

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
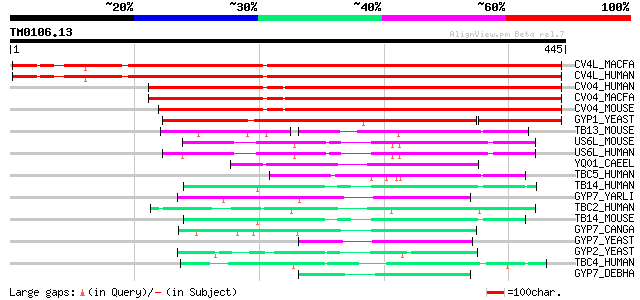
Query= TM0106.13
(445 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CV4L_MACFA (Q95LL3) TBC1 domain family protein C6orf197 homolog ... 344 3e-94
CV4L_HUMAN (Q9NU19) TBC1 domain family protein C6orf197 344 3e-94
CV04_HUMAN (Q8WUA7) TBC1 domain family protein C22orf4 322 1e-87
CV04_MACFA (Q95KI1) TBC1 domain family protein C22orf4 homolog (... 321 3e-87
CV04_MOUSE (Q8R5A6) TBC1 domain family protein C22orf4 homolog (... 314 3e-85
GYP1_YEAST (Q08484) GTPase-activating protein GYP1 (GAP for YPT1) 255 1e-67
TB13_MOUSE (Q8R3D1) TBC1 domain family member 13 80 8e-15
US6L_MOUSE (Q80XC3) USP6 N-terminal like protein 74 6e-13
US6L_HUMAN (Q92738) USP6 N-terminal like protein (Related to the... 74 6e-13
YQ01_CAEEL (Q09445) Hypothetical protein C04A2.1 in chromosome II 72 2e-12
TBC5_HUMAN (Q92609) TBC1 domain family member 5 69 3e-11
TB14_HUMAN (Q9P2M4) TBC1 domain family member 14 66 2e-10
GYP7_YARLI (P09379) GTPase-activating protein GYP7 (GAP for YPT7) 64 6e-10
TBC2_HUMAN (Q9BYX2) TBC1 domain family member 2 (Prostate antige... 63 1e-09
TB14_MOUSE (Q8CGA2) TBC1 domain family member 14 63 1e-09
GYP7_CANGA (Q6FWI1) GTPase-activating protein GYP7 (GAP for YPT7) 59 2e-08
GYP7_YEAST (P48365) GTPase-activating protein GYP7 (GAP for YPT7) 53 2e-06
GYP2_YEAST (P53258) GTPase-activating protein GYP2 (MAC1-depende... 50 1e-05
TBC4_HUMAN (O60343) TBC1 domain family member 4 49 3e-05
GYP7_DEBHA (Q6BU76) GTPase-activating protein GYP7 (GAP for YPT7) 48 6e-05
>CV4L_MACFA (Q95LL3) TBC1 domain family protein C6orf197 homolog
(QtsA-20424)
Length = 505
Score = 344 bits (882), Expect = 3e-94
Identities = 189/445 (42%), Positives = 275/445 (61%), Gaps = 21/445 (4%)
Query: 3 NNDEDNNNNPNIGIFDSRFSQTLRSVQGLLKGRSMPGKILLSQRVDPIDNSNLSSPT--- 59
+++E++ ++P+ +S+ + L + +L+ S RV P + + +S
Sbjct: 74 DDEEEDFSSPSFQTLNSKVA--LATAAQVLENHS-------KLRVKPERSQSTTSDVPAN 124
Query: 60 YTRSNSYNDAGTSDHASET-VEVEVHSSSGIPGENKLKISTSHVENPSEDVRKSSMGARA 118
Y S +DA S ++S+T + +H +P L+ V S+ +
Sbjct: 125 YKVIKSSSDAQLSRNSSDTCLRNPLHKQQSLP----LRPIIPLVARISDQNASGAPPMTV 180
Query: 119 TDSARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRR 178
+ R+ KF ++LS LD+LR+ +W GVP +RP WRLL GY P N++RR+ L+R
Sbjct: 181 REKTRLEKFRQLLSSHNTDLDELRKCSWPGVPREVRPVTWRLLSGYLPANTERRKLTLQR 240
Query: 179 KRLEYLDCVSQYYDIPDTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYA 238
KR EY + QYYD + E +D RQI +D PRT P + FQQ VQ+ ERIL+
Sbjct: 241 KREEYFGFIEQYYDSRNEEHHQDTY---RQIHIDIPRTNPLIPLFQQPLVQEIFERILFI 297
Query: 239 WAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSK 298
WAIRHPASGYVQGINDLVTPFFVVFLSEY+E ++N+ +++LS D + ++EAD +WC+SK
Sbjct: 298 WAIRHPASGYVQGINDLVTPFFVVFLSEYVEEDVENFDVTNLSQDMLRSIEADSFWCMSK 357
Query: 299 LLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIRE 358
LLDG+QD+YTFAQPGIQ+ V L+ELV RID+ V H +E+LQFAFRW N LL+RE
Sbjct: 358 LLDGIQDNYTFAQPGIQKKVKALEELVSRIDEQVHNHFRRYEVEYLQFAFRWMNNLLMRE 417
Query: 359 IPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTW-SDKLQKLDFQDLVMFLQHLPTQDW 417
+P RLWDTY +E + F +Y+ A+FL+ W + L + DFQ L+M LQ+LPT W
Sbjct: 418 LPLRCTIRLWDTYQSEPEGFSHFHLYVCAAFLIKWRKEILDEEDFQGLLMLLQNLPTIHW 477
Query: 418 THQELEMVLSRAFMWHSMFNNSPNH 442
++E+ ++L+ A+ MF ++PNH
