

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0103.12
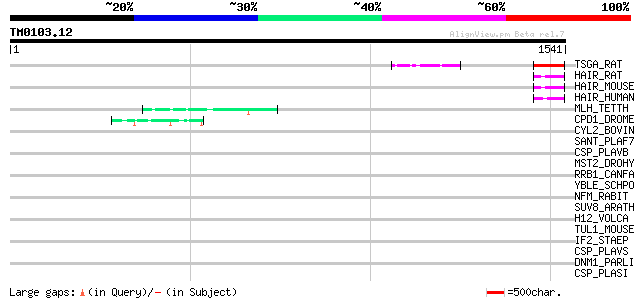
(1541 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TSGA_RAT (Q63679) Testis specific protein A (Zinc finger protein... 90 5e-17
HAIR_RAT (P97609) Hairless protein 55 1e-06
HAIR_MOUSE (Q61645) Hairless protein 55 2e-06
HAIR_HUMAN (O43593) Hairless protein 54 4e-06
MLH_TETTH (P40631) Micronuclear linker histone polyprotein (MIC ... 51 3e-05
CPD1_DROME (P22058) Chromosomal protein D1 47 4e-04
CYL2_BOVIN (Q28092) Cylicin II (Multiple-band polypeptide II) 46 0.001
SANT_PLAF7 (Q03400) S-antigen protein precursor 45 0.001
CSP_PLAVB (P08677) Circumsporozoite protein precursor (CS) 45 0.001
MST2_DROHY (Q08696) Axoneme-associated protein mst101(2) 44 0.003
RRB1_CANFA (Q28298) Ribosome-binding protein 1 (180 kDa ribosome... 44 0.003
YBLE_SCHPO (Q10342) Hypothetical protein C106.14c in chromosome II 44 0.004
NFM_RABIT (P54938) Neurofilament triplet M protein (160 kDa neur... 43 0.006
SUV8_ARATH (Q9C5P0) Histone-lysine N-methyltransferase, H3 lysin... 42 0.017
H12_VOLCA (Q08865) Histone H1-II 41 0.029
TUL1_MOUSE (Q9Z273) Tubby related protein 1 (Tubby-like protein 1) 40 0.038
IF2_STAEP (Q8CST4) Translation initiation factor IF-2 40 0.050
CSP_PLAVS (P13826) Circumsporozoite protein (CS) (Fragment) 40 0.050
DNM1_PARLI (Q27746) DNA (cytosine-5)-methyltransferase PliMCI (E... 40 0.065
CSP_PLASI (Q03110) Circumsporozoite protein precursor (CS) 40 0.065
>TSGA_RAT (Q63679) Testis specific protein A (Zinc finger protein
TSGA)
Length = 1214
Score = 89.7 bits (221), Expect = 5e-17
Identities = 61/197 (30%), Positives = 95/197 (47%), Gaps = 15/197 (7%)
Query: 1061 DNFLYCPDAVDMGDTEYEHFQRHWIRGEPVIVRNVFEKASGLSWHPMVMWRAFRGANKIL 1120
DN L C + + + F+ W +G+PV+V V K + W P + F G ++
Sbjct: 832 DNRLLCLQDPN-NKSNWNVFRECWKQGQPVMVSGVHHKLNTELWKPESFRKEF-GEQEVD 889
Query: 1121 KEEPTTFKAIDCLDWCEVQINIFQFFKGYLEG----RRYRNGWPEMLKLKDWPPSNSFEE 1176
T + I + F+ G+ + + + P +LKLKDWPP F +
Sbjct: 890 LVNCRTNEII-------TGATVGDFWDGFEDVPNRLKNEKEKEPMVLKLKDWPPGEDFRD 942
Query: 1177 CFPRHGAEFIAMLPFSDYTHPKFGILNLATKLPAV-LKPDLGPKTYIAYGSLEELSRGDS 1235
P + +A +P +YT + G LNLA++LP ++PDLGPK Y AYG + R
Sbjct: 943 MMPSRFDDLMANIPLPEYTR-RDGKLNLASRLPNYFVRPDLGPKMYNAYGLITPEDRKYG 1001
Query: 1236 VTKLHCDISDAVNILTH 1252
T LH D+SDA N++ +
Sbjct: 1002 TTNLHLDVSDAANVMVY 1018
Score = 78.2 bits (191), Expect = 2e-13
Identities = 39/86 (45%), Positives = 53/86 (61%), Gaps = 4/86 (4%)
Query: 1455 AVWDIFRRQDVPKLTEYLKKHHKEFRHANNLPVDSVTHPIHDQILYLNEKHKRQLKKEYG 1514
A+W I+ +D K+ E+LKK +E N P D PIHDQ YL+ +++L +EYG
Sbjct: 1058 ALWHIYAAKDTEKIREFLKKVSEEQGQEN--PADH--DPIHDQSWYLDRSLRKRLYQEYG 1113
Query: 1515 IEPWTFEQHLGEAVFIPAGCPHQVRN 1540
++ W Q LG+ VFIPAG PHQV N
Sbjct: 1114 VQGWAIVQFLGDVVFIPAGAPHQVHN 1139
>HAIR_RAT (P97609) Hairless protein
Length = 1181
Score = 55.1 bits (131), Expect = 1e-06
Identities = 31/87 (35%), Positives = 46/87 (52%), Gaps = 10/87 (11%)
Query: 1454 SAVWDIFRRQDVPKLTEYLKKHHKEFRHANNLPVDSVT-HPIHDQILYLNEKHKRQLKKE 1512
S VW +FR QD ++ +L+ P + T P YL+ +R+L++E
Sbjct: 1043 STVWHVFRAQDAQRIRRFLQMV---------CPAGAGTLEPGAPGSCYLDSGLRRRLREE 1093
Query: 1513 YGIEPWTFEQHLGEAVFIPAGCPHQVR 1539
+G+ WT Q GEAV +PAG PHQV+
Sbjct: 1094 WGVSCWTLLQAPGEAVLVPAGAPHQVQ 1120
>HAIR_MOUSE (Q61645) Hairless protein
Length = 1182
Score = 54.7 bits (130), Expect = 2e-06
Identities = 31/87 (35%), Positives = 46/87 (52%), Gaps = 10/87 (11%)
Query: 1454 SAVWDIFRRQDVPKLTEYLKKHHKEFRHANNLPVDSVT-HPIHDQILYLNEKHKRQLKKE 1512
S VW +FR QD ++ +L+ P + T P YL+ +R+L++E
Sbjct: 1044 STVWHVFRAQDAQRIRRFLQMV---------CPAGAGTLEPGAPGSCYLDAGLRRRLREE 1094
