

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0106.10
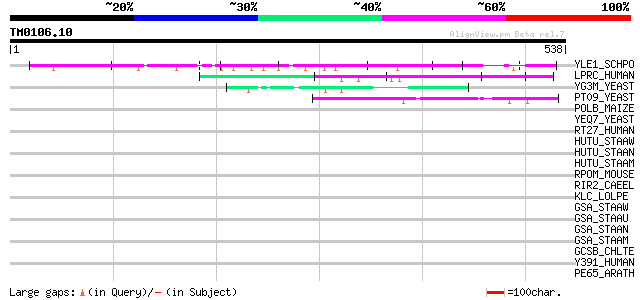
(538 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YLE1_SCHPO (Q10451) Hypothetical protein C1093.01 in chromosome I 76 2e-13
LPRC_HUMAN (P42704) 130 kDa leucine-rich protein (LRP 130) (GP13... 61 6e-09
YG3M_YEAST (P48237) Hypothetical 101.4 kDa protein in RPL24B-RSR... 45 5e-04
PT09_YEAST (P32522) PET309 protein, mitochondrial precursor 44 8e-04
POLB_MAIZE (P15718) Retrovirus-related Pol polyprotein from tran... 40 0.015
YEQ7_YEAST (P40050) Hypothetical 79.5 kDa protein in PTP3-SER3 i... 37 0.13
RT27_HUMAN (Q92552) Mitochondrial 28S ribosomal protein S27 (S27... 35 0.38
HUTU_STAAW (P67418) Urocanate hydratase (EC 4.2.1.49) (Urocanase... 33 1.4
HUTU_STAAN (P67417) Urocanate hydratase (EC 4.2.1.49) (Urocanase... 33 1.4
HUTU_STAAM (Q931G1) Urocanate hydratase (EC 4.2.1.49) (Urocanase... 33 1.4
RPOM_MOUSE (Q8BKF1) DNA-directed RNA polymerase, mitochondrial p... 33 2.4
RIR2_CAEEL (P42170) Ribonucleoside-diphosphate reductase small c... 32 5.4
KLC_LOLPE (P46825) Kinesin light chain (KLC) 32 5.4
GSA_STAAW (Q8NW75) Glutamate-1-semialdehyde 2,1-aminomutase (EC ... 32 5.4
GSA_STAAU (O34092) Glutamate-1-semialdehyde 2,1-aminomutase (EC ... 32 5.4
GSA_STAAN (P99096) Glutamate-1-semialdehyde 2,1-aminomutase (EC ... 32 5.4
GSA_STAAM (P63508) Glutamate-1-semialdehyde 2,1-aminomutase (EC ... 32 5.4
GCSB_CHLTE (Q8KAN3) Probable glycine dehydrogenase [decarboxylat... 32 5.4
Y391_HUMAN (O15091) Hypothetical protein KIAA0391 31 7.1
PE65_ARATH (Q9FJR1) Peroxidase 65 precursor (EC 1.11.1.7) (Atper... 31 7.1
>YLE1_SCHPO (Q10451) Hypothetical protein C1093.01 in chromosome I
Length = 1261
Score = 76.3 bits (186), Expect = 2e-13
Identities = 75/349 (21%), Positives = 155/349 (43%), Gaps = 29/349 (8%)
Query: 20 SSLHSHSDDVVSSFNRLLEMYP---TPCISKFNKNLTTLVKMKHYSTAISLYRQMEFSRI 76
S+ +DD ++ N E P + +N L+ L + + + L+++M+ S +
Sbjct: 898 STRRGDTDDATTALNIFEETKRHNVKPSVFLYNAVLSKLGRARRTTECWKLFQEMKESGL 957
Query: 77 MPDIFTFNILINCYCHIRQMNFAFSVFGKILKM-GYHPDTITFTSLIK---GLCINNEVQ 132
+P T+ +IN C I + A +F ++ Y P + ++I+ N E
Sbjct: 958 LPTSVTYGTVINAACRIGDESLAEKLFAEMENQPNYQPRVAPYNTMIQFEVQTMFNRE-- 1015
Query: 133 KALHLHDQLVAQGVQLNNVSYGTLVNGL-----CKMGETRAALKMLRQIEGRLVQSADVV 187
KAL +++L A ++ ++ +Y L++ +G +A L+++ + + ++ +
Sbjct: 1016 KALFYYNRLCATDIEPSSHTYKLLMDAYGTLKPVNVGSVKAVLELMERTDVPILS----M 1071
Query: 188 MYNAVIDGLCKGKLVSD---ACDLYSEMVLR----RISPDVYTYNALMYGFSTVGQLKEA 240
Y A I L G +VSD A Y + + I D + + + ++ E
Sbjct: 1072 HYAAYIHIL--GNVVSDVQAATSCYMNALAKHDAGEIQLDANLFQSQIESLIANDRIVEG 1129
Query: 241 VGLLNDMGLNNVDPNVYTFNILVDAFCKEGKVKEAKSIFAVMMKEGVE-PDVFTYDSLIE 299
+ +++DM NV N Y N L+ F K G + +A+ F ++ EG+ + TY++++
Sbjct: 1130 IQIVSDMKRYNVSLNAYIVNALIKGFTKVGMISKARYYFDLLECEGMSGKEPSTYENMVR 1189
Query: 300 GYFLVKKVNKAKDVFNSMTRMGV-APDVWSYNIMINGYCKRRMVHGALN 347
Y V KA ++ + R P V + ++N + ++ +LN
Sbjct: 1190 AYLSVNDGRKAMEIVEQLKRKRYPLPVVNRISSLVNSHMGQKPKRRSLN 1238
Score = 72.0 bits (175), Expect = 4e-12
Identities = 71/337 (21%), Positives = 142/337 (42%), Gaps = 47/337 (13%)
Query: 205 ACDLYSEMVLRRISPDVYTYNALMYGFSTVGQLKEAVGLLNDMGLNNVDPNVYTFNILVD 264
A +++ E + P V+ YNA++ + E L +M + + P T+ +++
Sbjct: 910 ALNIFEETKRHNVKPSVFLYNAVLSKLGRARRTTECWKLFQEMKESGLLPTSVTYGTVIN 969
Query: 265 AFCKEGKVKEAKSIFAVMMKE-GVEPDVFTYDSLIEGYFLVKKVNKAKDVF--NSMTRMG 321
A C+ G A+ +FA M + +P V Y+++I+ + + N+ K +F N +