Sbjct: 478 GNEEIGLLLAEAYRLKYMFADAPNH 502
>CV4L_HUMAN (Q9NU19) TBC1 domain family protein C6orf197
Length = 505
Score = 344 bits (882), Expect = 3e-94
Identities = 189/445 (42%), Positives = 275/445 (61%), Gaps = 21/445 (4%)
Query: 3 NNDEDNNNNPNIGIFDSRFSQTLRSVQGLLKGRSMPGKILLSQRVDPIDNSNLSSPT--- 59
+++E++ ++P+ +S+ + L + +L+ S RV P + + +S
Sbjct: 74 DDEEEDFSSPSFQTLNSKVA--LATAAQVLENHS-------KLRVKPERSQSTTSDVPAN 124
Query: 60 YTRSNSYNDAGTSDHASET-VEVEVHSSSGIPGENKLKISTSHVENPSEDVRKSSMGARA 118
Y S +DA S ++S+T + +H +P L+ V S+ +
Sbjct: 125 YKVIKSSSDAQLSRNSSDTCLRNPLHKQQSLP----LRPIIPLVARISDQNASGAPPMTV 180
Query: 119 TDSARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRR 178
+ R+ KF ++LS LD+LR+ +W GVP +RP WRLL GY P N++RR+ L+R
Sbjct: 181 REKTRLEKFRQLLSSQNTDLDELRKCSWPGVPREVRPITWRLLSGYLPANTERRKLTLQR 240
Query: 179 KRLEYLDCVSQYYDIPDTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYA 238
KR EY + QYYD + E +D RQI +D PRT P + FQQ VQ+ ERIL+
Sbjct: 241 KREEYFGFIEQYYDSRNEEHHQDTY---RQIHIDIPRTNPLIPLFQQPLVQEIFERILFI 297
Query: 239 WAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSK 298
WAIRHPASGYVQGINDLVTPFFVVFLSEY+E ++N+ +++LS D + ++EAD +WC+SK
Sbjct: 298 WAIRHPASGYVQGINDLVTPFFVVFLSEYVEEDVENFDVTNLSQDMLRSIEADSFWCMSK 357
Query: 299 LLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIRE 358
LLDG+QD+YTFAQPGIQ+ V L+ELV RID+ V H +E+LQFAFRW N LL+RE
Sbjct: 358 LLDGIQDNYTFAQPGIQKKVKALEELVSRIDEQVHNHFRRYEVEYLQFAFRWMNNLLMRE 417
Query: 359 IPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTW-SDKLQKLDFQDLVMFLQHLPTQDW 417
+P RLWDTY +E + F +Y+ A+FL+ W + L + DFQ L+M LQ+LPT W
Sbjct: 418 LPLRCTIRLWDTYQSEPEGFSHFHLYVCAAFLIKWRKEILDEEDFQGLLMLLQNLPTIHW 477
Query: 418 THQELEMVLSRAFMWHSMFNNSPNH 442
++E+ ++L+ A+ MF ++PNH
Sbjct: 478 GNEEIGLLLAEAYRLKYMFADAPNH 502
>CV04_HUMAN (Q8WUA7) TBC1 domain family protein C22orf4
Length = 517
Score = 322 bits (825), Expect = 1e-87
Identities = 159/333 (47%), Positives = 224/333 (66%), Gaps = 6/333 (1%)
Query: 112 SSMGARATDSARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDR 171
SS +++R+ KF ++L+G L++LR L+WSG+P +RP W+LL GY P N DR
Sbjct: 186 SSSALSEREASRLDKFKQLLAGPNTDLEELRRLSWSGIPKPVRPMTWKLLSGYLPANVDR 245
Query: 172 REGVLRRKRLEYLDCVSQYYDIPDTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKS 231
R L+RK+ EY + YYD + E +D RQI +D PR P+ + Q +V +
Sbjct: 246 RPATLQRKQKEYFAFIEHYYDSRNDEVHQDTY---RQIHIDIPRMSPE-ALILQPKVTEI 301
Query: 232 LERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGS-IDNWSMSDLSSDEISNVEA 290
ERIL+ WAIRHPASGYVQGINDLVTPFFVVF+ EY+E +D +S + ++ + N+EA
Sbjct: 302 FERILFIWAIRHPASGYVQGINDLVTPFFVVFICEYIEAEEVDTVDVSGVPAEVLCNIEA 361
Query: 291 DCYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRW 350
D YWC+SKLLDG+QD+YTFAQPGIQ V L+ELV RID+ V H++ + +LQFAFRW
Sbjct: 362 DTYWCMSKLLDGIQDNYTFAQPGIQMKVKMLEELVSRIDEQVHRHLDQHEVRYLQFAFRW 421
Query: 351 FNCLLIREIPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTW-SDKLQKLDFQDLVMFL 409
N LL+RE+P RLWDTY +E D F +Y+ A+FL+ W + L++ DFQ+L++FL
Sbjct: 422 MNNLLMREVPLRCTIRLWDTYQSEPDGFSHFHLYVCAAFLVRWRKEILEEKDFQELLLFL 481
Query: 410 QHLPTQDWTHQELEMVLSRAFMWHSMFNNSPNH 442
Q+LPT W +++ ++L+ A+ F ++PNH
Sbjct: 482 QNLPTAHWDDEDISLLLAEAYRLKFAFADAPNH 514
>CV04_MACFA (Q95KI1) TBC1 domain family protein C22orf4 homolog
(QtrA-11492) (Fragment)
Length = 497
Score = 321 bits (822), Expect = 3e-87