Query: 1513 YGIEPWTFEQHLGEAVFIPAGCPHQVR 1539
+G+ WT Q GEAV +PAG PHQV+
Sbjct: 1095 WGVSCWTLLQAPGEAVLVPAGAPHQVQ 1121
>HAIR_HUMAN (O43593) Hairless protein
Length = 1189
Score = 53.5 bits (127), Expect = 4e-06
Identities = 30/86 (34%), Positives = 44/86 (50%), Gaps = 8/86 (9%)
Query: 1454 SAVWDIFRRQDVPKLTEYLKKHHKEFRHANNLPVDSVTHPIHDQILYLNEKHKRQLKKEY 1513
S VW +FR QD ++ +L+ A P YL+ +R+L++E+
Sbjct: 1051 STVWHVFRAQDAQRIRRFLQMVCPAGAGA--------LEPGAPGSCYLDAGLRRRLREEW 1102
Query: 1514 GIEPWTFEQHLGEAVFIPAGCPHQVR 1539
G+ WT Q GEAV +PAG PHQV+
Sbjct: 1103 GVSCWTLLQAPGEAVLVPAGAPHQVQ 1128
>MLH_TETTH (P40631) Micronuclear linker histone polyprotein (MIC LH)
[Contains: Micronuclear linker histone-alpha;
Micronuclear linker histone-beta; Micronuclear linker
histone-delta; Micronuclear linker histone-gamma]
Length = 633
Score = 50.8 bits (120), Expect = 3e-05
Identities = 75/392 (19%), Positives = 139/392 (35%), Gaps = 42/392 (10%)
Query: 368 ETARAEKSGRPKVSKNKIRRVEHVGNVVAVKIVGPKKHGRPKSSKCRKKNIMEAGDEAAG 427
++ R+ G+ VSK + + + GR +SS ++ + D+ G
Sbjct: 184 KSRRSSSKGKSSVSKGRTKSTSSKRRADS-----SASQGRSQSSSSNRRKASSSKDQK-G 237
Query: 428 EIGGDKKLGRPKGSLNKLKNTVDCNNEGSGAGEIVRPKKRGRPRGSKNKVNNIMEVSKKA 487
+K KG N N S + + K + SKNK ++ + K +
Sbjct: 238 TRSSSRKASNSKGRKNSTSN----KRNSSSSSKRSSSSKNKKSSSSKNKKSSSSKGRKSS 293
Query: 488 ASGGDCKIAGPKKCGRPKGSKNKQKNIVQVSQEVAGSADCEIAGPKKCGRPKGSMKKRKS 547
+S G + A K + SK+++ + + + + S + GR S K RKS
Sbjct: 294 SSRG--RKASSSKNRKSSKSKDRKSSSSKGRKSSSSSKSNKRKASSSRGRKSSSSKGRKS 351
Query: 548 LVCASILEGAGGITREGLENKMLSNLCQEHIEYTQPVVRGGRPKGSRNKKIKLAFQDMVD 607
+ E K + +E R R++ K + ++ +
Sbjct: 352 --------------SKSQERKNSHADTSKQMEDEGQKRRQSSSSAKRDESSKKSRRNSMK 397
Query: 608 EVRFANKESDKATCAVGEEQKDHGSDIGKPIGLDNDKATLASDRDQETPNQTLAQ----- 662
E R + A+ A K G K G + K + R P AQ
Sbjct: 398 EARTKKANNKSASKASKSGSKSKGKSASKSKGKSSSKGKNSKSRSASKPKSNAAQNSNNT 457
Query: 663 ----DEVQNDKSSVKPK-RGRPKGSKNKMKSIANKARNKFGKVR---NMRGRPK---GSL 711
D +N S+ + + RGR + K+ + +N + GK N R + K S
Sbjct: 458 HQTADSSENASSTTQTRTRGRQREQKDMVNEKSNSKSSSKGKKNSKSNTRSKSKSKSASK 517
Query: 712 RKKNETAYCLDSQNERNSLDGRTSTEAAYRND 743
+KN + D+ N ++ +E+ +++
Sbjct: 518 SRKNASKSKKDTTNHGRQTRSKSRSESKSKSE 549
Score = 37.0 bits (84), Expect = 0.42
Identities = 61/314 (19%), Positives = 113/314 (35%), Gaps = 26/314 (8%)
Query: 465 KKRGRPRGSKNKVNNIMEVSKKAASGGDCKIAGPKKCGRPKGSKNKQKNIVQVSQEVAGS 524
KK+ + + S +K K + S G K K+ S+ + ++ ++ + S
Sbjct: 173 KKQTKRKNSTSKSRRSSSKGKSSVSKGRTKSTSSKRRADSSASQGRSQSSSSNRRKASSS 232
Query: 525 ADCEIAGPKKCGRPKGSMKKRKSLVCASILEGAGGITREGLENKMLSNLCQEHIEYTQ-- 582
D + G + R + K RK+ + +NK S+ + ++
Sbjct: 233 KDQK--GTRSSSRKASNSKGRKNSTSNKRNSSSSSKRSSSSKNKKSSSSKNKKSSSSKGR 290
Query: 583 --PVVRGGRPKGSRNKKIKLAFQDMVDEVRFANKESD-------KATCAVGEEQKDH-GS 632
RG + S+N+K + +D K S KA+ + G + G
Sbjct: 291 KSSSSRGRKASSSKNRKSSKS-KDRKSSSSKGRKSSSSSKSNKRKASSSRGRKSSSSKGR 349
Query: 633 DIGKPIGLDNDKATLASDRDQETPNQTLAQDEVQNDKSSVKPKRG-----RPKGSKNKMK 687
K N A + + E + + + D+SS K +R R K + NK
Sbjct: 350 KSSKSQERKNSHADTSKQMEDEGQKRRQSSSSAKRDESSKKSRRNSMKEARTKKANNKSA 409
Query: 688 SIANKARNKFGKVRNMRGRPKGSLRKKNETAYCLD------SQNERNSLDGRTSTEAAYR 741
S A+K+ +K + + K S + KN + +QN N+ S+E A
Sbjct: 410 SKASKSGSKSKGKSASKSKGKSSSKGKNSKSRSASKPKSNAAQNSNNTHQTADSSENASS 469
Query: 742 NDVDLHRGHCSQEE 755
RG +++
Sbjct: 470 TTQTRTRGRQREQK 483
Score = 35.4 bits (80), Expect = 1.2
Identities = 56/278 (20%), Positives = 98/278 (35%), Gaps = 35/278 (12%)
Query: 259 SNKEGPGSNYKMSALGFEEEIGLLLSRGGTTNEEARCEALRPLSKRGRP-KGSKNENDNK 317
SNK S+ K S+ ++ S + + + + S RGR SKN +K
