

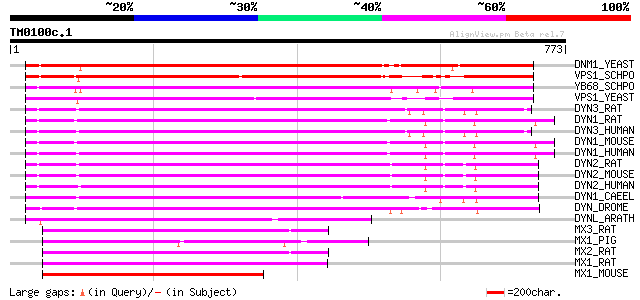
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0100c.1
(773 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
DNM1_YEAST (P54861) Dynamin-related protein DNM1 (EC 3.6.5.5) 566 e-161
VPS1_SCHPO (Q9URZ5) Vacuolar sorting protein 1 544 e-154
YB68_SCHPO (Q09748) Dynamin-like protein C12C2.08 527 e-149
VPS1_YEAST (P21576) Vacuolar sorting protein 1 503 e-141
DYN3_RAT (Q08877) Dynamin 3 (EC 3.6.5.5) (Dynamin, testicular) (... 485 e-136
DYN1_RAT (P21575) Dynamin-1 (EC 3.6.5.5) (D100) (Dynamin, brain)... 483 e-135
DYN3_HUMAN (Q9UQ16) Dynamin 3 (EC 3.6.5.5) (Dynamin, testicular)... 482 e-135
DYN1_MOUSE (P39053) Dynamin-1 (EC 3.6.5.5) 482 e-135
DYN1_HUMAN (Q05193) Dynamin-1 (EC 3.6.5.5) 481 e-135
DYN2_RAT (P39052) Dynamin 2 (EC 3.6.5.5) 478 e-134
DYN2_MOUSE (P39054) Dynamin 2 (EC 3.6.5.5) (Dynamin UDNM) 478 e-134
DYN2_HUMAN (P50570) Dynamin 2 (EC 3.6.5.5) 477 e-134
DYN1_CAEEL (P39055) Dynamin (EC 3.6.5.5) 451 e-126
DYN_DROME (P27619) Dynamin (EC 3.6.5.5) (dDyn) (Shibire protein) 450 e-126
DYNL_ARATH (P42697) Dynamin-like protein 322 2e-87
MX3_RAT (P18590) Interferon-induced GTP-binding protein Mx3 211 8e-54
MX1_PIG (P27594) Interferon-induced GTP-binding protein Mx1 210 1e-53
MX2_RAT (P18589) Interferon-induced GTP-binding protein Mx2 209 2e-53
MX1_RAT (P18588) Interferon-induced GTP-binding protein Mx1 204 5e-52
MX1_MOUSE (P09922) Interferon-induced GTP-binding protein Mx1 (I... 203 2e-51
>DNM1_YEAST (P54861) Dynamin-related protein DNM1 (EC 3.6.5.5)
Length = 757
Score = 567 bits (1460), Expect = e-161
Identities = 312/754 (41%), Positives = 471/754 (62%), Gaps = 58/754 (7%)
Query: 23 VISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTRR 82
+I VN+LQD+ G + +DLP +AVVGSQSSGKSS+LE+LVGRDFLPRGT I TRR
Sbjct: 7 LIPTVNKLQDVMYDSGIDT--LDLPILAVVGSQSSGKSSILETLVGRDFLPRGTGIVTRR 64
Query: 83 PLVLQLVHTKPSHP---------------------------------------EFGEFLH 103
PLVLQL + P+ P E+GEFLH
Sbjct: 65 PLVLQLNNISPNSPLIEEDDNSVNPHDEVTKISGFEAGTKPLEYRGKERNHADEWGEFLH 124
Query: 104 LPGRKFHDFTQIRHEIQAETDREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPV 163
+PG++F+DF I+ EI+ ET R AG++KG+S+ I LK+FSP+VL++TLVDLPGITKVP+
Sbjct: 125 IPGKRFYDFDDIKREIENETARIAGKDKGISKIPINLKVFSPHVLNLTLVDLPGITKVPI 184
Query: 164 GDQPSDIEARIRTMIMSYIKIPTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVIT 223