Sbjct: 970 AACRIGDESLAEKLFAEMENQPNYQPRVAPYNTMIQ-FEVQTMFNREKALFYYNRLCATD 1028
Query: 322 VAPDVWSYNIMINGYCKRRMVH-GALNLFEEMHSKNLIPD-TVTYSSLIDGLCKI----- 374
+ P +Y ++++ Y + V+ G++ E+ + +P ++ Y++ I L +
Sbjct: 1029 IEPSSHTYKLLMDAYGTLKPVNVGSVKAVLELMERTDVPILSMHYAAYIHILGNVVSDVQ 1088
Query: 375 GRISCAWELVGKMHRTGQ-QADIITYNSLLHALCKSHHVDEAIALFEKVKDKGIQPDMYI 433
SC + K H G+ Q D + S + +L + + E I + +K + + YI
Sbjct: 1089 AATSCYMNALAK-HDAGEIQLDANLFQSQIESLIANDRIVEGIQIVSDMKRYNVSLNAYI 1147
Query: 434 YNVLIDGLCKSGRLKDAQEVFQNLLTKGYPLDVVTYNIMINGLCIEGLSDEALALQSKME 493
N LI G K G + A+ F L EG+S +
Sbjct: 1148 VNALIKGFTKVGMISKARYYFDLLEC-------------------EGMSGK--------- 1179
Query: 494 DNGCVSDVVTYDTIMRALYRKNDNDKAQNLLREMNAR 530
+ TY+ ++RA ND KA ++ ++ +
Sbjct: 1180 ------EPSTYENMVRAYLSVNDGRKAMEIVEQLKRK 1210
Score = 62.4 bits (150), Expect = 3e-09
Identities = 63/328 (19%), Positives = 137/328 (41%), Gaps = 26/328 (7%)
Query: 99 AFSVFGKILKMGYHPDTITFTSLIKGLCINNEVQKALHLHDQLVAQGVQLNNVSYGTLVN 158
A ++F + + P + +++ L + L ++ G+ +V+YGT++N
Sbjct: 910 ALNIFEETKRHNVKPSVFLYNAVLSKLGRARRTTECWKLFQEMKESGLLPTSVTYGTVIN 969
Query: 159 GLCKMGETRAALKMLRQIEGRLVQSADVVMYNAVIDGLCKGKLVSD-ACDLYSEMVLRRI 217
C++G+ A K+ ++E + V YN +I + + A Y+ + I
Sbjct: 970 AACRIGDESLAEKLFAEMENQPNYQPRVAPYNTMIQFEVQTMFNREKALFYYNRLCATDI 1029
Query: 218 SPDVYTYNALMYGFST-----VGQLKEAVGLL--NDMGLNNVDPNVYTF---NILVDAFC 267
P +TY LM + T VG +K + L+ D+ + ++ Y N++ D
Sbjct: 1030 EPSSHTYKLLMDAYGTLKPVNVGSVKAVLELMERTDVPILSMHYAAYIHILGNVVSD--- 1086
Query: 268 KEGKVKEAKSIFAVMMKE----GVEPDVFTYDSLIEGYFLVKKVNKAKDVFNSMTRMGVA 323
V+ A S + + + ++ D + S IE ++ + + + M R V+
Sbjct: 1087 ----VQAATSCYMNALAKHDAGEIQLDANLFQSQIESLIANDRIVEGIQIVSDMKRYNVS 1142
Query: 324 PDVWSYNIMINGYCKRRMVHGALNLFEEMHSKNLI-PDTVTYSSLIDGLCKIGRISCAWE 382
+ + N +I G+ K M+ A F+ + + + + TY +++ + A E
Sbjct: 1143 LNAYIVNALIKGFTKVGMISKARYYFDLLECEGMSGKEPSTYENMVRAYLSVNDGRKAME 1202
Query: 383 LVGKMHRTGQQADIITYNSLLHALCKSH 410
+V ++ R ++ + + +L SH
Sbjct: 1203 IVEQLKRKRYPLPVV---NRISSLVNSH 1227
Score = 62.0 bits (149), Expect = 4e-09
Identities = 47/185 (25%), Positives = 89/185 (47%), Gaps = 9/185 (4%)
Query: 261 ILVDAFCKEGKVKEAKSIFAVMMKE-GVEPDVFTYDSLIEGYFL---VKKVNKAKDVFNS 316
IL +F ++ K + ++F MK G P T+ LI A ++F
Sbjct: 860 ILSSSFARQFK---SSNLFCDNMKMLGYIPRASTFAHLINNSTRRGDTDDATTALNIFEE 916
Query: 317 MTRMGVAPDVWSYNIMINGYCKRRMVHGALNLFEEMHSKNLIPDTVTYSSLIDGLCKIGR 376
R V P V+ YN +++ + R LF+EM L+P +VTY ++I+ C+IG
Sbjct: 917 TKRHNVKPSVFLYNAVLSKLGRARRTTECWKLFQEMKESGLLPTSVTYGTVINAACRIGD 976
Query: 377 ISCAWELVGKM-HRTGQQADIITYNSLLHALCKS-HHVDEAIALFEKVKDKGIQPDMYIY 434
S A +L +M ++ Q + YN+++ ++ + ++A+ + ++ I+P + Y
Sbjct: 977 ESLAEKLFAEMENQPNYQPRVAPYNTMIQFEVQTMFNREKALFYYNRLCATDIEPSSHTY 1036
Query: 435 NVLID 439
+L+D
Sbjct: 1037 KLLMD 1041
Score = 46.2 bits (108), Expect = 2e-04
Identities = 65/313 (20%), Positives = 125/313 (39%), Gaps = 47/313 (15%)
Query: 185 DVVMYNAVIDGLCKGKLVSDACDLYSEMVLRRISPDVYTYNALMYGFSTVGQLKEAVGLL 244
D ++ +V D + +V + D + +L IS Y + L + +++ L
Sbjct: 721 DEMLVASVRDDIVSEAVVGFSTDNNGQKILADISQVCYCLDDLEC-------IDQSINSL 773
Query: 245 NDMGLNNVDPNVYTFNILVDAFCKEGKVKEAKSIFAVMMKEGVEPDVFTYDSLIEGYFLV 304
L + P NIL F + GK+ E + V P + + S + + V
Sbjct: 774 VSKMLTSASPEQVDVNIL---FFQFGKLIETNKF----LHPEVYPTLISVLSKNKRFDAV 826
Query: 305 KKV-NKAKDVFNSMTRMGVAPDVWSYNIMINGYCKRRMVHGALNLFEEMHSKNLIPDTVT 363
++V +K ++ ++ + W ++++ + + + S NL D +
