +AL+ F + L+N + V++ + +++ VG
Sbjct: 354 QGSSETNISLVVSEEDVDKALKALKREFGDFGKKSFLNNNLIRDVSVDKDVCVISVVGAG 413
Query: 497 MASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRF 550
M G++ +F A++++ N++ IAQG SE NI+ V+ +D + +R +H +F
Sbjct: 414 MRGAKGIAGKIFTAVSESGANIKMIAQGSSEVNISFVIDEKDLLNCVRKLHEKF 467
>DHOM_YEAST (P31116) Homoserine dehydrogenase (EC 1.1.1.3) (HDH)
Length = 359
Score = 263 bits (672), Expect = 2e-69
Identities = 152/367 (41%), Positives = 228/367 (61%), Gaps = 15/367 (4%)
Query: 552 LSRTTIAMGIIGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGILGSKSMLLSDWGIDLA 611
+S + + +IG G++GS LDQL S + + L SK + G D
Sbjct: 1 MSTKVVNVAVIGAGVVGSAFLDQLLAMKSTITYNLVLLAEAERSLISKDFSPLNVGSD-- 58
Query: 612 RWRE-LRDESGEVANLEKFVQHVHGNHFIPNTAI-VDCTADSVIAGHYYDWLRKGIHVVT 669
W+ L + + L+ + H+ + P I VD T+ + IAG Y ++ GI + T
Sbjct: 59 -WKAALAASTTKTLPLDDLIAHLKTS---PKPVILVDNTSSAYIAGFYTKFVENGISIAT 114
Query: 670 PNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDRILQIEGIF 729
PNKKA S L + L + + + ++EATVGAGLPI+S LR +++TGD + +IEGIF
Sbjct: 115 PNKKAFSSDLATWKALFS-NKPTNGFVYHEATVGAGLPIISFLREIIQTGDEVEKIEGIF 173
Query: 730 SGTLSYIFNNFKDGRA----FSEVVQEAKEAGYTEPDPRDDLSGTDVARKVIILARESGL 785
SGTLSYIFN F +A FS+VV+ AK+ GYTEPDPRDDL+G DVARKV I+ R SG+
Sbjct: 174 SGTLSYIFNEFSTSQANDVKFSDVVKVAKKLGYTEPDPRDDLNGLDVARKVTIVGRISGV 233
Query: 786 KLEL-SNIPVESLVPEPLQACASAQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGVVDV 844
++E ++ PV+SL+P+PL++ SA EF+++L ++D+ + +EA +VLR++G VDV
Sbjct: 234 EVESPTSFPVQSLIPKPLESVKSADEFLEKLSDYDKDLTQLKKEAATENKVLRFIGKVDV 293
Query: 845 TSKKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIFSDI 904
+K V + +Y HPFA L GSDN+I+ T+RY + P++++G GAGA VTA G+ D+
Sbjct: 294 ATKSVSVGIEKYDYSHPFASLKGSDNVISIKTKRYTN-PVVIQGAGAGAAVTAAGVLGDV 352
Query: 905 LRLASYL 911
+++A L
Sbjct: 353 IKIAQRL 359
>AK_SCHPO (O60163) Probable aspartokinase (EC 2.7.2.4) (Aspartate
kinase)
Length = 519
Score = 170 bits (431), Expect = 1e-41
Identities = 132/486 (27%), Positives = 225/486 (46%), Gaps = 33/486 (6%)
Query: 86 WSVHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVV----SAMSKVTDMMYELINKAQSR 141
W V KFGGT +G IK D+ E S +++ +V S +K LI ++
Sbjct: 15 WVVQKFGGTSVGKFP-IKIAVDVAKEYLSTKRVALVCSARSTDTKAEGTTTRLIRATEAA 73
Query: 142 DESYVSSLDAVLE----KHSLTAHDLLDGDGLAT-FLSQLHHDISNLKAMLRAIYIAGHA 196
V S+ ++ H A D + G+ + H D L+ L AI +
Sbjct: 74 LRPAVGSVHDLVRIIETDHVQAARDFIQDVGIQDELIDAFHADCVELEQYLNAIRVLSEV 133
Query: 197 TESFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQVDPDYLESEKR 256
+ D V+G GE S + ++ V++ G D +++D ++ N Y +
Sbjct: 134 SPRTRDLVIGMGERLSCRFMAAVLKDQGIDSEFIDMSHIIDEQREWRNLDASFYAYLASQ 193