G+QP DIE +I+ +I+ YI P CLILAV+PAN DL NS++L++A+ DP G RTIGVIT
Sbjct: 185 GEQPPDIEKQIKNLILDYIATPNCLILAVSPANVDLVNSESLKLAREVDPQGKRTIGVIT 244
Query: 224 KLDIMDRGTDARNLLLGKVIPLRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVY 283
KLD+MD GT+A ++L GK+ PL+LG+VGVVNRSQ+DIQ+N++++++L EE++FR PVY
Sbjct: 245 KLDLMDSGTNALDILSGKMYPLKLGFVGVVNRSQQDIQLNKTVEESLDKEEDYFRKHPVY 304
Query: 284 SGLADSCGVPQLAKKLNKILAQHIKAVLPGLKARISTSLVALAKEHASYGEI-TESKAGQ 342
++ CG LAK LN+ L HI+ LP +K +++T + +E A YG + + +
Sbjct: 305 RTISTKCGTRYLAKLLNQTLLSHIRDKLPDIKTKLNTLISQTEQELARYGGVGATTNESR 364
Query: 343 GALVLNILSKYCDAFSSMLEGKNKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCEDLTD 402
+LVL +++K+ F S ++G + +++T EL GGARI+YI+ +++ SL+++DP +L+
Sbjct: 365 ASLVLQLMNKFSTNFISSIDGTSSDINTKELCGGARIYYIYNNVFGNSLKSIDPTSNLSV 424
Query: 403 DDIRTALQNATGPKSALFIPEVPFEIFVRRQISRLLDPSLQCARFIYDELMKISHRCMFT 462
D+RTA++N+TGP+ LF+PE+ F++ V+ QI LL+PS +C +Y+ELMKI H+C
Sbjct: 425 LDVRTAIRNSTGPRPTLFVPELAFDLLVKPQIKLLLEPSQRCVELVYEELMKICHKCGSA 484
Query: 463 ELQRFPFLRKHMDEVIGNFLRDGLEPSETMITHIIEMEMDYINTSHPNFIGGSKALEAAV 522
EL R+P L+ + EVI LR+ L+P+ + + +I++ YINT+HPNF+ ++A++ +
Sbjct: 485 ELARYPKLKSMLIEVISELLRERLQPTRSYVESLIDIHRAYINTNHPNFLSATEAMD-DI 543
Query: 523 QQTKPSRVSPPVSLDALESDKGLASERNGKSRGIIARQANGVVADHGVRGASDVEKVAPS 582
+T+ R + + L+S L+ + NG++ GI + D SD +
Sbjct: 544 MKTRRKR-----NQELLKSK--LSQQENGQTNGINGTSSISSNIDQDSAKNSDYDDDGID 596
Query: 583 GNAGGSSWRISSIFGGRDNRVYVKENTSTKPHA-------EPVHNVQSSSTIHLREPPPV 635
+ + + + F G+D + + S K + E N+Q S L + +
Sbjct: 597 AESKQTKDKFLNYFFGKDKKGQPVFDASDKKRSIAGDGNIEDFRNLQISD-FSLGDIDDL 655
Query: 636 LRPSESNSEMEGVEITVTKLLLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEK 695
+E E +E + K L+ SY+DI R+ IED VPKA+M LVN K + N + K
Sbjct: 656 ENAEPPLTEREELECELIKRLIVSYFDIIREMIEDQVPKAVMCLLVNYCKDSVQNRLVTK 715
Query: 696 LYREDLFEEMLQEPAEVAAKRKRCRELLRAYQQA 729
LY+E LFEE+L E +A R+ C + L Y++A
Sbjct: 716 LYKETLFEELLVEDQTLAQDRELCVKSLGVYKKA 749
>VPS1_SCHPO (Q9URZ5) Vacuolar sorting protein 1
Length = 678
Score = 544 bits (1402), Expect = e-154
Identities = 305/729 (41%), Positives = 456/729 (61%), Gaps = 82/729 (11%)
Query: 22 SVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTR 81
S+I +VN+LQ+ FS VG Q+ +IDLPQ+ VV SQSSGKSSVLE++VGRDFLPRGT I TR
Sbjct: 4 SLIKVVNQLQEAFSTVGVQN-LIDLPQITVVRSQSSGKSSVLENIVGRDFLPRGTGIVTR 62