Sbjct: 256 SNKRNSSSSSKRSSSSKNKK-----SSSSKNKKSSSSKGRKSSSSRGRKASSSKNRKSSK 310
Query: 318 ---QLSTALDGQSVGGDDNAG------TIGMSSVTDLGIEIAVLSGEKDKSSD---EVAD 365
+ S++ G+ + + G S + G + + K+ +D ++ D
Sbjct: 311 SKDRKSSSSKGRKSSSSSKSNKRKASSSRGRKSSSSKGRKSSKSQERKNSHADTSKQMED 370
Query: 366 LGETARAEKSGRPKVSKNKIRRVEHVGNVVAVKIVGPKKHGRPKSSKCRKKNIMEAGDEA 425
G+ R S + +K R ++K KK +SK K G A
Sbjct: 371 EGQKRRQSSSSAKRDESSKKSRRN------SMKEARTKKANNKSASKASKSGSKSKGKSA 424
Query: 426 AGEIGGDKKLGRPKGSLNKLK----------NTVDCNNEGSGAGEIVRPKKRGRPRGSKN 475
+ G G+ S + K NT + A + + RGR R K+
Sbjct: 425 SKSKGKSSSKGKNSKSRSASKPKSNAAQNSNNTHQTADSSENASSTTQTRTRGRQREQKD 484
Query: 476 KVNNIMEVSKKAASGGDCKIAGPKKCGRPKGSKNKQKN 513
VN SK ++ G + + + K + +KN
Sbjct: 485 MVNE-KSNSKSSSKGKKNSKSNTRSKSKSKSASKSRKN 521
>CPD1_DROME (P22058) Chromosomal protein D1
Length = 355
Score = 47.0 bits (110), Expect = 4e-04
Identities = 77/289 (26%), Positives = 104/289 (35%), Gaps = 71/289 (24%)
Query: 284 SRGGTTNEEARCEALRPLSKRGRPKGSKNENDNKQLSTALDGQSVGGDDNAGTIGMSSVT 343
S GG ++ A + KRGRP +K G S GG S +
Sbjct: 16 SVGGKSSTAAVAAISPGIKKRGRPAKNK-------------GSSGGGGQRGRPPKASKIQ 62
Query: 344 ------DLGIEIAVLSGEKDKSSDEVADLGETARAEKSGRPKVSKNKIRRVEHVGNVVAV 397
D G E G+ D S E+A+ + + GRPK S G+ +V
Sbjct: 63 NDEDPEDEGEE----DGDGDGSGAELANNSSPSPTKGRGRPKSSGGA-----GSGSGDSV 113
Query: 398 KIVGP---KKHGRPKS---SKCRKKNIMEAGDEAAGEIGGDKKLGRPK-GSLNK------ 444
K G +K GRPK S ++ + D+ I + +GRP GS+N
Sbjct: 114 KTPGSAKKRKAGRPKKHQPSDSENEDDQDEDDDGNSSIEERRPVGRPSAGSVNLNISRTG 173
Query: 445 --LKNTVDCNNEGSGAGEIVRPKKRGRPRGSKNKVNNIMEVSKKAASGGDCKIAGPKKCG 502
L E +G GE PKKRGRP K+ SGG G G
Sbjct: 174 RGLGRPKKRAVESNGDGEPQVPKKRGRP------------PQNKSGSGGS---TGYVPTG 218
Query: 503 RPKGSKNKQKNIVQVSQEVAGSADCEIAG-------------PKKCGRP 538
RP+G V+ ++ D E +G PKK GRP
Sbjct: 219 RPRGRPKANAAPVEKHEDNDDDQDDENSGEEEHSSPEKTVVAPKKRGRP 267
Score = 42.0 bits (97), Expect = 0.013
Identities = 64/255 (25%), Positives = 91/255 (35%), Gaps = 69/255 (27%)
Query: 249 ENPCFEGENDSNKEGPGSNYKMSALGFEEEIGLLLSRGGTTNEEARCEALRPLSKRGRPK 308
E+P EGE D + +G G+ E A + P RGRPK
Sbjct: 65 EDPEDEGEEDGDGDGSGA------------------------ELANNSSPSPTKGRGRPK 100
Query: 309 GSKNENDNKQLSTALDGQSVGGDDNAGTIGMSSVTDLGIEIAVLSGEKDKSSDEVADLGE 368
S G G D+ T G + G + + D+ D
Sbjct: 101 SS-------------GGAGSGSGDSVKTPGSAKKRKAGRPKKHQPSDSENEDDQDEDDDG 147
Query: 369 TARAEKS---GRPK---VSKNKIRRVEHVGNVV--AVKIVG------PKKHGRPKSSKCR 414
+ E+ GRP V+ N R +G AV+ G PKK GRP +K
Sbjct: 148 NSSIEERRPVGRPSAGSVNLNISRTGRGLGRPKKRAVESNGDGEPQVPKKRGRPPQNK-- 205
Query: 415 KKNIMEAGDEAAGEIGGDKKLGRPKGSLNKLK----NTVDCNNEGSGAGE-------IVR 463
+ G + + GRPK + ++ N D ++E SG E +V
Sbjct: 206 -----SGSGGSTGYVPTGRPRGRPKANAAPVEKHEDNDDDQDDENSGEEEHSSPEKTVVA 260
Query: 464 PKKRGRPRGSKNKVN 478
PKKRGRP + KV+
Sbjct: 261 PKKRGRPSLAAGKVS 275
>CYL2_BOVIN (Q28092) Cylicin II (Multiple-band polypeptide II)
Length = 488
Score = 45.8 bits (107), Expect = 0.001
Identities = 76/357 (21%), Positives = 125/357 (34%), Gaps = 39/357 (10%)
Query: 299 RPLSKRGRPKGSKNENDNKQLSTALDGQSVGGDDNA------GTIGMSSVTDLGIEIAVL 352
+P K+ K K+ K+ +T + + G + A G + +D G E
Sbjct: 154 KPDEKKDLKKERKDSKKGKESATESEDEKAGAEKGAKKDRKGSKKGKETPSDSGSEKG-- 211
Query: 353 SGEKDKSSDEVADLGETARAEKSGRP---KVSKNKIRRVEHVGNVVAVKIVGPKKHGRPK 409
+KD + G+ + E G K K ++ G A + G K +
Sbjct: 212 DAKKDSKKSKKDSKGKESATESEGEKGDAKKDDKKGKKGSKKGKESATESEGEKGDAKKD 271
Query: 410 SSKCRK--KNIMEAGDEAAGEIGGDKKLGRPKGSLNKLKNTVDCNNEGSGAGEIVRPKKR 467
K +K K E+ E+ GE GD K KG K G+ + K+
Sbjct: 272 DKKGKKGSKKGKESATESEGE-KGDAKKDDKKGKKGSKKGKESATESEGEKGDAKKDDKK 330
Query: 468 GRPRGSKNKVNNIMEVSKKAASGGDCKIAGPKKCGRPKGSKNKQKNIVQVSQEVAGSADC 527
G+ K K + +K + D K KG K +K S+ D