Query: 257 LETWFSLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWTDVDGV 316
L + + KV + TGF P + + + R +DF AA++ A ++ IW +VDG+
Sbjct: 194 LASKVTAVGNKVPVVTGFFGMVPGGLLSQIGRGYTDFCAALLAVGLNADELQIWKEVDGI 253
Query: 317 YSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPG 376
++ADPRKV A +L ++ +EA E++Y+G+ V+HP T+ V+ IPI I+N+ N G
Sbjct: 254 FTADPRKVPTARLLPLITPEEAAELTYYGSEVIHPFTMSQVVHARIPIRIKNVGNPRGKG 313
Query: 377 TKICHPSVNDN------------EDMMNVKNFVKGFATI---DNLALINVEGTGMAGVPG 421
T I +++ + D ++ KG + D + +IN++ G
Sbjct: 314 TVIFPDTISRHGSATPPHPPKIMPDDISASLANKGATAVTIKDTIMVINIQSNRKISAHG 373
Query: 422 TASAIFGAVKDVGANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQV 481
++IF A+ D + + S H E + + EA ++ RL +
Sbjct: 374 FLASIF-AILDKYKLAVDLITTSEVHVSMALYEESDDGNMHEAF-------VELRRLGTL 425
Query: 482 AIIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVK 541
I+ +IL+ VG+ M +T G + +F LA+A IN+ I+QG SE NI+ V+ + VK
Sbjct: 426 DILHGLAILSLVGKHMRNTTGYAGRMFCKLAEAQINIEMISQGASEINISCVIDEKMAVK 485
Query: 542 ALRAVH 547
AL +H
Sbjct: 486 ALNVIH 491
>AK3_ECOLI (P08660) Lysine-sensitive aspartokinase III (EC 2.7.2.4)
(Aspartate kinase III)
Length = 449
Score = 169 bits (427), Expect = 4e-41
Identities = 127/466 (27%), Positives = 222/466 (47%), Gaps = 25/466 (5%)
Query: 88 VHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVVSAMSKVTDMMYELINKAQSRDESYVS 147
V KFGGT + + DIVL D + R LVV+SA + +T+++ L + +
Sbjct: 6 VSKFGGTSVADFDAMNRSADIVLSDANVR-LVVLSASAGITNLLVALAEGLEPGER--FE 62
Query: 148 SLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDISNLKAMLRAIYIAGHATESFTDFVVGH 207
LDA+ L + + + +L + N+ + A +A + + TD +V H
Sbjct: 63 KLDAIRNIQFAILERLRYPNVIREEIERL---LENITVLAEAAALA--TSPALTDELVSH 117
Query: 208 GELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQVDPDY--LESEKRLETWFSLNP 265
GEL S + ++R+ +W D R V+ N + +PD L L+ LN
Sbjct: 118 GELMSTLLFVEILRERDVQAQWFDVRKVMRTNDRFG-RAEPDIAALAELAALQLLPRLNE 176
Query: 266 CKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWTDVDGVYSADPRKVS 325
++I GFI S + TTL R GSD++AA++ A +V IWTDV G+Y+ DPR VS
Sbjct: 177 -GLVITQGFIGSENKGRTTTLGRGGSDYTAALLAEALHASRVDIWTDVPGIYTTDPRVVS 235
Query: 326 EAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPGTKICHPSVN 385
A + +++ EA EM+ FGA VLHP T++P ++ IP+ + + + A GT +C+ + N
Sbjct: 236 AAKRIDEIAFAEAAEMATFGAKVLHPATLLPAVRSDIPVFVGSSKDPRAGGTLVCNKTEN 295
Query: 386 DNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDVGANVIMISQASS 445
+ A N L+ + M G + +FG + +V +I+ +S
Sbjct: 296 --------PPLFRALALRRNQTLLTLHSLNMLHSRGFLAEVFGILARHNISVDLIT--TS 345
Query: 446 EHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIPNCSILAAVGQKMASTPGVSA 505