Identities = 158/333 (47%), Positives = 225/333 (67%), Gaps = 6/333 (1%)
Query: 112 SSMGARATDSARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDR 171
SS +++R+ KF ++L+G L++LR+L+WSG+P +RP W+LL GY P N DR
Sbjct: 166 SSSALSEREASRLDKFEQLLAGPNTDLEELRKLSWSGIPKPVRPMTWKLLSGYLPANVDR 225
Query: 172 REGVLRRKRLEYLDCVSQYYDIPDTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKS 231
R L+RK+ EY + YYD + E +D RQI +D PR P+ + Q +V +
Sbjct: 226 RPATLQRKQKEYFAFIEHYYDSRNDEVHQDTY---RQIHIDIPRMSPE-ALILQPKVTEI 281
Query: 232 LERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGS-IDNWSMSDLSSDEISNVEA 290
ERIL+ WAIRHPASGYVQGINDLVTPFFVVF+ EY+E +D +S + ++ + N+EA
Sbjct: 282 FERILFIWAIRHPASGYVQGINDLVTPFFVVFICEYIEAEEVDTVDVSGVPAEVLRNIEA 341
Query: 291 DCYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRW 350
D YWC+SKLLDG+QD+YTFAQPGIQ V L+ELV RID+ V H++ + +LQFAFRW
Sbjct: 342 DTYWCMSKLLDGIQDNYTFAQPGIQMKVKMLEELVSRIDEQVHRHLDQHEVRYLQFAFRW 401
Query: 351 FNCLLIREIPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTW-SDKLQKLDFQDLVMFL 409
N LL+RE+P RLWDTY +E + F +Y+ A+FL+ W + L++ DFQ+L++FL
Sbjct: 402 MNNLLMREVPLRCTIRLWDTYQSEPEGFSHFHLYVCAAFLVRWRKEILEEKDFQELLLFL 461
Query: 410 QHLPTQDWTHQELEMVLSRAFMWHSMFNNSPNH 442
Q+LPT W +++ ++L+ A+ F ++PNH
Sbjct: 462 QNLPTAHWDDEDISLLLAEAYRLKFAFADAPNH 494
>CV04_MOUSE (Q8R5A6) TBC1 domain family protein C22orf4 homolog
(Fragment)
Length = 353
Score = 314 bits (804), Expect = 3e-85
Identities = 155/325 (47%), Positives = 221/325 (67%), Gaps = 6/325 (1%)
Query: 120 DSARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRK 179
+++R+ KF ++L+G L++LR+L+WSG+P +RP W+LL GY P N DRR L+RK
Sbjct: 30 ETSRLDKFKQLLAGPNTDLEELRKLSWSGIPKPVRPMTWKLLSGYLPANVDRRPATLQRK 89
Query: 180 RLEYLDCVSQYYDIPDTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAW 239
+ EY + YY + E +D RQI +D PR P+ + Q +V + ERIL+ W
Sbjct: 90 QKEYFAFIEHYYSSRNDEVHQDTY---RQIHIDIPRMSPE-ALILQPKVTEIFERILFIW 145
Query: 240 AIRHPASGYVQGINDLVTPFFVVFLSEYLEGS-IDNWSMSDLSSDEISNVEADCYWCLSK 298
AIRHPASGYVQGINDLVTPFFVVF+ EY + +D +S + ++ + N+EAD YWC+SK
Sbjct: 146 AIRHPASGYVQGINDLVTPFFVVFICEYTDREDVDKVDVSSVPAEVLRNIEADTYWCMSK 205
Query: 299 LLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIRE 358
LLDG+QD+YTFAQPGIQ V L+ELV RID+ V H++ + +LQFAFRW N LL+RE
Sbjct: 206 LLDGIQDNYTFAQPGIQMKVKMLEELVSRIDERVHRHLDGHEVRYLQFAFRWMNNLLMRE 265
Query: 359 IPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTW-SDKLQKLDFQDLVMFLQHLPTQDW 417
+P RLWDTY +E + F +Y+ A+FL+ W + L++ DFQ+L++FLQ+LPT W
Sbjct: 266 LPLRCTIRLWDTYQSEPEGFSHFHLYVCAAFLVRWRREILEERDFQELLLFLQNLPTARW 325
Query: 418 THQELEMVLSRAFMWHSMFNNSPNH 442
Q++ ++L+ A+ F ++PNH
Sbjct: 326 DDQDVSLLLAEAYRLKFAFADAPNH 350
>GYP1_YEAST (Q08484) GTPase-activating protein GYP1 (GAP for YPT1)
Length = 637
Score = 255 bits (652), Expect = 1e-67
Identities = 127/258 (49%), Positives = 176/258 (67%), Gaps = 10/258 (3%)
Query: 123 RVMKFTKVLSG-TVVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRKRL 181
R+ KF +L T++ LR+++W+G+P RP VW+LL+GY P N+ R+EG L+RKR
Sbjct: 254 RISKFDNILKDKTIINQQDLRQISWNGIPKIHRPVVWKLLIGYLPVNTKRQEGFLQRKRK 313
Query: 182 EYLDCVSQYYDIPDTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAI 241
EY D + + +++ +I QI +D PRT P + +Q + VQ SL+RILY WAI
Sbjct: 314 EYRDSLKHTF----SDQHSRDIPTWHQIEIDIPRTNPHIPLYQFKSVQNSLQRILYLWAI 369