Sbjct: 331 GKKGSKKGKESATESEGEKGDAKKDDK----------KGKKGSKKGKESDSKAEGDKGDA 380
Query: 528 EIAGPKKCGRPKGSMKKRKSLVCASILEGAGGITREGLENKMLSNLCQEHIEYTQPVVRG 587
+ K KGS K ++S A+ EG +++ K + T+ +G
Sbjct: 381 KKDDKKD---KKGSKKGKES---ATESEGEKKDSKKDKAGKK---------DPTKAGEKG 425
Query: 588 GRPKGSRNKKIKLAFQDMVDEVRFANKESDKATCAVGEEQKDHGSDIGKPIGLDNDK 644
K ++ K K + ++ DE + ES+ A + +KD D K D K
Sbjct: 426 DESKDKKDAKKKDSKKEKKDEKKPGEAESEPKDSAKKDAKKDAKKDAKKDAKKDAKK 482
Score = 43.1 bits (100), Expect = 0.006
Identities = 86/374 (22%), Positives = 144/374 (37%), Gaps = 53/374 (14%)
Query: 368 ETARAEKSGRPKVSKNKIRR------VEHVGNVVAVKIVGPKKHGRPKSSKCRKKNIMEA 421
+ +A K PKV K+K + E + + + + K +P K KK ++
Sbjct: 109 DAKQAAKPSSPKVKKSKEDKDKSDSEAESIVSKEKPRKLSKAKEEKPDEKKDLKKERKDS 168
Query: 422 --GDEAAGEIGGDKKLGRPKGSLNKLKNTVDCNNEGSGAGEIVRPKKRGRPRGSKNKVNN 479
G E+A E D+K G KG+ K GS G+ P G +G K +
Sbjct: 169 KKGKESATE-SEDEKAGAEKGAKKDRK--------GSKKGKET-PSDSGSEKGDAKKDS- 217
Query: 480 IMEVSKKAASGGDCKIAGPKKCGRPKGSKNKQKNIVQVSQEVAGSADCEIAGPKKCGRPK 539
+ SKK + G + + G K K K + +E A ++ E KK + K
Sbjct: 218 --KKSKKDSKGKESATESEGEKGDAKKDDKKGKKGSKKGKESATESEGEKGDAKKDDK-K 274
Query: 540 GSMKKRKSLVCASILEGA-GGITREGLENKMLSNLCQEHIEYTQPVVRGGRPKGSRNKKI 598
G +K A+ EG G ++ + K S +E ++ + + KK
Sbjct: 275 GKKGSKKGKESATESEGEKGDAKKDDKKGKKGSKKGKESATESEGEKGDAKKDDKKGKK- 333
Query: 599 KLAFQDMVDEVRFANKESDKATCAVGEEQKDHGSDIGKPIGLDNDKATLASDRDQETPNQ 658
+ K + AT + GE+ D K D+ K S + +E+ ++
Sbjct: 334 ------------GSKKGKESATESEGEK-----GDAKK----DDKKGKKGSKKGKESDSK 372
Query: 659 TLAQ--DEVQNDKSSVKPKRGRPKGSKNKMKSIANK---ARNKFGKVRNMRGRPKGSLRK 713
D ++DK K K+G KG ++ +S K ++K GK + KG K
Sbjct: 373 AEGDKGDAKKDDK---KDKKGSKKGKESATESEGEKKDSKKDKAGKKDPTKAGEKGDESK 429
Query: 714 KNETAYCLDSQNER 727
+ A DS+ E+
Sbjct: 430 DKKDAKKKDSKKEK 443
Score = 41.6 bits (96), Expect = 0.017
Identities = 82/375 (21%), Positives = 136/375 (35%), Gaps = 53/375 (14%)
Query: 352 LSGEKDKSSDEVADLGETARAEKSGRP--------KVSKNKIRRVEHVGNVVAVKIV--- 400
LS K++ DE DL + + K G+ K K + + G+ +
Sbjct: 147 LSKAKEEKPDEKKDLKKERKDSKKGKESATESEDEKAGAEKGAKKDRKGSKKGKETPSDS 206
Query: 401 GPKKHGRPKSSKCRKKNIM--EAGDEAAGEIGGDKKLGRPKGSLNKLKNTVDCNNEGSGA 458
G +K K SK KK+ E+ E+ GE GD K KG K
Sbjct: 207 GSEKGDAKKDSKKSKKDSKGKESATESEGE-KGDAKKDDKKGKKGSKKGKESATESEGEK 265
Query: 459 GEIVRPKKRGRPRGSKNKVNNIMEVSKKAASGGDCKIAGPKKCGRPKGSKNKQKNIVQVS 518
G+ + K+G+ K K + +K + D K KG K +K
Sbjct: 266 GDAKKDDKKGKKGSKKGKESATESEGEKGDAKKDDK----------KGKKGSKKG----- 310
Query: 519 QEVAGSADCEIAGPKKCGRPKGSMKKRKSLVCASILEGA-GGITREGLENKMLSNLCQEH 577
+E A ++ E KK + KG +K A+ EG G ++ + K S +E
Sbjct: 311 KESATESEGEKGDAKKDDK-KGKKGSKKGKESATESEGEKGDAKKDDKKGKKGSKKGKES 369
Query: 578 IEYTQPVVRGGRPKGSRNKKIKLAFQDMVDEVRFANKESDKATCAVGEEQKDHGSDIGKP 637
+ +G + A +D + + + K + AT + GE++ GK
Sbjct: 370 ---------DSKAEGDKGD----AKKDDKKDKKGSKKGKESATESEGEKKDSKKDKAGKK 416
Query: 638 IGLDNDKATLASDRDQETPN------QTLAQDEVQNDKSSVKPKRGRPKGSKNKMKSIAN 691
D KA D ++ + + +DE + ++ +PK K +K K A
Sbjct: 417 ---DPTKAGEKGDESKDKKDAKKKDSKKEKKDEKKPGEAESEPKDSAKKDAKKDAKKDAK 473
Query: 692 KARNKFGKVRNMRGR 706
K K K +G+
Sbjct: 474 KDAKKDAKKDAKKGK 488
>SANT_PLAF7 (Q03400) S-antigen protein precursor
Length = 593
Score = 45.4 bits (106), Expect = 0.001
Identities = 96/477 (20%), Positives = 183/477 (38%), Gaps = 36/477 (7%)
Query: 226 VTGFEGLSGENTHGFRDEVGGFVENPCFEGENDSNKEGPGSNYKMSALGFEEEIGLLLSR 285
+ G EG+ E THG DEV E+ G D G + G E+++ S
Sbjct: 88 IKGQEGVEQE-THGTEDEVSNGREDKVSNGGEDEVSNGGEDEV---SNGREDKV----SN 139
Query: 286 GGTTNEEARCEALRPLSKRGRPKGSKNENDNKQLSTALDGQSVGGDDNAGTIGMSSVTDL 345
GG E+ G N ++K + D S G +D G V++