E S+ + + + L ++ ++ L +V + +++A +G ++ GV
Sbjct: 346 EVSVALTLDTTGSTSTGDTLLTQ-SLLMELSALCRVEVEEGLALVALIGNDLSKACGVGK 404
Query: 506 TLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFY 551
+F L N+R I G S +N+ ++ ED + ++ +HS +
Sbjct: 405 EVFGVLEP--FNIRMICYGASSHNLCFLVPGEDAEQVVQKLHSNLF 448
>AK_YEAST (P10869) Aspartokinase (EC 2.7.2.4) (Aspartate kinase)
Length = 527
Score = 163 bits (413), Expect = 2e-39
Identities = 133/510 (26%), Positives = 235/510 (46%), Gaps = 42/510 (8%)
Query: 78 QQLQKGETWSVHKFGGTCMGSSQRIKNVGDIVL-----EDDSERKLVVVSAMSK------ 126
Q W V KFGGT +G ++ V DIV + + VV SA S
Sbjct: 6 QPTSSHSNWVVQKFGGTSVGKFP-VQIVDDIVKHYSKPDGPNNNVAVVCSARSSYTKAEG 64
Query: 127 VTDMMYELINKAQSRDESYVSSLDAVLEKHSLTAHDLLDGDGL-ATFLSQLHHDISNLKA 185
T + + + A S++ + ++ + + H A + L A + + ++ +K
Sbjct: 65 TTSRLLKCCDLA-SQESEFQDIIEVIRQDHIDNADRFILNPALQAKLVDDTNKELELVKK 123
Query: 186 MLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQ 245
L A + G + D V+ GE S ++ + G K++D + + + S++
Sbjct: 124 YLNASKVLGEVSSRTVDLVMSCGEKLSCLFMTALCNDRGCKAKYVDLSHI-VPSDFSASA 182
Query: 246 VDPDYLES-----EKRLETWFSLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGA 300
+D + +++L + S V + TGF P + + R +D AA++
Sbjct: 183 LDNSFYTFLVQALKEKLAPFVSAKERIVPVFTGFFGLVPTGLLNGVGRGYTDLCAALIAV 242
Query: 301 LFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQY 360
A ++ +W +VDG+++ADPRKV EA +L +++ +EA E++Y+G+ V+HP T+ V++
Sbjct: 243 AVNADELQVWKEVDGIFTADPRKVPEARLLDSVTPEEASELTYYGSEVIHPFTMEQVIRA 302
Query: 361 GIPILIRNIFNLSAPGTKIC-------------HPSVNDNEDMMNV-KNFVKGFATIDNL 406
IPI I+N+ N GT I HP N + K T +++
Sbjct: 303 KIPIRIKNVQNPLGNGTIIYPDNVAKKGESTPPHPPENLSSSFYEKRKRGATAITTKNDI 362
Query: 407 ALINVEGTGMAGVPGTASAIFGAVKDVGANVIMISQASSEHSICFAVPEKEVKAVAEALQ 466
+IN+ G + IF + V +IS + S+ +P+ A++L+
Sbjct: 363 FVINIHSNKKTLSHGFLAQIFTILDKYKLVVDLISTSEVHVSMALPIPD------ADSLK 416
Query: 467 SRFRHALDNGR-LSQVAIIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGC 525
S R A + R L V I SI++ VG+ M G++ T+F LA+ IN+ I+QG
Sbjct: 417 S-LRQAEEKLRILGSVDITKKLSIVSLVGKHMKQYIGIAGTMFTTLAEEGINIEMISQGA 475
Query: 526 SEYNITVVLKREDCVKALRAVHSRFYLSRT 555
+E NI+ V+ D +KAL+ +H++ RT
Sbjct: 476 NEINISCVINESDSIKALQCIHAKLLSERT 505
>AK2_BACSU (P08495) Aspartokinase 2 (EC 2.7.2.4) (Aspartokinase II)
(Aspartate kinase 2) [Contains: Aspartokinase II alpha
subunit; Aspartokinase II beta subunit]
Length = 408
Score = 147 bits (371), Expect = 1e-34
Identities = 133/471 (28%), Positives = 213/471 (44%), Gaps = 76/471 (16%)
Query: 88 VHKFGGTCMGSSQRIKNVGD--IVLEDDSERKLVVVSAMSKVTDMMYELINKAQSRDESY 145
V KFGGT +GS ++I+N + I + + +VVVSAM K TD EL++ A+