Sbjct: 827 QRVFEHSKHLYRKISTKSLEKANWFMALILDAMIL------SSSFARQFKSSNLFCDNMK 880
Query: 364 YSSLIDGLCKIGRISCAWELVGKMHRTGQQADIITYNSLLHALCKSHHVDEAIALFEKVK 423
L I R S L+ R G D T A+ +FE+ K
Sbjct: 881 M------LGYIPRASTFAHLINNSTRRGDTDDATT----------------ALNIFEETK 918
Query: 424 DKGIQPDMYIYNVLIDGLCKSGRLKDAQEVFQNLLTKGYPLDVVTYNIMINGLCIEGLSD 483
++P +++YN ++ L ++ R + ++FQ + G VTY +IN C + D
Sbjct: 919 RHNVKPSVFLYNAVLSKLGRARRTTECWKLFQEMKESGLLPTSVTYGTVINAAC--RIGD 976
Query: 484 EALA--LQSKMED 494
E+LA L ++ME+
Sbjct: 977 ESLAEKLFAEMEN 989
Score = 36.6 bits (83), Expect = 0.17
Identities = 35/163 (21%), Positives = 64/163 (38%), Gaps = 14/163 (8%)
Query: 385 GKMHRTGQQADIITYNSLLHALCKSHHVDEAIALFE-------KVKDKGIQPDMYIYNVL 437
GK+ T + Y +L+ L K+ D +FE K+ K ++ + ++
Sbjct: 796 GKLIETNKFLHPEVYPTLISVLSKNKRFDAVQRVFEHSKHLYRKISTKSLEKANWFMALI 855
Query: 438 IDGLCKSG----RLKDAQEVFQNLLTKGYPLDVVTYNIMINGLCIEGLSDEALALQSKME 493
+D + S + K + N+ GY T+ +IN G +D+A + E
Sbjct: 856 LDAMILSSSFARQFKSSNLFCDNMKMLGYIPRASTFAHLINNSTRRGDTDDATTALNIFE 915
Query: 494 D---NGCVSDVVTYDTIMRALYRKNDNDKAQNLLREMNARGLL 533
+ + V Y+ ++ L R + L +EM GLL
Sbjct: 916 ETKRHNVKPSVFLYNAVLSKLGRARRTTECWKLFQEMKESGLL 958
Score = 32.0 bits (71), Expect = 4.2
Identities = 26/91 (28%), Positives = 41/91 (44%), Gaps = 13/91 (14%)
Query: 196 LCKGKLVSDACDLYSEMVLRRISPDVYT---YNALM-------YGFSTVGQLKEAVGLLN 245
L KL DLY L R SP++ T YN ++ Y S+ +++ + +
Sbjct: 190 LASSKLSIHTFDLYK---LVRNSPELLTLEAYNIVLQCMSTDDYFLSSSKNIQKIIKVYV 246
Query: 246 DMGLNNVDPNVYTFNILVDAFCKEGKVKEAK 276
DM + + PNV TF ++ A C+ K K
Sbjct: 247 DMLNSFISPNVTTFETVIFALCRRAKFVHQK 277
>LPRC_HUMAN (P42704) 130 kDa leucine-rich protein (LRP 130) (GP130)
(Leucine-rich PPR-motif containing protein)
Length = 1273
Score = 61.2 bits (147), Expect = 6e-09
Identities = 74/354 (20%), Positives = 139/354 (38%), Gaps = 49/354 (13%)
Query: 185 DVVMYNAVIDGLCKGKLVSDACDLYSEMVLRRISPDVYTYNALMYGFSTVGQLKEAVGLL 244
DV YNA++ + + D ++M I P+ TY L+ + VG ++ A +L
Sbjct: 40 DVSHYNALLKVYLQNEYKFSPTDFLAKMEEANIQPNRVTYQRLIASYCNVGDIEGASKIL 99
Query: 245 NDMGLNNVDPNVYTFNILVDAFCKEGKVKEAKSIFAVMMKEGVEPDVFTYDSLIEGYFLV 304
M ++ F+ LV + G ++ A++I VM G+EP TY +L+ Y
Sbjct: 100 GFMKTKDLPVTEAVFSALVTGHARAGDMENAENILTVMRDAGIEPGPDTYLALLNAYAEK 159
Query: 305 KKVNKAKDVFNSMTRM-------GVAPDVWSYNIMINGY------------CKRRMVHGA 345
++ K + + + ++S++ GY C+RR + A
Sbjct: 160 GDIDHVKQTLEKVEKFELHLMDRDLLQIIFSFSKA--GYLSMSQKFWKKFTCERRYIPDA 217
Query: 346 LNLFEEMHSKNLIPDTVTYSSLI-------DGLCKIGR------------ISCAWELVGK 386
+NL + ++ L + V L+ DG G + + K
Sbjct: 218 MNLILLLVTEKL--EDVALQILLACPVSKEDGPSVFGSFFLQHCVTMNTPVEKLTDYCKK 275
Query: 387 MHRTGQQADIITYNSLLHALCKSHHVDEAIALFEKVKDKGIQPDMYIYNVLIDGLCKSGR 446
+ + + + LH ++ D A AL + VK++G + + L+ G K
Sbjct: 276 LKEVQMHSFPLQFT--LHCALLANKTDLAKALMKAVKEEGFPIRPHYFWPLLVGRRKEKN 333
Query: 447 LKDAQEVFQNLLTKGYPLDVVTYNIMINGLCIEGLSDEALALQSKMEDNGCVSD 500
++ E+ + + G D TY + C + ++ LQ +NGC+SD
Sbjct: 334 VQGIIEILKGMQELGVHPDQETYTDYVIP-CFDSVNSARAILQ----ENGCLSD 382
Score = 59.3 bits (142), Expect = 2e-08
Identities = 33/162 (20%), Positives = 74/162 (45%)
Query: 296 SLIEGYFLVKKVNKAKDVFNSMTRMGVAPDVWSYNIMINGYCKRRMVHGALNLFEEMHSK 355
SL+ L ++ A +++++ ++G DV YN ++ Y + + +M
Sbjct: 11 SLLPELKLEERTEFAHRIWDTLQKLGAVYDVSHYNALLKVYLQNEYKFSPTDFLAKMEEA 70
Query: 356 NLIPDTVTYSSLIDGLCKIGRISCAWELVGKMHRTGQQADIITYNSLLHALCKSHHVDEA 415
N+ P+ VTY LI C +G I A +++G M +++L+ ++ ++ A
Sbjct: 71 NIQPNRVTYQRLIASYCNVGDIEGASKILGFMKTKDLPVTEAVFSALVTGHARAGDMENA 130