Query: 242 RHPASGYVQGINDLVTPFFVVFLSEYLEGS-IDNWSMSDLSS----DEISNVEADCYWCL 296
RHPASGYVQGINDLVTPFF FL+EYL S ID+ + D S+ ++I+++EAD +WCL
Sbjct: 370 RHPASGYVQGINDLVTPFFETFLTEYLPPSQIDDVEIKDPSTYMVDEQITDLEADTFWCL 429
Query: 297 SKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLI 356
+KLL+ + D+Y QPGI R V L +LV+RID + H +N+ +EF+QFAFRW NCLL+
Sbjct: 430 TKLLEQITDNYIHGQPGILRQVKNLSQLVKRIDADLYNHFQNEHVEFIQFAFRWMNCLLM 489
Query: 357 REIPFNMVTRLWDTYLAE 374
RE V R+WDTYL+E
Sbjct: 490 REFQMGTVIRMWDTYLSE 507
Score = 78.6 bits (192), Expect = 3e-14
Identities = 30/66 (45%), Positives = 52/66 (78%)
Query: 377 ALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQELEMVLSRAFMWHSMF 436
+L +F V++ A+FL+ WSD+L ++DFQ+ + FLQ+ PT+DWT ++EM+LS AF+W S++
Sbjct: 570 SLNEFHVFVCAAFLIKWSDQLMEMDFQETITFLQNPPTKDWTETDIEMLLSEAFIWQSLY 629
Query: 437 NNSPNH 442
++ +H
Sbjct: 630 KDATSH 635
>TB13_MOUSE (Q8R3D1) TBC1 domain family member 13
Length = 400
Score = 80.5 bits (197), Expect = 8e-15
Identities = 53/189 (28%), Positives = 96/189 (50%), Gaps = 18/189 (9%)
Query: 232 LERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEAD 291
+ERIL+ +A +P YVQG+N++V P + F + D +S+ + EAD
Sbjct: 211 VERILFIYAKLNPGIAYVQGMNEIVGPLYYTFAT-------------DPNSEWKEHAEAD 257
Query: 292 CYWCLSKLLDGMQDHYTFA----QPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFA 347
++C + L+ ++D++ + Q GI + K+ ++ D + ++ Q ++ FA
Sbjct: 258 TFFCFTNLMAEIRDNFIKSLDDSQCGITYKMEKVYSTLKDKDVELYLKLQEQSIKPQFFA 317
Query: 348 FRWFNCLLIREIPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQDLVM 407
FRW LL +E V R+WD+ A+G+ DFL+ + + L+ ++L + DF +
Sbjct: 318 FRWLTLLLSQEFLLPDVIRIWDSLFADGNRF-DFLLLVCCAMLILIREQLLEGDFTVNMR 376
Query: 408 FLQHLPTQD 416
LQ P D
Sbjct: 377 LLQDYPITD 385
Score = 45.4 bits (106), Expect = 3e-04
Identities = 36/133 (27%), Positives = 57/133 (42%), Gaps = 29/133 (21%)
Query: 122 ARVMKFTKVLSGTVVILDKLRELAWSGVP--DYMRPTVWRLLLGYAPPNSDRREGVLRRK 179
+R+ F VL ++L+KLREL++SG+P +R W++LL Y P +L ++
Sbjct: 7 SRIADFQDVLKEPSIVLEKLRELSFSGIPCEGGLRCLCWKILLNYLPLERASWTSILAKQ 66
Query: 180 RLEYLDCVSQ--------------------YYDIPDTERSEDEIN-------MLRQIAVD 212
R Y + + + D P + N +L QI D
Sbjct: 67 RGLYSQFLREMIIQPGIAKANMGVFREDVTFEDHPLNPNPDSRWNTYFKDNEVLLQIDKD 126
Query: 213 CPRTVPDVSFFQQ 225
R PD+SFFQ+
Sbjct: 127 VRRLCPDISFFQR 139
>US6L_MOUSE (Q80XC3) USP6 N-terminal like protein
Length = 819
Score = 74.3 bits (181), Expect = 6e-13
Identities = 66/289 (22%), Positives = 120/289 (40%), Gaps = 50/289 (17%)
Query: 139 DKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRKRLEYLDCVSQYYDIPDTER 198
+K + G+P +R VW LLL + R+ + K R
Sbjct: 91 EKFHRRIYKGIPLQLRGEVWALLLEIPKMKEETRDLYSKLKH-----------------R 133
Query: 199 SEDEINMLRQIAVDCPRTVPDVSFFQQQQ--VQKSLERILYAWAIRHPASGYVQGINDLV 256
+ +RQI +D RT D F+ + Q+SL +L A++I + GY QG++ +
Sbjct: 134 ARGCSPDIRQIDLDVNRTFRDHIMFRDRYGVKQQSLFHVLAAYSIYNTEVGYCQGMSQ-I 192
Query: 257 TPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDGMQD--HYTFAQ--P 312
T +++++E D +W L KL G + H F Q P
Sbjct: 193 TALLLMYMNE-----------------------EDAFWALVKLFSGPKHAMHGFFVQGFP 229
Query: 313 GIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTYL 372
+ R ++++ + + H+++Q + + +WF + PF + R+WD Y+
Sbjct: 230 KLLRFQEHHEKILNKFLSKLKQHLDSQEIYTSFYTMKWFFQCFLDRTPFRLNLRIWDIYI 289
Query: 373 AEGDALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQE 421