Sbjct: 5 VQKFGGTSVGSVEKIQNAANRAIAEKQKGHQVVVVVSAMGKSTD---ELVSLAK------ 55
Query: 146 VSSLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDISNLKAMLRAIYIAGHATESFTDFVV 205
A+ ++ S D+L G +S L A+ G+ S+T
Sbjct: 56 -----AISDQPSKREMDMLLATGEQVTISLLS----------MALQEKGYDAVSYTG--- 97
Query: 206 GHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQVDPDYLESEKRLETWFSLNP 265
W A + + I N +D L + L
Sbjct: 98 -----WQAGIRTEAIHGNAR-----------------ITDIDTSVLADQ--------LEK 127
Query: 266 CKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWTDVDGVYSADPRKVS 325
K++I GF T TTL R GSD +A + A +A + I+TDV GV++ DPR V
Sbjct: 128 GKIVIVAGFQGMTEDCEITTLGRGGSDTTAVALAAALKADKCDIYTDVPGVFTTDPRYVK 187
Query: 326 EAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPGTKICHPSVN 385
A L+ +SY E E++ GA VLHPR + Y +P+ +R+ A GT I
Sbjct: 188 SARKLEGISYDEMLELANLGAGVLHPRAVEFAKNYQVPLEVRSSTETEA-GTLI------ 240
Query: 386 DNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFG--AVKDVGANVIMISQA 443
+ E M V+G A D + + + G+ T S IF A +++ ++I+ +QA
Sbjct: 241 EEESSMEQNLIVRGIAFEDQITRVTI--YGLTSGLTTLSTIFTTLAKRNINVDIIIQTQA 298
Query: 444 SSEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIPNCSILAAVGQKMASTPGV 503
+ I F+V ++ L+ ++ AL+ ++ + + SI VG M S PGV
Sbjct: 299 EDKTGISFSVKTEDADQTVAVLE-EYKDALEFEKIETESKLAKVSI---VGSGMVSNPGV 354
Query: 504 SATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFYLSR 554
+A +F LA+ NI ++ ++ SE ++ V+ D VKA+ ++H F LS+
Sbjct: 355 AAEMFAVLAQKNILIKMVS--TSEIKVSTVVSENDMVKAVESLHDAFELSK 403
>AK_THET2 (P61488) Aspartokinase (EC 2.7.2.4) (Aspartate kinase)
[Contains: Aspartokinase alpha subunit (ASK-alpha);
Aspartokinase beta subunit (ASK-beta)]
Length = 405
Score = 140 bits (352), Expect = 2e-32
Identities = 134/475 (28%), Positives = 203/475 (42%), Gaps = 83/475 (17%)
Query: 88 VHKFGGTCMGSSQRIKNVGDIV--LEDDSERKLVVVSAMSKVTDMMYELINKAQSRDESY 145
V K+GGT +G +RI V + + R VVVSAM TD + L + R
Sbjct: 5 VQKYGGTSVGDLERIHKVAQRIAHYREKGHRLAVVVSAMGHTTDELIALAKRVNPRP--- 61
Query: 146 VSSLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDISNLKAMLRAIYIAGHATESFTDFVV 205
DLL G + ++ + ++ G + F +
Sbjct: 62 -----------PFRELDLLTTTG----------EQVSVALLSMQLWAMGIPAKGFVQHQI 100
Query: 206 GHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQVDPDYLESEKRLETWFSLNP 265
G D ++ D R +L VNP + LE F
Sbjct: 101 G----------------ITTDGRYGDAR-ILEVNPARIREA----------LEQGF---- 129
Query: 266 CKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWTDVDGVYSADPRKVS 325
V + GF+ +TP+ TTL R GSD +A + A A++ I+TD +GVY+ DP +
Sbjct: 130 --VAVIAGFMGTTPEGEITTLGRGGSDTTAVAIAAALGAKECEIYTDTEGVYTTDPHLIP 187
Query: 326 EAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPGTKICHPSVN 385