Query: 416 IALFEKVKDKGIQPDMYIYNVLIDGLCKSGRLKDAQEVFQNL 457
+ ++D GI+P Y L++ + G + ++ + +
Sbjct: 131 ENILTVMRDAGIEPGPDTYLALLNAYAEKGDIDHVKQTLEKV 172
Score = 54.3 bits (129), Expect = 8e-07
Identities = 37/162 (22%), Positives = 68/162 (41%)
Query: 366 SLIDGLCKIGRISCAWELVGKMHRTGQQADIITYNSLLHALCKSHHVDEAIALFEKVKDK 425
SL+ L R A + + + G D+ YN+LL ++ + K+++
Sbjct: 11 SLLPELKLEERTEFAHRIWDTLQKLGAVYDVSHYNALLKVYLQNEYKFSPTDFLAKMEEA 70
Query: 426 GIQPDMYIYNVLIDGLCKSGRLKDAQEVFQNLLTKGYPLDVVTYNIMINGLCIEGLSDEA 485
IQP+ Y LI C G ++ A ++ + TK P+ ++ ++ G G + A
Sbjct: 71 NIQPNRVTYQRLIASYCNVGDIEGASKILGFMKTKDLPVTEAVFSALVTGHARAGDMENA 130
Query: 486 LALQSKMEDNGCVSDVVTYDTIMRALYRKNDNDKAQNLLREM 527
+ + M D G TY ++ A K D D + L ++
Sbjct: 131 ENILTVMRDAGIEPGPDTYLALLNAYAEKGDIDHVKQTLEKV 172
Score = 43.1 bits (100), Expect = 0.002
Identities = 60/327 (18%), Positives = 131/327 (39%), Gaps = 10/327 (3%)
Query: 45 ISKFNKNLTTLVKMKHYSTAISLYRQMEFSRIMPDIFTFNILINCYCHIRQMNFAFSVFG 104
+S +N L ++ ++ + +ME + I P+ T+ LI YC++ + A + G
Sbjct: 41 VSHYNALLKVYLQNEYKFSPTDFLAKMEEANIQPNRVTYQRLIASYCNVGDIEGASKILG 100
Query: 105 KILKMGYHPDTITFTSLIKGLCINNEVQKALHLHDQLVAQGVQLNNVSYGTLVNGLCKMG 164
+ F++L+ G +++ A ++ + G++ +Y L+N + G
Sbjct: 101 FMKTKDLPVTEAVFSALVTGHARAGDMENAENILTVMRDAGIEPGPDTYLALLNAYAEKG 160
Query: 165 ETRAALKMLRQIEGRLVQSADVVMYNAVIDGLCKGKLVSDACDLYSEMVL-RRISPDVYT 223
+ + L ++E + D + +I K +S + + + RR PD
Sbjct: 161 DIDHVKQTLEKVEKFELHLMDRDLLQ-IIFSFSKAGYLSMSQKFWKKFTCERRYIPDAMN 219
Query: 224 YNALMYGFSTVGQLKE-AVGLLNDMGLNNVD-PNVYTFNILVDAFCKEGKVKEAKSIFAV 281
L+ +L++ A+ +L ++ D P+V+ L V++ +
Sbjct: 220 LILLL----VTEKLEDVALQILLACPVSKEDGPSVFGSFFLQHCVTMNTPVEKLTD-YCK 274
Query: 282 MMKEGVEPDVFTYDSLIEGYFLVKKVNKAKDVFNSMTRMGVAPDVWSYNIMINGYCKRRM 341
+KE V+ F + L K + AK + ++ G + ++ G K +
Sbjct: 275 KLKE-VQMHSFPLQFTLHCALLANKTDLAKALMKAVKEEGFPIRPHYFWPLLVGRRKEKN 333
Query: 342 VHGALNLFEEMHSKNLIPDTVTYSSLI 368
V G + + + M + PD TY+ +
Sbjct: 334 VQGIIEILKGMQELGVHPDQETYTDYV 360
Score = 36.6 bits (83), Expect = 0.17
Identities = 79/394 (20%), Positives = 148/394 (37%), Gaps = 66/394 (16%)
Query: 116 ITFTSLIKGLCINNEVQKALHLHDQLVAQGVQ--LNNVSYGTLVNGLCKMGETRAALKML 173
++F ++ G + E++ LH+ +V G+ N+S+ LV + G+ AL++
Sbjct: 666 LSFFHMLNGAALRGEIETVKQLHEAIVTLGLAEPSTNISF-PLVTVHLEKGDLSTALEVA 724
Query: 174 RQIEGRLVQSADVVMYNAVIDGLCK--GKLVSDACDLYSEMVLRRISPDVYTYNALMYGF 231
+ + + D LCK K +D + V + V Y+ L + F
Sbjct: 725 IDCYEKYK------VLPRIHDVLCKLVEKGETDLIQKAMDFVSQEQGEMVMLYD-LFFAF 777
Query: 232 STVGQLKEAVGLLNDMGLNN-------------VDPNVYTFNILVDAFCK---------- 268
G KEA ++ G+ + V T LV+ K
Sbjct: 778 LQTGNYKEAKKIIETPGIRARSARLQWFCDRCVANNQVETLEKLVELTQKLFECDRDQMY 837
Query: 269 ---------EGKVKEAKSIFAVMMKEGVEPDVFTYDSLIEGYFLVKKVNKA--------- 310
G + A +++ + +E V P T L E ++++ N+
Sbjct: 838 YNLLKLYKINGDWQRADAVWNKIQEENVIPREKTLRLLAE---ILREGNQEVPFDVPELW 894
Query: 311 ----KDVFNSMTRMGVAPDVWSYNIMINGYCKRRMVHGALNLFEEMHSKNLIPDTVTYSS 366
K NS + PD + +I+I C+ GA ++F +N++ + TYS+
Sbjct: 895 YEDEKHSLNSSSASTTEPD-FQKDILIA--CRLNQKKGAYDIFLNAKEQNIVFNAETYSN 951
Query: 367 LIDGLCKIGRISCAWEL--VGKMHRTGQQADIITYNSLLHALCKSHHVDEAIALFEKVKD 424
LI L + A E+ + H G + + L+ + ++ EA+ + V D
Sbjct: 952 LIKLLMSEDYFTQAMEVKAFAETHIKGFTLNDAANSRLIITQVRRDYLKEAVTTLKTVLD 1011
Query: 425 KGIQPDMYIYNVLIDGLCKSGRLKDAQEVFQNLL 458
+ P +I L G +++ EV Q +L
Sbjct: 1012 QQQTPSRLAVTRVIQALAMKGDVENI-EVVQKML 1044
Score = 33.9 bits (76), Expect = 1.1