EG+ + + Y + L L KL ++LV FLQ +D+ ++
Sbjct: 290 FEGERVLTAMSY---TILKLHKKHLMKLSMEELVEFLQETLAKDFFFED 335
>US6L_HUMAN (Q92738) USP6 N-terminal like protein (Related to the N
terminus of tre) (RN-tre)
Length = 828
Score = 74.3 bits (181), Expect = 6e-13
Identities = 70/307 (22%), Positives = 126/307 (40%), Gaps = 52/307 (16%)
Query: 123 RVMKFTKVLSGTVVI--LDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRKR 180
R K+ K+L G +K + G+P +R VW LLL + R+ + K
Sbjct: 73 RTTKWLKMLKGWEKYKNTEKFHRRIYKGIPLQLRGEVWALLLEIPKMKEETRDLYSKLKH 132
Query: 181 LEYLDCVSQYYDIPDTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQ--VQKSLERILYA 238
R+ +RQI +D RT D F+ + Q+SL +L A
Sbjct: 133 -----------------RARGCSPDIRQIDLDVNRTFRDHIMFRDRYGVKQQSLFHVLAA 175
Query: 239 WAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSK 298
++I + GY QG++ +T +++++E D +W L K
Sbjct: 176 YSIYNTEVGYCQGMSQ-ITALLLMYMNE-----------------------EDAFWALVK 211
Query: 299 LLDGMQD--HYTFAQ--PGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCL 354
L G + H F Q P + R ++++ + + H+++Q + + +WF
Sbjct: 212 LFSGPKHAMHGFFVQGFPKLLRFQEHHEKILNKFLSKLKQHLDSQEIYTSFYTMKWFFQC 271
Query: 355 LIREIPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPT 414
+ PF + R+WD Y+ EG+ + + Y + L L KL ++LV F Q
Sbjct: 272 FLDRTPFTLNLRIWDIYIFEGERVLTAMSY---TILKLHKKHLMKLSMEELVEFFQETLA 328
Query: 415 QDWTHQE 421
+D+ ++
Sbjct: 329 KDFFFED 335
>YQ01_CAEEL (Q09445) Hypothetical protein C04A2.1 in chromosome II
Length = 330
Score = 72.4 bits (176), Expect = 2e-12
Identities = 54/199 (27%), Positives = 83/199 (41%), Gaps = 25/199 (12%)
Query: 178 RKRLEYLDCVSQYYDIPDTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILY 237
RK L C S+ + DE+ ++QI +D PRT PD F + + +K+L R L+
Sbjct: 84 RKELWLRSCPSRADGVWQRHEVPDEV--IKQIKLDLPRTFPDNKFLKTEGTRKTLGRALF 141
Query: 238 AWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLS 297
A A P+ GY QG+N + +V N E+ L
Sbjct: 142 AVAEHIPSVGYCQGLNFVAGVILLVV-----------------------NDESRAIDLLV 178
Query: 298 KLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIR 357
L+ Q++Y G++R + L L+R V +E + +WF C +
Sbjct: 179 HLVSQRQEYYGKNMIGLRRDMHVLHSLLREHCPRVVVTLEKLDVGLDMLVGKWFVCWFVE 238
Query: 358 EIPFNMVTRLWDTYLAEGD 376
+P V RLWD + EGD
Sbjct: 239 SLPMETVLRLWDCLIYEGD 257
>TBC5_HUMAN (Q92609) TBC1 domain family member 5
Length = 795
Score = 68.6 bits (166), Expect = 3e-11
Identities = 64/237 (27%), Positives = 106/237 (44%), Gaps = 35/237 (14%)
Query: 209 IAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYL 268
I D RT P++ FFQQ+ V+K L +L+ +A + Y QG+++L+ P +VF+
Sbjct: 163 IEQDVKRTFPEMQFFQQENVRKILTDVLFCYARENEQLLYKQGMHELLAP--IVFVLHCD 220
Query: 269 EGSIDNWSMSDLSSDEISNV------EADCYWCLSKLL------------DGMQDHYT-- 308
+ + S S S+E+ V E D Y S+L+ DG + T
Sbjct: 221 HQAFLHASESAQPSEEMKTVLNPEYLEHDAYAVFSQLMETAEPWFSTFEHDGQKGKETLM 280
Query: 309 ----FAQP-------GIQRLVFKLKE-LVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLI 356
FA+P I V ++++ L+++ D + H+ + + RW L
Sbjct: 281 TPIPFARPQDLGPTIAIVTKVNQIQDHLLKKHDIELYMHLNRLEIAPQIYGLRWVRLLFG 340
Query: 357 REIPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLP 413
RE P + +WD A+G +L + YIF + LL D L ++Q + L H P
Sbjct: 341 REFPLQDLLVVWDALFADGLSL-GLVDYIFVAMLLYIRDALISSNYQTCLGLLMHYP 396
>TB14_HUMAN (Q9P2M4) TBC1 domain family member 14
Length = 678