Query: 82 RPLVLQLVHTKPS-------------------HPEFGEFLHLPGRKFHDFTQIRHEIQAE 122
RPLVLQL++ +PS E+GEFLHLPG+KF +F +IR EI E
Sbjct: 63 RPLVLQLIN-RPSASGKNEETTTDSDGKDQNNSSEWGEFLHLPGQKFFEFEKIREEIVRE 121
Query: 123 TDREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYI 182
T+ + G+N G+S I L+I+SP+VL +TLVDLPG+TKVPVGDQP DIE +IR M++ YI
Sbjct: 122 TEEKTGKNVGISSVPIYLRIYSPHVLTLTLVDLPGLTKVPVGDQPRDIEKQIREMVLKYI 181
Query: 183 KIPTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKV 242
+ILAV AN+DLANSD L++A+ DP+G RTIGV+TK+D+MD+GTD ++L G+V
Sbjct: 182 SKNNAIILAVNAANTDLANSDGLKLAREVDPEGLRTIGVLTKVDLMDKGTDVVDILAGRV 241
Query: 243 IPLRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKI 302
IPLRLGYV V+NR Q+DI+ +SI+ AL AE FF + P Y A CG P LA+KLN I
Sbjct: 242 IPLRLGYVPVINRGQKDIEGKKSIRIALEAERNFFETHPSYGSKAQYCGTPFLARKLNMI 301
Query: 303 LAQHIKAVLPGLKARISTSLVALAKEHASYGEITESKAG-QGALVLNILSKYCDAFSSML 361
L HI+ LP +K RI+ ALAK A + ++ G ++VLN+++ +C+ + +++
Sbjct: 302 LMHHIRNTLPEIKVRIN---AALAKYQAELHSLGDTPVGDNSSIVLNLITDFCNEYRTVV 358
Query: 362 EGKNKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFI 421
+G+++E+S ELSGGARI ++F I+ ++A+DP +++ D DIRT L N++GP +LF+
Sbjct: 359 DGRSEELSATELSGGARIAFVFHEIFSNGIQAIDPFDEVKDSDIRTILYNSSGPSPSLFM 418
Query: 422 PEVPFEIFVRRQISRLLDPSLQCARFIYDELMKISHRCMFTEL-QRFPFLRKHMDEVIGN 480
FE+ V++QI RL DPSL+C IYDEL++I ++ + + +R+P L+ +V+
Sbjct: 419 GTAAFEVIVKQQIKRLEDPSLKCVSLIYDELVRILNQLLQRPIFKRYPLLKDEFYKVVIG 478
Query: 481 FLRDGLEPSETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVSLDALE 540
F R ++P+ T++ ++ ME YINT HP+F+ G +A+ A VQ S+ S P+ +D +
Sbjct: 479 FFRKCMQPTNTLVMDMVAMEGSYINTVHPDFLSGHQAM-AIVQ----SQNSKPIPVDP-K 532
Query: 541 SDKGLASERNGKSRGIIARQANGVVADHGVRGASDVEKVAPSGNAGGSSWRISSIFGGRD 600
+ K L + + V V S ++G + + S FG ++
Sbjct: 533 TGKALTN--------------------------NPVPPVETSSSSGQNFF--GSFFGSKN 564
Query: 601 NRVYVKENTSTKPHAEPVHNVQSSSTIHLREPPPVLRPSESNSEMEGVEITVTKLLLRSY 660
K + +P PPPVLR S + S+ E + V KLL+ SY
Sbjct: 565 K----KRLAAMEP------------------PPPVLRASTTLSDREKTDTEVIKLLIMSY 602
Query: 661 YDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAAKRKRCR 720
++I ++ + D+VPK+I ++ +K + + +E+LY+ F+++LQE +RK C
Sbjct: 603 FNIVKRTLADMVPKSISLKMIKYSKEHIQHELLEQLYKSQAFDKLLQESEVTVQRRKECE 662
Query: 721 ELLRAYQQA 729
+++ + QA
Sbjct: 663 QMVESLLQA 671
>YB68_SCHPO (Q09748) Dynamin-like protein C12C2.08