Identities = 32/131 (24%), Positives = 56/131 (42%), Gaps = 6/131 (4%)
Query: 329 YNIMINGYCKRRMVHGALNLFEEMH--SKNLIPDTVTYSSLIDGLCKIGRISCAWELVGK 386
Y +IN C+ V ALNL EE + + DT Y L+ L K G++ A +++ +
Sbjct: 593 YAALINLCCRHDKVEDALNLKEEFDRLDSSAVLDTGNYLGLVRVLAKHGKLQDAIKILKE 652
Query: 387 MHRTG---QQADIITYNSLLHALCKSHHVDEAIALFEKVKDKGI-QPDMYIYNVLIDGLC 442
M + +++ +L+ ++ L E + G+ +P I L+
Sbjct: 653 MKEKDVLIKDTTALSFFHMLNGAALRGEIETVKQLHEAIVTLGLAEPSTNISFPLVTVHL 712
Query: 443 KSGRLKDAQEV 453
+ G L A EV
Sbjct: 713 EKGDLSTALEV 723
>YG3M_YEAST (P48237) Hypothetical 101.4 kDa protein in RPL24B-RSR1
intergenic region
Length = 864
Score = 45.1 bits (105), Expect = 5e-04
Identities = 57/242 (23%), Positives = 93/242 (37%), Gaps = 49/242 (20%)
Query: 211 EMVLRRISPDVYTYNALMYG--FSTVGQLKEAVGLLNDMGLNNVDPNVYTFNILVDAFCK 268
EM+L +I+ DV +N MY F+ +G LK N G N D + L++ + K
Sbjct: 205 EMLLEQINGDVVKFNTHMYECIFNNLGNLKPTN--FNQDGTN--DKVILKMKELLERYDK 260
Query: 269 EGKVKEAKSIFAVMMKEGVEPDVFTYDSLIEGYFLV--KKVNKAKDVFNSMTRM----GV 322
K+ E + + KEG V I L K + D+ +T+ G+
Sbjct: 261 ALKITEER----INKKEGFPSKVPKMTQAILNNCLKYSTKCSSFHDMDYFITKFRDDYGI 316
Query: 323 APDVWSYNIMINGYCKRRMVHGALNLFEEMHSKNLIPDTVTYSSLIDGLCKIGRISCAWE 382
P+ + +I Y ++ M A N F+ M
Sbjct: 317 TPNKQNLTTVIQFYSRKEMTKQAWNTFDTM------------------------------ 346
Query: 383 LVGKMHRTGQQADIITYNSLLHALCKSHHVDEAIALFEKVKDKGIQPDMYIYNVLIDGLC 442
K T DI TYN++L K + +A+ LF++++D I+P Y ++ L
Sbjct: 347 ---KFLSTKHFPDICTYNTMLRICEKERNFPKALDLFQEIQDHNIKPTTNTYIMMARVLA 403
Query: 443 KS 444
S
Sbjct: 404 SS 405
>PT09_YEAST (P32522) PET309 protein, mitochondrial precursor
Length = 965
Score = 44.3 bits (103), Expect = 8e-04
Identities = 55/254 (21%), Positives = 105/254 (40%), Gaps = 26/254 (10%)
Query: 294 YDSLIEGYFLVKKVNKAKDVFNSMTRMGVAPDVWSYNIMINGYCKRRMVHGALNLFEEMH 353
+D L+ + + + + FN++ +G P + Y I++ + + L+ ++
Sbjct: 314 FDYLLVAHSRLHNWDALQQQFNALFGIGKLPSIQHYGILMYTMARIGELDSVNKLYTQLL 373
Query: 354 SKNLIPDTVTYSSLIDGLCKIGRISCA---WELVGKMHRTGQQADIITYNSLLHALCKSH 410
+ +IP SL+ K+G + +EL K T A T+ +L +
Sbjct: 374 RRGMIPTYAVLQSLLYAHYKVGDFAACFSHFELFKKYDITPSTA---THTIMLKVYRGLN 430
Query: 411 HVDEAIALFEKV-KDKGIQPDMYIYNVLIDGLCKSGRLKDAQEVFQNLLTKGYPLDVVTY 469
+D A + +++ +D ++ + +LI CK+ AQE+F NL+T+ Y
Sbjct: 431 DLDGAFRILKRLSEDPSVEITEGHFALLIQMCCKTTNHLIAQELF-NLMTE-------HY 482
Query: 470 NIMINGLCIEGLSD---------EALALQSKMEDNGCVSD--VVTYDTIMRALYRKNDND 518
NI G I L D EA+AL K N D + Y+ ++A + +
Sbjct: 483 NIQHTGKSISALMDVYIESNRPTEAIALFEKHSKNLSWRDGLISVYNKAIKAYIGLRNAN 542
Query: 519 KAQNLLREMNARGL 532
K + L ++ L
Sbjct: 543 KCEELFDKITTSKL 556
Score = 39.3 bits (90), Expect = 0.026
Identities = 68/346 (19%), Positives = 136/346 (38%), Gaps = 38/346 (10%)
Query: 17 LSFSSLHSHSDDVVSSFNRLLEMYPTPCISKFNKNLTTLVKMKHYSTAISLYRQMEFSRI 76
++ S LH+ D + FN L + P I + + T+ ++ + LY Q+ +
Sbjct: 319 VAHSRLHNW-DALQQQFNALFGIGKLPSIQHYGILMYTMARIGELDSVNKLYTQLLRRGM 377
Query: 77 MPDIFTFNILINCYCHIRQMNFA--FSVFGKILKMGYHPDTITFTSLIKGLCINNEVQKA 134
+P L+ Y H + +FA FS F K P T T T ++K N++ A
Sbjct: 378 IPTYAVLQSLL--YAHYKVGDFAACFSHFELFKKYDITPSTATHTIMLKVYRGLNDLDGA 435
Query: 135 LHLHDQLVAQ-GVQLNNVSYGTLVNGLCKMGETRAALKMLRQIEGRLVQSADVVMYNAVI 193
+ +L V++ + L+ CK A ++ + +A++
Sbjct: 436 FRILKRLSEDPSVEITEGHFALLIQMCCKTTNHLIAQELFNLMTEHYNIQHTGKSISALM 495
Query: 194 DGLCKGKLVSDACDLYSEMVLRRISPDVYTYNALMYGFSTVGQLKEAVGLLNDMGLNNVD 253
D + ++A L+ + S ++ + L+ ++ +K +GL N +
Sbjct: 496 DVYIESNRPTEAIALF-----EKHSKNLSWRDGLISVYNKA--IKAYIGLRNANKCEELF 548