EA L + Y + EM+ GA VLHPR + +YG+ + +R+ F+ + PGT +
Sbjct: 188 EARKLSVIGYDQMLEMAALGARVLHPRAVYYAKRYGVVLHVRSSFSYN-PGTLV------ 240
Query: 386 DNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDVGANVIMISQA-- 443
E M + V G A + A I + G+ PG A+ +F A+ + G V MI Q
Sbjct: 241 -KEVAMEMDKAVTGVALDLDHAQIGL--IGIPDQPGIAAKVFQALAERGIAVDMIIQGVP 297
Query: 444 ---SSEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAII-PNCSILAAVGQKMAS 499
S + F V + + EAL+ + AI+ P+ + ++ VG +AS
Sbjct: 298 GHDPSRQQMAFTVKKDFAQEALEALEPVL------AEIGGEAILRPDIAKVSIVGVGLAS 351
Query: 500 TPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFYLSR 554
TP V A +F A+A N+ IA SE I+V++ E ALRAVH F L +
Sbjct: 352 TPEVPAKMFQAVASTGANIEMIA--TSEVRISVIIPAEYAEAALRAVHQAFELDK 404
>AK_THETH (P61489) Aspartokinase (EC 2.7.2.4) (Aspartate kinase)
[Contains: Aspartokinase alpha subunit (ASK-alpha);
Aspartokinase beta subunit (ASK-beta)]
Length = 405
Score = 139 bits (350), Expect = 3e-32
Identities = 132/475 (27%), Positives = 203/475 (41%), Gaps = 83/475 (17%)
Query: 88 VHKFGGTCMGSSQRIKNVGDIV--LEDDSERKLVVVSAMSKVTDMMYELINKAQSRDESY 145
V K+GGT +G +RI V + + R VVVSAM TD + L + R
Sbjct: 5 VQKYGGTSVGDLERIHKVAQRIAHYREKGHRLAVVVSAMGHTTDELIALAKRVNPRP--- 61
Query: 146 VSSLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDISNLKAMLRAIYIAGHATESFTDFVV 205
DLL G + ++ + ++ G + F +
Sbjct: 62 -----------PFRELDLLTTTG----------EQVSVALLSMQLWAMGIPAKGFVQHQI 100
Query: 206 GHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQVDPDYLESEKRLETWFSLNP 265
G D ++ D R +L VNP + +L+
Sbjct: 101 G----------------ITTDGRYGDAR-ILEVNPARIRE----------------ALDQ 127
Query: 266 CKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWTDVDGVYSADPRKVS 325
V + GF+ +TP+ TTL R GSD +A + A A++ I+TD +GVY+ DP +
Sbjct: 128 GFVAVIAGFMGTTPEGEITTLGRGGSDTTAVAIAAALGAKECEIYTDTEGVYTTDPHLIP 187
Query: 326 EAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPGTKICHPSVN 385
EA L + Y + EM+ GA VLHPR + +YG+ + +R+ F+ + PGT +
Sbjct: 188 EARKLSVIGYDQMLEMAALGARVLHPRAVYYAKRYGVVLHVRSSFSYN-PGTLV------ 240
Query: 386 DNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDVGANVIMISQA-- 443
E M + V G A + A I + G+ PG A+ +F A+ + G V MI Q
Sbjct: 241 -KEVAMEMDKAVTGVALDLDHAQIGL--IGIPDQPGIAAKVFQALAERGIAVDMIIQGVP 297
Query: 444 ---SSEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAII-PNCSILAAVGQKMAS 499
S + F V + + EAL+ + AI+ P+ + ++ VG +AS
Sbjct: 298 GHDPSRQQMAFTVKKDFAQEALEALEPVL------AEIGGEAILRPDIAKVSIVGVGLAS 351
Query: 500 TPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFYLSR 554
TP V A +F A+A N+ IA SE I+V++ E ALRAVH F L +
Sbjct: 352 TPEVPAKMFQAVASTGANIEMIA--TSEVRISVIIPAEYAEAALRAVHQAFELDK 404