Length = 781
Score = 527 bits (1357), Expect = e-149
Identities = 300/774 (38%), Positives = 459/774 (58%), Gaps = 71/774 (9%)
Query: 23 VISLVNRLQD-IFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTR 81
+I LVN+LQD +++ +G S +DLP + VVGSQS GKSSVLE++VG+DFLPRGT I TR
Sbjct: 4 LIPLVNQLQDLVYNTIG--SDFLDLPSIVVVGSQSCGKSSVLENIVGKDFLPRGTGIVTR 61
Query: 82 RPLVLQLVH----TKPSHPE--------------------------------FGEFLHLP 105
RPL+LQL++ TK +H E + EFLH+P
Sbjct: 62 RPLILQLINLKRKTKNNHDEESTSDNNSEETSAAGETGSLEGIEEDSDEIEDYAEFLHIP 121
Query: 106 GRKFHDFTQIRHEIQAETDREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGD 165
KF D ++R EI+ ET R AG NKG+++ I LKI+S VL++TL+DLPG+TK+PVGD
Sbjct: 122 DTKFTDMNKVRAEIENETLRVAGANKGINKLPINLKIYSTRVLNLTLIDLPGLTKIPVGD 181
Query: 166 QPSDIEARIRTMIMSYIKIPTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKL 225
QP+DIEA+ R++IM YI P +ILAV+PAN D+ NS+ L++A+ DP G RTIGV+TKL
Sbjct: 182 QPTDIEAQTRSLIMEYISRPNSIILAVSPANFDIVNSEGLKLARSVDPKGKRTIGVLTKL 241
Query: 226 DIMDRGTDARNLLLGKVIPLRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSG 285
D+MD+GT+A ++L G+V PL+LG+V VNRSQ DI ++S++DAL +E FF P Y
Sbjct: 242 DLMDQGTNAMDILSGRVYPLKLGFVATVNRSQSDIVSHKSMRDALQSERSFFEHHPAYRT 301
Query: 286 LADSCGVPQLAKKLNKILAQHIKAVLPGLKARISTSLVALAKEHASYGEITESKAGQ-GA 344
+ D CG P LAK L+ +L HI+ LP +KAR+ST + ++ +YG+ S Q G
Sbjct: 302 IKDRCGTPYLAKTLSNLLVSHIRERLPDIKARLSTLISQTQQQLNNYGDFKLSDQSQRGI 361
Query: 345 LVLNILSKYCDAFSSMLEGKNKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDD 404
++L ++++ + F + ++G + + T ELSGGAR++ IF +++ +L ++DP ++L+ D
Sbjct: 362 ILLQAMNRFANTFIASIDGNSSNIPTKELSGGARLYSIFNNVFTTALNSIDPLQNLSTVD 421
Query: 405 IRTALQNATGPKSALFIPEVPFEIFVRRQISRLLDPSLQCARFIYDELMKISHRCMFTEL 464
IRTA+ N+TG ++ LF+ E+ F+I V+ Q++ L P QC +Y+ELMKI H +++
Sbjct: 422 IRTAILNSTGSRATLFLSEMAFDILVKPQLNLLAAPCHQCVELVYEELMKICHYSGDSDI 481
Query: 465 QRFPFLRKHMDEVIGNFLRDGLEPSETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQ 524
FP L+ + E + + LR+ L P+ + + +I ++ YINT+HP+F+G A+ + +
Sbjct: 482 SHFPKLQTALVETVSDLLRENLTPTYSFVESLIAIQSAYINTNHPDFLGVQGAMAVVLSR 541
Query: 525 TKPSRV------SPPVS--LDALESD--KGLASERNGKSRGIIARQANGVVA----DHGV 570
+ +R+ P+S LD ++ D + +S+ + + I + N + D
Sbjct: 542 KEQNRLMLSQENDEPISSALDTVKPDGIELYSSDPDTSVKSITNKATNEITTLKSDDSAK 601
Query: 571 RGASDVEKVAPSGNAGGSSWR-----ISSIFGGRDNRVYVKENTSTKPHAEPVHNVQSSS 625