Sbjct: 140 GG---EDEVSNGREDKVSNGGEDEVSNGREDKVSNGGEDEVSNGREDKVSNGGEDEVSN- 195
Query: 346 GIEIAVLSGEKDKSSDEVADLGETARAEKSGRPKVSKNKIRRVEHVGNVVAVKIVGPKKH 405
G E V +G +DK S+ D R ++ + K + V N K+
Sbjct: 196 GREDEVSNGREDKVSNGGEDEVSNGREDEVSNGREDKVSNGGEDEVSNGREDKV---SNG 252
Query: 406 GRPKSSKCRKKNIMEAGDEAAGEIGGDKKLGRPKGSLNKLKNTVDCNNEGSGAGEIVRPK 465
G + S R+ + G++ E+ ++ G +++ N + G E+ +
Sbjct: 253 GEDEVSNGREDKVSNGGED---EVSNGREDEVSNGREDEVSNGREDKVSNGGEDEVSNGR 309
Query: 466 KRGRPRGSKNKVNNIM--EVSK----KAASGGDCKIAGPKKCGRPKGSKNKQKNIVQVSQ 519
+ G +++V+N EVS K ++GG+ +++ ++ G +++ N
Sbjct: 310 EDKVSNGGEDEVSNGREDEVSNGREDKVSNGGEDEVSNGREDKVSNGGEDEVSN--GRED 367
Query: 520 EVAGSADCEIAGPKKCGRPKGSMKKRKSLVCASILEGAGGITREGLENKMLSNLCQEHIE 579
EV+ + E++ ++ G K + + G G E+K +SN ++ +
Sbjct: 368 EVSNGREDEVSNGREDEVSNGREDKVSNGGEDEVSNGREDEVSNGREDK-VSNGGEDEVS 426
Query: 580 --YTQPVVRGGRPKGSRNKKIKLAFQDMVDEVRFANKESDKATCAVGEEQKDHGSDIGKP 637
V GG + S ++ +++ DEV +N DK + GE++ +G +
Sbjct: 427 NGREDKVSNGGEDEVSNGREDEVS-NGREDEV--SNGREDKVSNG-GEDEVSNGGEDEVS 482
Query: 638 IGLDNDKATLASD---RDQETPNQTLAQDEVQNDKSSVKPKRGRPKGSKNKMKSIAN 691
G ++ + D +E +DEV N + G + S + ++N
Sbjct: 483 NGREDKVSNGGEDEVSNGREDKVSNGGEDEVSNGREDKVSNGGEDEVSNGREDEVSN 539
>CSP_PLAVB (P08677) Circumsporozoite protein precursor (CS)
Length = 378
Score = 45.4 bits (106), Expect = 0.001
Identities = 55/231 (23%), Positives = 97/231 (41%), Gaps = 21/231 (9%)
Query: 316 NKQLSTALDGQSVGGDD-NAGTIGMSSVTDLGIEIAVLSGEKDKSSDEVADLGETARAEK 374
N LS A++ V ++ +A ++G + V G + G + DE D A+ +K
Sbjct: 25 NVDLSKAINLNGVNFNNVDASSLGAAHV---GQSASRGRGLGENPDDEEGD----AKKKK 77
Query: 375 SGRPKVSKN-KIRRVEHVGNVVAVKIVGPKKHGRPKSSKCRKKNIMEAGDEAAGEIGGDK 433
G+ KN + +++ G+ + G + G+P + + AGD AAG+ GD+
Sbjct: 78 DGKKAEPKNPRENKLKQPGDRADGQPAGDRADGQPAGDRADGQ---PAGDRAAGQPAGDR 134
Query: 434 KLGRPKGSLNKLKNTVDCNNEGSGAGEIVRPKKRGRPRGSK--NKVNNIMEVSKKAASGG 491
G+P G + D +G AG+ + G+P G + + + A
Sbjct: 135 ADGQPAGDRADGQPAGD-RADGQPAGD----RADGQPAGDRAAGQPAGDRAAGQPAGDRA 189
Query: 492 DCKIAGPKKCGRPKGSK--NKQKNIVQVSQEVAGSADCEIAGPKKCGRPKG 540
D + AG + G+P G + + Q AD + AG + G+P G
Sbjct: 190 DGQPAGDRAAGQPAGDRADGQPAGDRAAGQPAGDRADGQPAGDRAAGQPAG 240
>MST2_DROHY (Q08696) Axoneme-associated protein mst101(2)
Length = 1391
Score = 44.3 bits (103), Expect = 0.003
Identities = 80/432 (18%), Positives = 157/432 (35%), Gaps = 40/432 (9%)
Query: 353 SGEKDKSSDEVADLGETARA-EKSGRPKVSKNKIRRVEHVGNVVAVKIVGPKKHGRPKSS 411
+ +K+K + E GE A+ +K+ K K ++ + + K+ +
Sbjct: 626 AAKKEKEAAEREKCGELAKKIKKAAEKKKCKKLAKKEKETAEKKKCEKAAKKRKEAAEKK 685
Query: 412 KCR---KKNIMEAGDEAAGEIGGDKKLGRPKGSLNKLKNTVDCNNEGSGAGEIVRPKKRG 468
KC KK A + E +K + +L + E ++ + KK G
Sbjct: 686 KCAEAAKKEKEAAEKKKCEEAAKKEKEAAERKKCEELAKKIKKAAEKKKCKKLAKKKKAG 745
Query: 469 RPRGSKN---KVNNIMEVSKKAASGGDCKIAGPKKCGRPKGSKNKQKNIVQVSQ-EVAGS 524
K K ++ KK K A KKC K + K+K + + E
Sbjct: 746 EKNKLKKGNKKGKKALKEKKKCRELAKKKAAEKKKC---KEAAKKEKEAAEKKKCEKTAK 802
Query: 525 ADCEIAGPKKCGRPKGSMKKRKSLVCASILEGAGGITREGLENKMLSNLCQEHIEYTQPV 584
E A KKC + + KKRK E A +E E K C++ + +
Sbjct: 803 KRKEEAEKKKCEK---TAKKRKEAAEKKKCEKAAKKRKEEAEKKK----CEKTAKKRKET 855
Query: 585 VRGGRPKGSRNKKIKLAFQDMVDEVRFANKES-DKATCAVGEEQKDHGSDIGKPIGLDND 643
+ + + K+ + A + ++ KE+ +K CA +++ ++
Sbjct: 856 AEKKKCEKAAKKRKQAAEKKKCEKAAKKRKEAAEKKKCAEAAKKEKELAE--------KK 907
Query: 644 KATLASDRDQETPNQTLAQDEVQNDKSSVKPKR-------GRPKGSKNKMKSIANKARNK 696
K A+ +++E + ++ + K + + K+ + G KNK+K A K + K
Sbjct: 908 KCEEAAKKEKEVAERKKCEELAKKIKKAAEKKKCKKLAKKEKKAGEKNKLKKKAGKGKKK 967
Query: 697 FGKVRNMRGRPKGSLRKKNETAYCLDSQNERNSLDGRTSTEAAYRNDVDLHRGHCSQEEL 756
K+ G+ +K + A ++ E+ + + E A + + C +
