

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
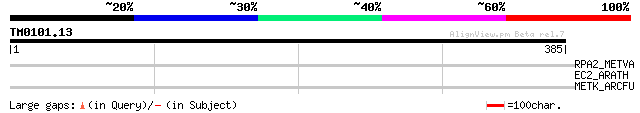
Query= TM0101.13
(385 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
RPA2_METVA (P14247) DNA-directed RNA polymerase subunit A'' (EC ... 31 4.6
EC2_ARATH (Q42377) EC protein homolog 2 31 4.6
METK_ARCFU (O30186) S-adenosylmethionine synthetase (EC 2.5.1.6)... 31 6.1
>RPA2_METVA (P14247) DNA-directed RNA polymerase subunit A'' (EC
2.7.7.6)
Length = 386
Score = 31.2 bits (69), Expect = 4.6
Identities = 18/55 (32%), Positives = 30/55 (53%), Gaps = 2/55 (3%)
Query: 122 DVNNSTSPEFRKVFVRG--KCVNFSPAIINQTLERSVVEFVEEELSLDTVAEELT 174
D S +++RG K S AI+++ +E + VE V E++S+D V E +T
Sbjct: 102 DARKEPSTPTMSIYLRGDYKYDRESAAIVSKNIESTTVESVSEDISVDLVNECIT 156
>EC2_ARATH (Q42377) EC protein homolog 2
Length = 85
Score = 31.2 bits (69), Expect = 4.6
Identities = 17/60 (28%), Positives = 24/60 (39%), Gaps = 2/60 (3%)
Query: 307 DICSCLYVCLRNSQCHC*T*GHFQGTSRDHQDCCC*ETKG--GCSHPNSQGGSWSRG*AC 364
D C C C C C G ++H C C E G C+ P +Q + ++G C
Sbjct: 15 DRCGCPSPCPGGESCRCKMMSEASGGDQEHNTCPCGEHCGCNPCNCPKTQTQTSAKGCTC 74
>METK_ARCFU (O30186) S-adenosylmethionine synthetase (EC 2.5.1.6)
(Methionine adenosyltransferase) (AdoMet synthetase)
Length = 399
Score = 30.8 bits (68), Expect = 6.1
Identities = 18/78 (23%), Positives = 36/78 (46%)
Query: 98 RKTVMNIGKCYDKLVKEFIVNLSEDVNNSTSPEFRKVFVRGKCVNFSPAIINQTLERSVV 157
+ + ++GK Y+ L + ++E+V R + GK +N A+ Q + +S
Sbjct: 305 KNPINHVGKIYNLLANQIAARIAEEVEGVEEVYVRILSQIGKPINEPKALSVQVIPKSGY 364
Query: 158 EFVEEELSLDTVAEELTA 175
+ + E +AEE+ A
Sbjct: 365 DISKLERPARDIAEEMIA 382
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.339 0.147 0.497
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 41,182,325
Number of Sequences: 164201
Number of extensions: 1648014
Number of successful extensions: 5929
Number of sequences better than 10.0: 3
Number of HSP's better than 10.0 without gapping: 1
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 5928
Number of HSP's gapped (non-prelim): 3
length of query: 385
length of database: 59,974,054
effective HSP length: 112
effective length of query: 273
effective length of database: 41,583,542
effective search space: 11352306966
effective search space used: 11352306966
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0101.13