DV NA + +S +FG N K + K + P+++ S
Sbjct: 602 MQPLDVLASKRYNNAFSTETAERKTFLSYVFGA--NNATRKAMSIDKSSSYPLNDSLSGG 659
Query: 626 TIHLREPPPVLRPSESN----------SEMEGVEITVTKLLLRSYYDIARKNIEDLVPKA 675
+ + P+ SN SE E VE+ + K L+ SY+++ RK I D VPK
Sbjct: 660 DTNHKNNHPLKMTDLSNEVETMALEDMSEREEVEVDLIKELITSYFNLTRKIIIDQVPKV 719
Query: 676 IMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAAKRKRCRELLRAYQQA 729
IMH LVN +K + N + KLYRED F+ +L E V ++R++C LL Y QA
Sbjct: 720 IMHLLVNASKDAIQNRLVSKLYREDFFDTLLIEDENVKSEREKCERLLSVYNQA 773
>VPS1_YEAST (P21576) Vacuolar sorting protein 1
Length = 704
Score = 503 bits (1294), Expect = e-141
Identities = 284/751 (37%), Positives = 430/751 (56%), Gaps = 102/751 (13%)
Query: 23 VISLVNRLQDIFSRVGNQS-TIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTR 81
+IS +N+LQD + +G S + IDLPQ+ VVGSQSSGKSSVLE++VGRDFLPRGT I TR
Sbjct: 5 LISTINKLQDALAPLGGGSQSPIDLPQITVVGSQSSGKSSVLENIVGRDFLPRGTGIVTR 64
Query: 82 RPLVLQLVHTKP-----------------------------------------SHPEFGE 100
RPLVLQL++ +P + E+GE
Sbjct: 65 RPLVLQLINRRPKKSEHAKVNQTANELIDLNINDDDKKKDESGKHQNEGQSEDNKEEWGE 124
Query: 101 FLHLPGRKFHDFTQIRHEIQAETDREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITK 160
FLHLPG+KF++F +IR EI ETD+ G N G+S I L+I+SP+VL +TLVDLPG+TK
Sbjct: 125 FLHLPGKKFYNFDEIRKEIVKETDKVTGANSGISSVPINLRIYSPHVLTLTLVDLPGLTK 184
Query: 161 VPVGDQPSDIEARIRTMIMSYIKIPTCLILAVTPANSDLANSDALQMAQVADPDGNRTIG 220
VPVGDQP DIE +I+ M++ YI P +IL+V AN+DLANSD L++A+ DP+G RTIG
Sbjct: 185 VPVGDQPPDIERQIKDMLLKYISKPNAIILSVNAANTDLANSDGLKLAREVDPEGTRTIG 244
Query: 221 VITKLDIMDRGTDARNLLLGKVIPLRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSR 280
V+TK+D+MD+GTD ++L G+VIPLR GY+ V+NR Q+DI+ ++I++AL E +FF +
Sbjct: 245 VLTKVDLMDQGTDVIDILAGRVIPLRYGYIPVINRGQKDIEHKKTIREALENERKFFENH 304
Query: 281 PVYSGLADSCGVPQLAKKLNKILAQHIKAVLPGLKARISTSLVALAKEHASYGEITESKA 340
P YS A CG P LAKKLN IL HI+ LP +KA+I +L E + G T A
Sbjct: 305 PSYSSKAHYCGTPYLAKKLNSILLHHIRQTLPEIKAKIEATLKKYQNELINLGPETMDSA 364
Query: 341 GQGALVLNILSKYCDAFSSMLEGKNKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCEDL 400
++VL++++ + + ++ +L+G+ KE+S+ ELSGGARI Y+F + ++++DP + +
Sbjct: 365 --SSVVLSMITDFSNEYAGILDGEAKELSSQELSGGARISYVFHETFKNGVDSLDPFDQI 422
Query: 401 TDDDIRTALQNATGPKSALFIPEVPFEIFVRRQISRLLDPSLQCARFIYDELMKISHRCM 460
D DIRT + N++G +LF+ FE+ V++QI R +PSL+ ++DEL+++ + +