>AK3_BACSU (P94417) Probable aspartokinase (EC 2.7.2.4) (Aspartate
kinase)
Length = 454
Score = 133 bits (334), Expect = 2e-30
Identities = 121/475 (25%), Positives = 218/475 (45%), Gaps = 40/475 (8%)
Query: 88 VHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVVSAM-------SKVTDMMYELINKAQS 140
V KFGG+ + S ++ V IV D + RK VVVSA +KVTD++ + +
Sbjct: 3 VVKFGGSSLASGAQLDKVFHIVTSDPA-RKAVVVSAPGKHYAEDTKVTDLLIACAEQYLA 61
Query: 141 RDESYVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDISNLKAMLRAIYIAGHAT--E 198
S +AV+E+++L A++L G + + ++ D+ L + G + E
Sbjct: 62 TG-SAPELAEAVVERYALIANELQLGQSI---IEKIRDDLFTL--------LEGDKSNPE 109
Query: 199 SFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRD--VLIVNPTSSNQVDPDYLESEKR 256
+ D V GE +A++++ R G ++++ +D + + N + QV P+ ++ R
Sbjct: 110 QYLDAVKASGEDNNAKLIAAYFRYKGVKAEYVNPKDAGLFVTNEPGNAQVLPESYQNLYR 169
Query: 257 LETWFSLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWTDVDGV 316
L L II GF + T R GSD + +I+ +A +TDVD V
Sbjct: 170 LRERDGL-----IIFPGFFGFSKDGDVITFSRSGSDITGSILANGLQADLYENFTDVDAV 224
Query: 317 YSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPG 376
YS +P V + L+Y+E E+SY G +V H +IP + GIP+ I+N N SA G
Sbjct: 225 YSVNPSFVENPKEISELTYREMRELSYAGFSVFHDEALIPAFRAGIPVQIKNTNNPSAEG 284
Query: 377 TKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDVGAN 436
T++ N N ++ G A+ I + M G +++ G
Sbjct: 285 TRVVSKRDNTNGPVV-------GIASDTGFCSIYISKYLMNREIGFGRRALQILEEHGLT 337
Query: 437 VIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIPNCSILAAVGQK 496
+ + +I + ++ A E +S + ++ +V + + +++ VG+
Sbjct: 338 YEHVPSGIDDMTIILR--QGQMDAATE--RSVIKRIEEDLHADEVIVEHHLALIMVVGEA 393
Query: 497 MASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFY 551
M G +A AL++A +N+ I QG SE ++ +K + KA++A++ F+
Sbjct: 394 MRHNVGTTARAAKALSEAQVNIEMINQGSSEVSMMFGVKEAEERKAVQALYQEFF 448
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.134 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 104,250,458
Number of Sequences: 164201
Number of extensions: 4415803
Number of successful extensions: 12189
Number of sequences better than 10.0: 170
Number of HSP's better than 10.0 without gapping: 130
Number of HSP's successfully gapped in prelim test: 40
Number of HSP's that attempted gapping in prelim test: 11855
Number of HSP's gapped (non-prelim): 210
length of query: 915
length of database: 59,974,054
effective HSP length: 119
effective length of query: 796
effective length of database: 40,434,135
effective search space: 32185571460
effective search space used: 32185571460
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0108.1