Sbjct: 968 CKKL----GKKSKRAAEKKKCAEA--AKKEKEAATKKKCEERAKKQKEAAEKKQCEERAK 1021
Query: 757 LRMLSVEHKNIQ 768
+ E K +
Sbjct: 1022 KLKEAAEQKQCE 1033
>RRB1_CANFA (Q28298) Ribosome-binding protein 1 (180 kDa ribosome
receptor) (RRp)
Length = 1534
Score = 43.9 bits (102), Expect = 0.003
Identities = 87/395 (22%), Positives = 152/395 (38%), Gaps = 25/395 (6%)
Query: 355 EKDKSSDEVADLGETARAEKSGRPKVSKNKIRRVEHVGNVVAVKIVGP--KKHGRPKSSK 412
+K K +VA + E A + +V +K +E V + +V P K G P +S
Sbjct: 138 DKKKKEKKVAKV-EPAVSSVVNSVQVLASKAAILETAPKEVPMVVVPPVGAKAGTPATST 196
Query: 413 CRKKNIMEAGDEAAGEIGGDKKLGRPKGSLNKLKNTVDCNNEGSGAGEIVRPKKRGRPRG 472
+ K A +++ G + + +G+LN+ K N+G V P + + G
Sbjct: 197 AQGKKAEGAQNQSRKAEGAPNQGKKAEGALNQGKKAEGAQNQGKKVE--VAPNQGKKAEG 254
Query: 473 SKN---KVNNIMEVSKKA---ASGGDCKIAGPKKCGRPKGSKNKQKNIVQVSQEVAGSAD 526
+N KV KKA + G P + + G+ N+ K + +
Sbjct: 255 GQNQGKKVEGAQNQGKKAEGTPNQGKKAEGAPNQGKKTDGAPNQGKKSEGAPNQGKKAEG 314
Query: 527 CEIAGPKKCGRPKGSMKKRKSLVCASILEGA--GGITREGLEN--KMLSNLCQEHIEYTQ 582
+ G K P K +EGA G EG N K + +
Sbjct: 315 AQNQGKKVEVAPNQGKKAEGGQNQGKKVEGAQNQGKKAEGTPNQGKKAEGAPNQGKKTDG 374
Query: 583 PVVRGGRPKGSRN--KKIKLAFQDMVDEVRFANKESDKATCAVGEEQKDHGSDIGKPIGL 640
+G + +G+ N KK++ A Q+ +V + KA A + +K G+ G
Sbjct: 375 APNQGKKSEGAPNQGKKVEGA-QNQGKKVEGVQNQGKKAEGAQNQGKKAEGT---SSQGR 430
Query: 641 DNDKATLASDRDQETPNQTLAQDEVQNDKSSVK--PKRG-RPKGSKNKMKSIANKARNKF 697
+ + + +PNQ + VQN V+ P +G + +GS+N+ K A N+
Sbjct: 431 KEEGTPNLGKKAEGSPNQGKKVEVVQNQSKKVEGAPNQGKKAEGSQNQGKK-TEGASNQG 489
Query: 698 GKVRNMRGRPKGSLRKKNETAYCLDSQNERNSLDG 732
KV + + K + N+ +QN+ +G
Sbjct: 490 KKVDGAQNQGKKAEGAPNQGKKVEGAQNQGKKAEG 524
>YBLE_SCHPO (Q10342) Hypothetical protein C106.14c in chromosome II
Length = 719
Score = 43.5 bits (101), Expect = 0.004
Identities = 61/268 (22%), Positives = 112/268 (41%), Gaps = 44/268 (16%)
Query: 283 LSRGGTTNEEARCEALRPLSKRGRPKGSKNEN----DNKQLSTALDGQSVGGDDNAGTIG 338
L G N + L L++ G + EN DN ++S DGQ+ DD+ G I
Sbjct: 490 LKYGEELNVTHGIQGLELLAQYKAEHGEEGENGDDWDNWEVSE--DGQN--SDDSGGWID 545
Query: 339 MSSVTDLGIEIAVLSGEKDKSSDEVADLGETARAEKSGRPKVSKNKIRRVEHVGNVVAVK 398
+ S D IE++ E++K+ TAR E + S+ ++ V+ + + +
Sbjct: 546 VDS--DDNIELSDSDEEEEKA---------TARKESDEKGSSSQKEL--VDRMTELASQS 592
Query: 399 IVGPKKHGRPKSSKCRKKNIMEAGDEAAGEIGGDKKLGRPKGSLNKLKNTVDCNNEGSGA 458
I+ P N ++ +E + G D+ + PK SL + + V+ A
Sbjct: 593 ILTP--------------NDLKKLEELREQAGVDRLVNGPKRSLKRPDDAVE-------A 631
Query: 459 GEIVRPKKRGRPRGSKNKVNNIMEVSKKAASGGDCKIAGPKKCGRPKGSKNKQKNIVQVS 518
EI P+K+ + + ++ ++ME ++ K AG K + KN + +
Sbjct: 632 DEIEGPRKKAK-NDREARIASVME-GREGRDKFSSKKAGFNPTSLSNKRKQRNKNFMMIK 689
Query: 519 QEVAGSADCEIAGPKKCGRPKGSMKKRK 546
++ G A + +K R + +KRK
Sbjct: 690 HKLKGKAGRSLVQKQKVLREHVAREKRK 717
>NFM_RABIT (P54938) Neurofilament triplet M protein (160 kDa
neurofilament protein) (Neurofilament medium
polypeptide) (NF-M) (Fragment)
Length = 644
Score = 43.1 bits (100), Expect = 0.006
Identities = 63/269 (23%), Positives = 107/269 (39%), Gaps = 26/269 (9%)
Query: 256 ENDSNKEGPGSNYKMSALGFEEEIGLLLSRGGTTNEEARCEALRPLSKRGRPKGSKNEND 315
E + +E G + G E+ G + G EE EA + + + K E
Sbjct: 320 EEEEEEEDEGVKSDQAEEGGSEKEGSSKNEG--EQEEGETEAEGEVEEAEAKEEKKTEEK 377
Query: 316 NKQLSTA---LDGQSVGGDDNAGT-IGMSSVTDLG--IEIAVLSGEKDKSSDEVADLGET 369
+++++ + VG + A + + S V ++ E GE+ + ++V + +
Sbjct: 378 SEEVAAKEEPVTEAKVGKPEKAKSPVPKSPVEEVKPKAEATAGKGEQKEEEEKVEEEKKK 437
Query: 370 ARAEKSGRPKVSKNKIRR-----------VEHVGNVVAVKIVGPKKHGRPKSSKCRKKNI 418
A E KV K + + V+ A I P K G K +K +K +
Sbjct: 438 AAKESPKEEKVEKKEEKPKDVPKKKAESPVKEEAAEEAATITKPTKVGLEKETKEGEKPL 497
Query: 419 MEAGD-EAAGEIGGDKKLGRPKGSLNKLKNTVDCNNEGSG-AGEIVRPKKRGRPR-GSKN 475