Query: 254 PNVYTFNILVDAFCKEGKVKEAKSIFAVMMKEGVEPDVFTYDSLIEGYFLVKKVNKAKDV 313
+ T + V++ + +K F V + E E + D LI+
Sbjct: 549 DKITTSKLAVNSEFYKMMIK-----FLVTLNEDCETALSIIDQLIK-------------- 589
Query: 314 FNSMTRMGVAPDVWSYNIMINGYCKRRMVHGALNLFEEMHSKNLIP 359
+S+ ++ D + I++ Y K G +NL++ M S+N +P
Sbjct: 590 -HSVIKV----DATHFEIIMEAYDKEGYRDGIINLYKTM-SQNKVP 629
>POLB_MAIZE (P15718) Retrovirus-related Pol polyprotein from
transposon element BS1 (ORF 1) [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 740
Score = 40.0 bits (92), Expect = 0.015
Identities = 20/67 (29%), Positives = 37/67 (54%), Gaps = 9/67 (13%)
Query: 447 LKDAQEVFQNLLTKGYP---------LDVVTYNIMINGLCIEGLSDEALALQSKMEDNGC 497
++DA +VF +LTKG + V +N+++ L ++G + +A + +M++NG
Sbjct: 442 VEDAAKVFDKMLTKGEEAPSVQRGVVMTAVAHNLLVQALFMDGRASDAYVVLEEMQNNGP 501
Query: 498 VSDVVTY 504
DV TY
Sbjct: 502 FPDVFTY 508
Score = 35.0 bits (79), Expect = 0.49
Identities = 20/73 (27%), Positives = 34/73 (46%), Gaps = 9/73 (12%)
Query: 410 HHVDEAIALFEKVKDKG---------IQPDMYIYNVLIDGLCKSGRLKDAQEVFQNLLTK 460
+HV++A +F+K+ KG + +N+L+ L GR DA V + +
Sbjct: 440 YHVEDAAKVFDKMLTKGEEAPSVQRGVVMTAVAHNLLVQALFMDGRASDAYVVLEEMQNN 499
Query: 461 GYPLDVVTYNIMI 473
G DV TY + +
Sbjct: 500 GPFPDVFTYTLKV 512
Score = 31.6 bits (70), Expect = 5.4
Identities = 14/35 (40%), Positives = 19/35 (54%)
Query: 260 NILVDAFCKEGKVKEAKSIFAVMMKEGVEPDVFTY 294
N+LV A +G+ +A + M G PDVFTY
Sbjct: 474 NLLVQALFMDGRASDAYVVLEEMQNNGPFPDVFTY 508
>YEQ7_YEAST (P40050) Hypothetical 79.5 kDa protein in PTP3-SER3
intergenic region
Length = 688
Score = 37.0 bits (84), Expect = 0.13
Identities = 38/151 (25%), Positives = 71/151 (46%), Gaps = 15/151 (9%)
Query: 263 VDAFCKEGKVKEAKSIFAVMMKEGVEPDVFTYDSLIEGYFL--VKKVNKAKDVFNSMTRM 320
V F K G++++A + + K+GV +LI Y++ V+ A D+FN +
Sbjct: 174 VQKFLKRGQLEKAVFLARLAKKKGV-----VGMNLIMKYYIEVVQSQQSAVDIFNWRKKW 228
Query: 321 GVAPDVWSYNIMINGYCKRRMV----HGALNL--FEEMHSKNLIPDTVTYSSLIDGLCKI 374
GV D S I+ NG K+ + +G L L + + KN + + + Y++ + L
Sbjct: 229 GVPIDQHSITILFNGLSKQENLVSKKYGELVLKTIDSLCDKNELTE-IEYNTALAALINC 287
Query: 375 GRISCAWELVGKMHRTGQQADIITYNSLLHA 405
+ ++L+ K G + D ITY ++ +
Sbjct: 288 TDETLVFKLLNK-KCPGLKKDSITYTLMIRS 317
>RT27_HUMAN (Q92552) Mitochondrial 28S ribosomal protein S27 (S27mt)
(MRP-S27)
Length = 414
Score = 35.4 bits (80), Expect = 0.38
Identities = 17/36 (47%), Positives = 26/36 (72%), Gaps = 1/36 (2%)
Query: 254 PNVYTFNILVDAFCKEGKVKEAKS-IFAVMMKEGVE 288
P+ +TFN+L+D+F K+ K+A S +F VMM+E E
Sbjct: 139 PDNFTFNLLMDSFIKKENYKDALSVVFEVMMQEAFE 174
>HUTU_STAAW (P67418) Urocanate hydratase (EC 4.2.1.49) (Urocanase)
(Imidazolonepropionate hydrolase)
Length = 553
Score = 33.5 bits (75), Expect = 1.4
Identities = 26/89 (29%), Positives = 46/89 (51%), Gaps = 18/89 (20%)
Query: 408 KSHHVDEAIALFEKVKDKGIQPDMYIYNVLIDGLCKSGRLKDAQEVFQNLLTKGYPLDVV 467
K+ +DEA+ L E+ K++G +GL G + +A ++ Q +L KG+ +D++
Sbjct: 207 KTADLDEALKLAEEAKERG------------EGL-SIGLVGNAVDIHQAILEKGFKIDII 253
Query: 468 TYNIM----INGLCIEGLS-DEALALQSK 491
T +NG +G S +EA L+ K
Sbjct: 254 TDQTSAHDPLNGYVPQGYSVEEAKVLREK 282
>HUTU_STAAN (P67417) Urocanate hydratase (EC 4.2.1.49) (Urocanase)
(Imidazolonepropionate hydrolase)
Length = 553
Score = 33.5 bits (75), Expect = 1.4
Identities = 26/89 (29%), Positives = 46/89 (51%), Gaps = 18/89 (20%)
Query: 408 KSHHVDEAIALFEKVKDKGIQPDMYIYNVLIDGLCKSGRLKDAQEVFQNLLTKGYPLDVV 467
K+ +DEA+ L E+ K++G +GL G + +A ++ Q +L KG+ +D++