Score = 65.9 bits (159), Expect = 2e-10
Identities = 71/290 (24%), Positives = 115/290 (39%), Gaps = 35/290 (12%)
Query: 140 KLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRKRLEYLDCVSQYYDIPDTE-- 197
K+R+L W G+P +R VW L +G + + + E +S + E
Sbjct: 378 KVRDLWWQGIPPSVRGKVWSLAIGNELNITHELFDICLARAKERWRSLSTGGSEVENEDA 437
Query: 198 --RSEDEINMLRQIAVDCPRTVPDVSFFQQ-QQVQKSLERILYAWAIRHPASGYVQGIND 254
+ D L I +D RT P++ FQQ L IL A+ P GYVQG++
Sbjct: 438 GFSAADREASLELIKLDISRTFPNLCIFQQGGPYHDMLHSILGAYTCYRPDVGYVQGMS- 496
Query: 255 LVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLD-GMQDHYTFAQPG 313
F++ L ++D AD + S LL+ Q + G
Sbjct: 497 --------FIAAVLILNLDT---------------ADAFIAFSNLLNKPCQMAFFRVDHG 533
Query: 314 IQRLVFKLKELVRRIDDP-VSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTYL 372
+ F E+ + P + H + L + W L + +P ++ R+WD +
Sbjct: 534 LMLTYFAAFEVFFEENLPKLFAHFKKNNLTPDIYLIDWIFTLYSKSLPLDLACRIWDVFC 593
Query: 373 AEGDALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQEL 422
+G+ +FL L + D L K+DF + FL LP +D +EL
Sbjct: 594 RDGE---EFLFRTALGILKLFEDILTKMDFIHMAQFLTRLP-EDLPAEEL 639
>GYP7_YARLI (P09379) GTPase-activating protein GYP7 (GAP for YPT7)
Length = 730
Score = 64.3 bits (155), Expect = 6e-10
Identities = 59/263 (22%), Positives = 111/263 (41%), Gaps = 51/263 (19%)
Query: 135 VVILDKLRELAW-SGVPDYMRPTVWRLLLGYAPPNSD--RREGVLRRKRLEYLDCVSQYY 191
+V +++++E + G+ +RP W LLG P +S R+ ++ + R++Y +++
Sbjct: 374 IVTVNEVKERIFHGGLAPAVRPEGWLFLLGVYPWDSTAAERKELVSKLRVDYNRLKKEWW 433
Query: 192 DIPDTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKS-------------------- 231
D ER + + L +I D RT +++FF + +K
Sbjct: 434 VQEDKERDDFWRDQLSRIEKDVHRTDRNITFFAECDAKKDGDDDNYDKDEFGFSSQINSN 493
Query: 232 -----LERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEIS 286
L +L + + GYVQG++DL++P +VV + L
Sbjct: 494 IHLIQLRDMLITYNQHNKNLGYVQGMSDLLSPLYVVLQDDTL------------------ 535
Query: 287 NVEADCYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQF 346
+W S ++ M+ +Y Q G++ + L LV+ + + H+E L F
Sbjct: 536 -----AFWAFSAFMERMERNYLRDQSGMRNQLLCLDHLVQFMLPSLYKHLEKTESTNLFF 590
Query: 347 AFRWFNCLLIREIPFNMVTRLWD 369
FR RE+ ++ V RLW+
Sbjct: 591 FFRMLLVWFKRELLWDDVLRLWE 613
>TBC2_HUMAN (Q9BYX2) TBC1 domain family member 2 (Prostate antigen
recognized and indentified by SEREX) (PARIS-1)
Length = 917
Score = 63.2 bits (152), Expect = 1e-09
Identities = 71/325 (21%), Positives = 126/325 (37%), Gaps = 58/325 (17%)
Query: 114 MGARATDSARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRRE 173
+G A D R ++ G +V +L++L +GVP RP VWR L+
Sbjct: 593 LGLEAVD--RPLRERWAALGDLVPSAELKQLLRAGVPREHRPRVWRWLV----------- 639
Query: 174 GVLRRKRLEYLDCVSQYYDIPDTERSEDEINMLRQIAVDCPRTVPDVSFFQ--QQQVQKS 231
R+++L Y ++ ++ + RQI +D RT P+ F
Sbjct: 640 ----HLRVQHLHTPGCYQELLSRGQAREH-PAARQIELDLNRTFPNNKHFTCPTSSFPDK 694
Query: 232 LERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEAD 291
L R+L A++ ++P GY QG+N L +V E
Sbjct: 695 LRRVLLAFSWQNPTIGYCQGLNRLAAIALLVL-----------------------EEEES 731
Query: 292 CYWCLSKLLDGMQ--DHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFR 349
+WCL +++ + D+Y Q L++L+ + H+ ++ F
Sbjct: 732 AFWCLVAIVETIMPADYYCNTLTASQVDQRVLQDLLSEKLPRLMAHLGQHHVDLSLVTFN 791
Query: 350 WFNCLLIREIPFNMVTRLWDTYLAEG------------DALPDFLVYIFASFLLTWSDKL 397
WF + + N++ R+WD +L EG + L + F + ++ S KL