+ + E AGE GG ++ G +GS K + N EG G E K++G R K
Sbjct: 498 QQEKEKEKAGEEGGSEEEGSDQGSKRAKKEDIAVNGEGEGKEEEEPETKEKGSGREEEKG 557
Query: 476 KVNNIMEVS----KKAASGGDCKIAGPKK 500
V N +++S KK + K+ KK
Sbjct: 558 VVTNGLDLSPADEKKGGDRSEEKVVVTKK 586
>SUV8_ARATH (Q9C5P0) Histone-lysine N-methyltransferase, H3 lysine-9
specific 8 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 8) (H3-K9-HMTase 8) (Suppressor of
variegation 3-9 homolog 8) (Su(var)3-9 homolog 8)
Length = 755
Score = 41.6 bits (96), Expect = 0.017
Identities = 25/64 (39%), Positives = 33/64 (51%), Gaps = 7/64 (10%)
Query: 456 SGAGEIVRPKKRGR--PRGSKN-----KVNNIMEVSKKAASGGDCKIAGPKKCGRPKGSK 508
+ AG P KRGR P+GSKN K + + AS G G ++CGRPKG K
Sbjct: 164 ASAGPSTGPGKRGRGRPKGSKNGSRKPKKPKAYDNNSTDASAGPSSGLGKRRCGRPKGLK 223
Query: 509 NKQK 512
N+ +
Sbjct: 224 NRSR 227
Score = 34.7 bits (78), Expect = 2.1
Identities = 21/58 (36%), Positives = 26/58 (44%), Gaps = 5/58 (8%)
Query: 488 ASGGDCKIAGPKKCGRPKGSKNKQKNIVQVSQEVAGSADCEIA-----GPKKCGRPKG 540
AS G G + GRPKGSKN + + S D G ++CGRPKG
Sbjct: 164 ASAGPSTGPGKRGRGRPKGSKNGSRKPKKPKAYDNNSTDASAGPSSGLGKRRCGRPKG 221
>H12_VOLCA (Q08865) Histone H1-II
Length = 240
Score = 40.8 bits (94), Expect = 0.029
Identities = 48/207 (23%), Positives = 79/207 (37%), Gaps = 45/207 (21%)
Query: 345 LGIEIAVLSGE--KDKSSDEVADLGETARAEKSGRPKVSKNKIRRVEHVGNVVAVKIVGP 402
L ++ V +G+ K K+S +++D + ++A+ + +PK + K P
Sbjct: 72 LALKTFVKNGKLVKVKNSYKLSD-AQKSKAKAAAKPKAAPKKA--------------AAP 116
Query: 403 KKHGRPKSSKCRKKNIMEAGDEAAGEIGGDKKLGRPKGSLNKLKNTVDCNNEGSGAGEIV 462
KK PK +K KK G++ A + +KK +PK
Sbjct: 117 KKAAAPKKAKAPKKE----GEKKAVKPKSEKKAAKPKTE--------------------K 152
Query: 463 RPKKRGRPRGSKNKVNNIMEVSKKAA-SGGDCKIAGPKKCGRPKGSK---NKQKNIVQVS 518
+PK +P+ +K K AA K A PKK PK +K K+
Sbjct: 153 KPKAAKKPKAAKKPAAKKPAAKKPAAKKATPKKAAAPKKAAAPKKAKAATPKKAKAATPK 212
Query: 519 QEVAGSADCEIAGPKKCGRPKGSMKKR 545
+ A + A PK +PK K+
Sbjct: 213 KAKAAAKPKAAAKPKAAAKPKAKAAKK 239
>TUL1_MOUSE (Q9Z273) Tubby related protein 1 (Tubby-like protein 1)
Length = 543
Score = 40.4 bits (93), Expect = 0.038
Identities = 38/149 (25%), Positives = 61/149 (40%), Gaps = 11/149 (7%)
Query: 403 KKHGRPKSSKCRKKNIMEAGDEAAGEIGGDKKLGRPKGSLNKLKNTVDCNNEGSGAGEIV 462
KK G+ + S K + ++ A +G +G P L+ EG+ ++
Sbjct: 133 KKEGKKEKSSLPPKKAPKEREKKAKALGPRGDVGSPDAPRKPLRTKKKEVGEGT---KLR 189
Query: 463 RPKKRGR------PRGSKNKVNNIMEVSKKAASGGDCKIAGPKKCGRPKGSKNKQKNIVQ 516
+ KK+G P GS + + G G KK G PKGS+ ++K +
Sbjct: 190 KAKKKGPGETDKDPAGSPAALRKEFPAAMFLVGEGGAAEKGVKKKGPPKGSEEEKKEEEE 249
Query: 517 VSQEVAGSADCEIAGPKKCGRPKGSMKKR 545
+E SA + + K GR KG KK+
Sbjct: 250 EVEEEVASAVMKNSNQK--GRAKGKGKKK 276
>IF2_STAEP (Q8CST4) Translation initiation factor IF-2
Length = 720
Score = 40.0 bits (92), Expect = 0.050
Identities = 28/126 (22%), Positives = 54/126 (42%), Gaps = 5/126 (3%)
Query: 596 KKIKLAFQDMVDEVRFANKESDKATCAVGEEQ-KDHGSDIGKPIGLDNDKATLASDRDQE 654
K++ L ++++DE++ N E A+ EEQ K D +K ++ +
Sbjct: 11 KELNLKSKEIIDELKSMNVEVSNHMQALEEEQIKALDKKFKASQAKDTNKQNTQNNHQKS 70
Query: 655 TPNQTLAQDEVQNDKSSVKPKRGRPKGSKNKMKSIANKA----RNKFGKVRNMRGRPKGS 710
Q E Q K++ KP + + + +K K ++ NK +N K N +P+
Sbjct: 71 NNKQNSNDKEKQQSKNNSKPTKKKEQNNKGKQQNKNNKTNKNQKNNKNKKNNKNNKPQNE 130
Query: 711 LRKKNE 716
+ + E
Sbjct: 131 VEETKE 136
>CSP_PLAVS (P13826) Circumsporozoite protein (CS) (Fragment)
Length = 343
Score = 40.0 bits (92), Expect = 0.050
Identities = 66/280 (23%), Positives = 103/280 (36%), Gaps = 48/280 (17%)
Query: 266 SNYKMSALGFEEEIGLLLSRG---GTTNEEARCEALRPLS-KRGRPKGSKNENDNKQLST 321
+N S+LG +G SRG G ++ +A + K+ PK + EN KQ
Sbjct: 6 NNVDASSLG-AAHVGQSASRGRGLGENPDDEEGDAKKKKDGKKAEPKNPR-ENKLKQPGD 63