Sbjct: 207 KTADLDEALKLAEEAKERG------------EGL-SIGLVGNAVDIHQAILEKGFKIDII 253
Query: 468 TYNIM----INGLCIEGLS-DEALALQSK 491
T +NG +G S +EA L+ K
Sbjct: 254 TDQTSAHDPLNGYVPQGYSVEEAKVLREK 282
>HUTU_STAAM (Q931G1) Urocanate hydratase (EC 4.2.1.49) (Urocanase)
(Imidazolonepropionate hydrolase)
Length = 553
Score = 33.5 bits (75), Expect = 1.4
Identities = 26/89 (29%), Positives = 46/89 (51%), Gaps = 18/89 (20%)
Query: 408 KSHHVDEAIALFEKVKDKGIQPDMYIYNVLIDGLCKSGRLKDAQEVFQNLLTKGYPLDVV 467
K+ +DEA+ L E+ K++G +GL G + +A ++ Q +L KG+ +D++
Sbjct: 207 KTADLDEALKLAEEAKERG------------EGL-SIGLVGNAVDIHQAILEKGFKIDII 253
Query: 468 TYNIM----INGLCIEGLS-DEALALQSK 491
T +NG +G S +EA L+ K
Sbjct: 254 TDQTSAHDPLNGYVPQGYSVEEAKVLREK 282
>RPOM_MOUSE (Q8BKF1) DNA-directed RNA polymerase, mitochondrial
precursor (EC 2.7.7.6) (MtRPOL)
Length = 1207
Score = 32.7 bits (73), Expect = 2.4
Identities = 20/94 (21%), Positives = 43/94 (45%), Gaps = 8/94 (8%)
Query: 375 GRISCAWELVGKMHRTGQQADIIT---YNSLLHALCKSHHVDEAIALFEKVKDKGIQPDM 431
G++ A ++ H G + ++T YN+++ + E + +F +KD G+ PD+
Sbjct: 209 GQVPLAHHVLVTHHNNGDRQQVLTLHMYNTVMLGWARKGSFRELVYVFLMLKDAGLSPDL 268
Query: 432 YIYNVLIDGLCKSGRLKDA---QEVFQNLLTKGY 462
Y + C R +D Q + ++ +G+
Sbjct: 269 CSYAAALQ--CMGRRDQDVRTIQRCLKQMMEEGF 300
>RIR2_CAEEL (P42170) Ribonucleoside-diphosphate reductase small
chain (EC 1.17.4.1) (Ribonucleotide reductase)
Length = 381
Score = 31.6 bits (70), Expect = 5.4
Identities = 24/90 (26%), Positives = 44/90 (48%), Gaps = 12/90 (13%)
Query: 397 ITYNSLLHALCKSHHVDEAIALFEKVKDKGIQPDMYIYNVLIDGLCKSGRLKDAQEVFQN 456
+T+++ L + + H D A L+ K++ K Q IY+++ KDA + Q
Sbjct: 247 LTHSNELISRDEGLHRDFACLLYSKLQKKLTQ--QRIYDII----------KDAVAIEQE 294
Query: 457 LLTKGYPLDVVTYNIMINGLCIEGLSDEAL 486
LT+ P+D++ N + IE ++D L
Sbjct: 295 FLTEALPVDMIGMNCRLMSQYIEFVADHLL 324
>KLC_LOLPE (P46825) Kinesin light chain (KLC)
Length = 571
Score = 31.6 bits (70), Expect = 5.4
Identities = 42/133 (31%), Positives = 60/133 (44%), Gaps = 21/133 (15%)
Query: 173 LRQIEGRLVQSADVVMYNAVIDGLCKGKLVSDACDLYSEMVLRRISPDVYTY-NALMYGF 231
LR + ++Q A Y + LCK L DL E PDV T N L +
Sbjct: 220 LRTLHNLVIQYASQGRYEVAVP-LCKQALE----DL--EKTSGHDHPDVATMLNILALVY 272
Query: 232 STVGQLKEAVGLLND-MGL--NNVDPN----VYTFNILVDAFCKEGKVKEAKSIF--AVM 282
G+ KEA LLND +G+ + P+ T N L + K GK K+A+ + A++
Sbjct: 273 RDQGKYKEAANLLNDALGIREKTLGPDHPAVAATLNNLAVLYGKRGKYKDAEPLCKRALV 332
Query: 283 MKEGV----EPDV 291
++E V PDV
Sbjct: 333 IREKVLGKDHPDV 345
>GSA_STAAW (Q8NW75) Glutamate-1-semialdehyde 2,1-aminomutase (EC
5.4.3.8) (GSA) (Glutamate-1-semialdehyde
aminotransferase) (GSA-AT)
Length = 428
Score = 31.6 bits (70), Expect = 5.4
Identities = 28/97 (28%), Positives = 41/97 (41%), Gaps = 14/97 (14%)
Query: 214 LRRISPDVYTYNALMYGFSTVGQLKEAVGLLNDMGLNNVDPNVYTFNILVDAFCKEGKV- 272
L +++P+ Y Y F+ +G + E GL +NV V ++ F EG V
Sbjct: 314 LSQLTPETYEY------FNMLGDILED-GLKRVFAKHNVPITVNRAGSMIGYFLNEGPVT 366
Query: 273 ------KEAKSIFAVMMKEGVEPDVFTYDSLIEGYFL 303
K +FA M +E + VF S EG FL
Sbjct: 367 NFEQANKSDLKLFAEMYREMAKEGVFLPPSQFEGTFL 403
>GSA_STAAU (O34092) Glutamate-1-semialdehyde 2,1-aminomutase (EC
5.4.3.8) (GSA) (Glutamate-1-semialdehyde
aminotransferase) (GSA-AT)
Length = 428
Score = 31.6 bits (70), Expect = 5.4
Identities = 28/97 (28%), Positives = 41/97 (41%), Gaps = 14/97 (14%)
Query: 214 LRRISPDVYTYNALMYGFSTVGQLKEAVGLLNDMGLNNVDPNVYTFNILVDAFCKEGKV- 272
L +++P+ Y Y F+ +G + E GL +NV V ++ F EG V