Sbjct: 792 WFLVVFADSLISNILLRVWDAFLYEGTKYNEKEILRLQNGLEIYQYLRFFTKTISNSRKL 851
Query: 398 QKLDFQDLVMF-LQHLPTQDWTHQE 421
+ F D+ F ++ L H+E
Sbjct: 852 MNIAFNDMNPFRMKQLRQLRMVHRE 876
>TB14_MOUSE (Q8CGA2) TBC1 domain family member 14
Length = 679
Score = 63.2 bits (152), Expect = 1e-09
Identities = 67/281 (23%), Positives = 109/281 (37%), Gaps = 34/281 (12%)
Query: 140 KLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRKRLEYLDCVSQYYDIPDTE-- 197
K+R+L W G+P +R VW L +G + + + E +S + E
Sbjct: 379 KVRDLWWQGIPPSVRGKVWSLAIGNELNITHELFDICLARAKERWRSLSTGGSEVENEDA 438
Query: 198 --RSEDEINMLRQIAVDCPRTVPDVSFFQQ-QQVQKSLERILYAWAIRHPASGYVQGIND 254
+ D L I +D RT P++ FQQ L IL A+ P GYVQG++
Sbjct: 439 GFSAADREASLELIKLDISRTFPNLCIFQQGGPYHDMLHSILGAYTCYRPDVGYVQGMS- 497
Query: 255 LVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLD-GMQDHYTFAQPG 313
F++ L ++D AD + S LL+ Q + G
Sbjct: 498 --------FIAAVLILNLDT---------------ADAFIAFSNLLNKPCQMAFFRVDHG 534
Query: 314 IQRLVFKLKELVRRIDDP-VSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTYL 372
+ F E+ + P + H + L + W L + +P ++ R+WD +
Sbjct: 535 LMLTYFAAFEVFFEENLPKLFAHFKKNNLTADIYLIDWIFTLYSKSLPLDLACRIWDVFC 594
Query: 373 AEGDALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLP 413
+G+ +FL L + D L ++DF FL LP
Sbjct: 595 RDGE---EFLFRTALGILKLFEDILTRMDFIHSAQFLTRLP 632
>GYP7_CANGA (Q6FWI1) GTPase-activating protein GYP7 (GAP for YPT7)
Length = 745
Score = 59.3 bits (142), Expect = 2e-08
Identities = 56/269 (20%), Positives = 104/269 (37%), Gaps = 53/269 (19%)
Query: 136 VILDKLRELAWSG--VPDYMRPTVWRLLLGYAPPNSDRREGVLRRKRL--EYLDCVSQYY 191
V ++++++ + G D R VW LLG P +S E RK L EY++ ++
Sbjct: 377 VTVNEVKDYIFHGGLADDATRKEVWPFLLGVYPWDSSEDERKQLRKALHDEYMELKQKWV 436
Query: 192 DIP---DTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQ------------------- 229
D D + E + L +I D R ++ ++
Sbjct: 437 DREVNLDNDEEEYWKDQLFRIEKDVKRNDRNIDIYKYNTSDNLPFPEDTAPTTDDDDSIK 496
Query: 230 ----KSLERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEI 285
K L IL + I +P GYVQG+ DL++P + + E
Sbjct: 497 NPNLKKLADILTTYNIFNPNLGYVQGMTDLLSPLYYIIRDE------------------- 537
Query: 286 SNVEADCYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQ 345
+WC + ++ M+ ++ Q GI+ + L +L + + +S H++ L
Sbjct: 538 ----ETTFWCFTNFMERMERNFLRDQSGIRDQMLALTDLCQLMLPRLSAHLQKCDSSDLF 593
Query: 346 FAFRWFNCLLIREIPFNMVTRLWDTYLAE 374
F FR RE ++ + +W+ + +
Sbjct: 594 FCFRMLLVWFKREFNYDDIFNIWEVFFTD 622
>GYP7_YEAST (P48365) GTPase-activating protein GYP7 (GAP for YPT7)
Length = 746
Score = 52.8 bits (125), Expect = 2e-06
Identities = 33/140 (23%), Positives = 58/140 (40%), Gaps = 23/140 (16%)
Query: 232 LERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEAD 291
L+ IL + + + GYVQG+ DL++P +V+ E+
Sbjct: 513 LQNILITYNVYNTNLGYVQGMTDLLSPIYVIMKEEW-----------------------K 549
Query: 292 CYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWF 351
+WC + +D M+ ++ Q GI + L ELV+ + +S H+ L F FR
Sbjct: 550 TFWCFTHFMDIMERNFLRDQSGIHEQMLTLVELVQLMLPELSEHLNKCDSGNLFFCFRML 609
Query: 352 NCLLIREIPFNMVTRLWDTY 371
RE + +W+ +
Sbjct: 610 LVWFKREFEMEDIMHIWENF 629
>GYP2_YEAST (P53258) GTPase-activating protein GYP2 (MAC1-dependent
regulator) (MIC1 protein)
Length = 950
Score = 50.1 bits (118), Expect = 1e-05
Identities = 57/249 (22%), Positives = 97/249 (38%), Gaps = 55/249 (22%)