Query: 322 ALDGQSVGGDDNAGTIGMSSVTDLGIEIAVLSGEKDKSSDEVADLGETARAEKSGRPKVS 381
DGQ G D A G + + A + + D A G+ A G+P
Sbjct: 64 RADGQPAG--DRAD--GQPAGDRADGQPAGDRADGQPAGDRAA--GQPAGDRADGQP--- 114
Query: 382 KNKIRRVEHVGNVVAVKIVGPKKHGRPKSSKCRKKNIMEAGDEAAGEIGGDKKLGRPKGS 441
G+ + G + G+P + + AGD AAG+ GD+ G+P G
Sbjct: 115 ---------AGDRADGQPAGDRADGQPAGDRADGQ---PAGDRAAGQPAGDRAAGQPAG- 161
Query: 442 LNKLKNTVDCNNEGS-GAGEIVRPKKRGRPRGSKNKVNNIMEVSKKAASGGDCKIAGPKK 500
+ D G AG+ + G+P G + + A D + AG +
Sbjct: 162 -----DRADGQPAGDRAAGQPAGDRADGQPAGDR-------AAGQPAGDRADGQPAGDRA 209
Query: 501 CGRPKGSKNKQKNIVQVSQEVAGSADCEIAGPKKCGRPKG 540
G+P G + Q A + AG + G+ G
Sbjct: 210 AGQPAGDR-------AAGQPAGDRAAGQAAGDRAAGQAAG 242
>DNM1_PARLI (Q27746) DNA (cytosine-5)-methyltransferase PliMCI (EC
2.1.1.37) (Dnmt1) (DNA methyltransferase PliMCI) (DNA
MTase PliMCI) (MCMT) (M.PliMCI)
Length = 1612
Score = 39.7 bits (91), Expect = 0.065
Identities = 62/259 (23%), Positives = 104/259 (39%), Gaps = 19/259 (7%)
Query: 296 EALRPLSKRGRPKGSKN-ENDNKQLSTALDGQSVGGDDNAGTIGMSSVTDLGIEIAVLSG 354
E P++ + + S N ++D K ST +G + G G+ G S V S
Sbjct: 86 ETCSPVNGDTKEEASSNGKDDEKAESTVANGTTSNGSTTNGSSGSSKANGHTNGGYVQSS 145
Query: 355 EKDKS----SDEVADLGETARAEKSGRPKVSKNKIRRVEHVGNVVAVKIVGPKKHGRPKS 410
++++ S+E D+ +T + K G K K+K G K++G + +
Sbjct: 146 SQEETGTSQSEEEMDM-DTPTSGKGGSKKKKKSKGSGGGDAGKGRKRKVLGDDERDGVEK 204
Query: 411 SKCRKKNIM-EAGDEAAGEIGG-DKKLGRPKGSLNKLKNTVDCNNEGSGAGEIVRPKKRG 468
+ KK++ E G+EA E D+K R S K D + +P K+
Sbjct: 205 KEGEKKDVEGEEGEEAKEESATPDEKTLRT--SKRKRSPKADAKQPSIMSMFTKKPAKKE 262
Query: 469 RPRGSKNKVNNIMEVSKKAASGGDCKIAGPKKCGRPKGSKNKQKNIVQVSQEVAGSADCE 528
K + ++ MEV KK GD G K+ P G K+ ++ +E A E
Sbjct: 263 E---EKMEESSSMEVDKKEMENGD---NGKKEEEEPSGPGGKR---IKKEEEEEEKAKVE 313
Query: 529 IAGPKKCGRPKGSMKKRKS 547
P + R K + + +S
Sbjct: 314 PMSPSRDLRHKANHETAES 332
>CSP_PLASI (Q03110) Circumsporozoite protein precursor (CS)
Length = 386
Score = 39.7 bits (91), Expect = 0.065
Identities = 64/279 (22%), Positives = 103/279 (35%), Gaps = 55/279 (19%)
Query: 266 SNYKMSALGFEEEIGLLLSRG---GTTNEEARCEALRPLS-KRGRPKGSKNENDNKQLST 321
+N S+LG +G SRG G ++ +A + K+ PK + EN KQ
Sbjct: 40 NNVDASSLG-AAHVGQSASRGRGLGENPDDEEGDAKKKKDGKKAEPKNPR-ENKLKQPGD 97
Query: 322 ALDGQSVGGDDNAGTIGMSSVTDLGIEIAVLSGEKDKSSDEVADLGETARAEKSGRPKVS 381
DGQ G + G + + + + AD G+ A G+P
Sbjct: 98 RADGQPAGDRADGQPAGDRA--------------DGQPAGDRAD-GQPAGDRADGQP--- 139
Query: 382 KNKIRRVEHVGNVVAVKIVGPKKHGRPKSSKCRKKNIMEAGDEAAGEIGGDKKLGRPKGS 441
G+ A + G + G+P + + AGD AAG+ GD+ G+P G
Sbjct: 140 ---------AGDRAAGQPAGDRADGQPAGDRADGQ---PAGDRAAGQPAGDRAAGQPAGD 187
Query: 442 LNKLKNTVDCNNEGSGAGEIVRPKKRGRPRGSKNKVNNIMEVSKKAASGGDCKIAGPKKC 501
+ D +G AG+ + G+P G + + A + AG +
Sbjct: 188 RADGQPAGD-RADGQPAGD----RADGQPAGDR-------AAGQPAGDRAAGQPAGDRAA 235
Query: 502 GRPKGSKNKQKNIVQVSQEVAGSADCEIAGPKKCGRPKG 540
G+P G + Q A + AG + G+P G
Sbjct: 236 GQPAGDR-------AAGQPAGDRAAGQPAGDRAAGQPAG 267
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.135 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 196,617,849
Number of Sequences: 164201
Number of extensions: 9296712
Number of successful extensions: 17267
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 66
Number of HSP's that attempted gapping in prelim test: 17020
Number of HSP's gapped (non-prelim): 224
length of query: 1541
length of database: 59,974,054
effective HSP length: 123
effective length of query: 1418
effective length of database: 39,777,331
effective search space: 56404255358
effective search space used: 56404255358
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0103.12