Sbjct: 314 LSQLTPETYEY------FNMLGDILED-GLKRVFAKHNVPITVNRAGSMIGYFLNEGPVT 366
Query: 273 ------KEAKSIFAVMMKEGVEPDVFTYDSLIEGYFL 303
K +FA M +E + VF S EG FL
Sbjct: 367 NFEQANKSDLKLFAEMYREMAKEGVFLPPSQFEGTFL 403
>GSA_STAAN (P99096) Glutamate-1-semialdehyde 2,1-aminomutase (EC
5.4.3.8) (GSA) (Glutamate-1-semialdehyde
aminotransferase) (GSA-AT)
Length = 428
Score = 31.6 bits (70), Expect = 5.4
Identities = 28/97 (28%), Positives = 41/97 (41%), Gaps = 14/97 (14%)
Query: 214 LRRISPDVYTYNALMYGFSTVGQLKEAVGLLNDMGLNNVDPNVYTFNILVDAFCKEGKV- 272
L +++P+ Y Y F+ +G + E GL +NV V ++ F EG V
Sbjct: 314 LSQLTPETYEY------FNMLGDILED-GLKRVFAKHNVPITVNRAGSMIGYFLNEGPVT 366
Query: 273 ------KEAKSIFAVMMKEGVEPDVFTYDSLIEGYFL 303
K +FA M +E + VF S EG FL
Sbjct: 367 NFEQANKSDLKLFAEMYREMAKEGVFLPPSQFEGTFL 403
>GSA_STAAM (P63508) Glutamate-1-semialdehyde 2,1-aminomutase (EC
5.4.3.8) (GSA) (Glutamate-1-semialdehyde
aminotransferase) (GSA-AT)
Length = 428
Score = 31.6 bits (70), Expect = 5.4
Identities = 28/97 (28%), Positives = 41/97 (41%), Gaps = 14/97 (14%)
Query: 214 LRRISPDVYTYNALMYGFSTVGQLKEAVGLLNDMGLNNVDPNVYTFNILVDAFCKEGKV- 272
L +++P+ Y Y F+ +G + E GL +NV V ++ F EG V
Sbjct: 314 LSQLTPETYEY------FNMLGDILED-GLKRVFAKHNVPITVNRAGSMIGYFLNEGPVT 366
Query: 273 ------KEAKSIFAVMMKEGVEPDVFTYDSLIEGYFL 303
K +FA M +E + VF S EG FL
Sbjct: 367 NFEQANKSDLKLFAEMYREMAKEGVFLPPSQFEGTFL 403
>GCSB_CHLTE (Q8KAN3) Probable glycine dehydrogenase
[decarboxylating] subunit 2 (EC 1.4.4.2) (Glycine
decarboxylase subunit 2) (Glycine cleavage system
P-protein subunit 2)
Length = 486
Score = 31.6 bits (70), Expect = 5.4
Identities = 24/84 (28%), Positives = 38/84 (44%), Gaps = 10/84 (11%)
Query: 16 HLSFSSLHSHS---DDVVSSFNRLLEMYPTPCISKFNKNLTTLVKMKHYSTAISLYRQME 72
H +FS+ H V RL+E P P I KF K+ T ++ + S ++ R M
Sbjct: 268 HKTFSAPHGGGGPGSGPVGVSERLVEFLPVPVIEKFEKDGQTRYRL-NSSKPNTIGRMMN 326
Query: 73 FSRIMPDIFTFNILINCYCHIRQM 96
F F++L+ Y +IR +
Sbjct: 327 F------YGNFSVLVRAYTYIRML 344
>Y391_HUMAN (O15091) Hypothetical protein KIAA0391
Length = 567
Score = 31.2 bits (69), Expect = 7.1
Identities = 27/103 (26%), Positives = 46/103 (44%), Gaps = 3/103 (2%)
Query: 125 LCI-NNEVQKALHLHDQLVAQGVQLNNVSYGTLVNGLCKMGETRAALKMLRQIEGRLVQS 183
LC+ + + + + + + + A+ L Y L+ GL R AL +L I+ + S
Sbjct: 184 LCVFHMQTSEVIDVFEIMKARYKTLEPRGYSLLIRGLIHSDRWREALLLLEDIKKVITPS 243
Query: 184 ADVVMYNAVIDGLCKGKLVSDACDLYSEMVLRRISPDVYTYNA 226
YN I G + V+ A +LY E++ I P + T A
Sbjct: 244 KK--NYNDCIQGALLHQDVNTAWNLYQELLGHDIVPMLETLKA 284
>PE65_ARATH (Q9FJR1) Peroxidase 65 precursor (EC 1.11.1.7) (Atperox
P65) (ATP43)
Length = 334
Score = 31.2 bits (69), Expect = 7.1
Identities = 15/38 (39%), Positives = 21/38 (54%)
Query: 399 YNSLLHALCKSHHVDEAIALFEKVKDKGIQPDMYIYNV 436
+ + L LCK+H VD+ IA F V G +MY N+
Sbjct: 228 FAAALKDLCKNHTVDDTIAAFNDVMTPGKFDNMYFKNL 265
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.138 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 60,134,184
Number of Sequences: 164201
Number of extensions: 2491411
Number of successful extensions: 6024
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 5953
Number of HSP's gapped (non-prelim): 71
length of query: 538
length of database: 59,974,054
effective HSP length: 115
effective length of query: 423
effective length of database: 41,090,939
effective search space: 17381467197
effective search space used: 17381467197
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0106.10