Query: 135 VVILDKLRELAWSGVPDYMRPTVWRLLLG-----YAPPNSDRREGVLRRKRLEYLDCVSQ 189
VV R+L GVP+ MR +W L G YA NS E +L
Sbjct: 231 VVQTPMFRKLIRIGVPNRMRGEIWELCSGAMYMRYA--NSGEYERILNE----------- 277
Query: 190 YYDIPDTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYV 249
+ T ++ DEI D R++P+ S +Q ++ + L +L A++ ++P GY
Sbjct: 278 --NAGKTSQAIDEIEK------DLKRSLPEYSAYQTEEGIQRLRNVLTAYSWKNPDVGYC 329
Query: 250 QGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLD-GMQDHYT 308
Q +N +V F++F+S E +WCL L D + +Y+
Sbjct: 330 QAMN-IVVAGFLIFMS-----------------------EEQAFWCLCNLCDIYVPGYYS 365
Query: 309 FAQPG--IQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTR 366
G + + VF + V + ++ ++ + WF L +P R
Sbjct: 366 KTMYGTLLDQRVF--ESFVEDRMPVLWEYILQHDIQLSVVSLPWFLSLFFTSMPLEYAVR 423
Query: 367 LWDTYLAEG 375
+ D + G
Sbjct: 424 IMDIFFMNG 432
>TBC4_HUMAN (O60343) TBC1 domain family member 4
Length = 1299
Score = 48.5 bits (114), Expect = 3e-05
Identities = 66/298 (22%), Positives = 115/298 (38%), Gaps = 46/298 (15%)
Query: 138 LDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRKRLEYLDCVSQYYDIPDTE 197
++ + L GVP R +W+ L L+ + L Q DI E
Sbjct: 909 MEDIHTLLKEGVPKSRRGEIWQFL-------------ALQYRLRHRLPNKQQPPDISYKE 955
Query: 198 RSEDEINMLRQIAVDCPRTVPDVSFFQQQ--QVQKSLERILYAWAIRHPASGYVQGINDL 255
+ I VD RT P +F Q Q SL +L A+++ GY QGI+
Sbjct: 956 LLKQLTAQQHAILVDLGRTFPTHPYFSVQLGPGQLSLFNLLKAYSLLDKEVGYCQGIS-F 1014
Query: 256 VTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDGMQDHYTFAQPGIQ 315
V ++ +SE + + M DL G + Y +Q
Sbjct: 1015 VAGVLLLHMSEEQAFEMLKFLMYDL---------------------GFRKQYRPDMMSLQ 1053
Query: 316 RLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTYLAEG 375
+++L L+ + H+E + +A WF L + V R++D +G
Sbjct: 1054 IQMYQLSRLLHDYHRDLYNHLEENEISPSLYAAPWFLTLFASQFSLGFVARVFDIIFLQG 1113
Query: 376 DALPDFLVYIFASFLLTWSDKL--QKLDFQDLVMFLQH-LPTQDWTHQELEMVLSRAF 430
+++ A LL+ + L + F+++V FL++ LP D E+E ++++ F
Sbjct: 1114 TE----VIFKVALSLLSSQETLIMECESFENIVEFLKNTLP--DMNTSEMEKIITQVF 1165
>GYP7_DEBHA (Q6BU76) GTPase-activating protein GYP7 (GAP for YPT7)
Length = 757
Score = 47.8 bits (112), Expect = 6e-05
Identities = 34/138 (24%), Positives = 56/138 (39%), Gaps = 23/138 (16%)
Query: 232 LERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEAD 291
+ IL + + GYVQG+ DL++P +V F E L
Sbjct: 520 MREILLTYNEHNVNLGYVQGMTDLLSPLYVTFQDESLT---------------------- 557
Query: 292 CYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWF 351
+W +D M+ ++ Q G++ + L ELV+ + + H+E L F FR
Sbjct: 558 -FWAFVNFMDRMERNFLRDQSGMKNQMLTLNELVQFMLPDLFKHLEKCESTDLYFFFRML 616
Query: 352 NCLLIREIPFNMVTRLWD 369
RE ++ V LW+
Sbjct: 617 LVWFKREFEWSSVLSLWE 634
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 53,317,898
Number of Sequences: 164201
Number of extensions: 2207495
Number of successful extensions: 5925
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 24
Number of HSP's successfully gapped in prelim test: 27
Number of HSP's that attempted gapping in prelim test: 5811
Number of HSP's gapped (non-prelim): 74
length of query: 445
length of database: 59,974,054
effective HSP length: 113
effective length of query: 332
effective length of database: 41,419,341
effective search space: 13751221212
effective search space used: 13751221212
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0106.13