Sbjct: 423 KDSDIRTIMYNSSGSAPSLFVGTEAFEVLVKQQIRRFEEPSLRLVTLVFDELVRMLKQII 482
Query: 461 F-TELQRFPFLRKHMDEVIGNFLRDGLEPSETMITHIIEMEMDYINTSHPNFIGGSKALE 519
+ R+P LR+ + FL+D P+ + II+ E YINT+HP+ + GS+A+
Sbjct: 483 SQPKYSRYPALREAISNQFIQFLKDATIPTNEFVVDIIKAEQTYINTAHPDLLKGSQAMV 542
Query: 520 AAVQQTKPSRVSPPVSLDALESDKGLASERNGKSRGIIARQANGVVADHGVRGASDVEKV 579
++ P +V+ + GK
Sbjct: 543 MVEEKLHPRQVA--------------VDPKTGKPL-----------------------PT 565
Query: 580 APSGNAGGSSWRISSIFGGRDNRVYVKENTSTKPHAEPVHNVQSSSTIHLREPP-PVLRP 638
PS + S FGG + ++ + E P PVL+
Sbjct: 566 QPSSSKAPVMEEKSGFFGG-------------------FFSTKNKKKLAALESPPPVLKA 606
Query: 639 SESNSEMEGVEITVTKLLLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYR 698
+ +E E +E V KLL+ SY+ I ++ I D++PKA+M L+ +K ++ V +EKLY
Sbjct: 607 TGQMTERETMETEVIKLLISSYFSIVKRTIADIIPKALMLKLIVKSKTDIQKVLLEKLYG 666
Query: 699 EDLFEEMLQEPAEVAAKRKRCRELLRAYQQA 729
+ EE+ +E +RK C++++ + A
Sbjct: 667 KQDIEELTKENDITIQRRKECKKMVEILRNA 697
>DYN3_RAT (Q08877) Dynamin 3 (EC 3.6.5.5) (Dynamin, testicular)
(T-dynamin)
Length = 848
Score = 485 bits (1249), Expect = e-136
Identities = 289/725 (39%), Positives = 425/725 (57%), Gaps = 30/725 (4%)
Query: 23 VISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTRR 82
+I LVNRLQD FS +G QS +++LPQ+AVVG QS+GKSSVLE+ VGRDFLPRG+ I TRR
Sbjct: 9 LIPLVNRLQDAFSALG-QSCLLELPQIAVVGGQSAGKSSVLENFVGRDFLPRGSGIVTRR 67
Query: 83 PLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENKGVSEKQIRLKI 142
PLVLQLV +K E+ EFLH G+KF DF ++RHEI+AETDR G NKG+S I L++
Sbjct: 68 PLVLQLVTSKA---EYAEFLHCKGKKFTDFDEVRHEIEAETDRVTGMNKGISSVPINLRV 124
Query: 143 FSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAVTPANSDLANS 202
+SP+VL++TL+DLPGITKVPVGDQP DIE +IR MIM +I CLILAVTPAN+DLANS
Sbjct: 125 YSPHVLNLTLIDLPGITKVPVGDQPPDIEYQIRDMIMQFITRENCLILAVTPANTDLANS 184
Query: 203 DALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVGVVNRSQEDIQM 262
DAL++A+ DP G RTIGVITKLD+MD GTDAR++L K++PLR GYVGVVNRSQ+DI
Sbjct: 185 DALKLAKEVDPQGLRTIGVITKLDLMDEGTDARDVLENKLLPLRRGYVGVVNRSQKDIDG 244
Query: 263 NRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLPGLKARISTSL 322
+ IK A++AE +FF S P Y +AD G P L K LN+ L HI+ LP + ++ L
Sbjct: 245 KKDIKAAMLAERKFFLSHPAYRHIADRMGTPHLQKVLNQQLTNHIRDTLPNFRNKLQGQL 304
Query: 323 VALAKEHASYGEIT-ESKAGQGALVLNILSKYCDAFSSMLEGKNKEMSTCELSGGARIHY 381
+++ E ++ E + +L ++ ++ F +EG ++ T ELSGGA+I+
Sbjct: 305 LSIEHEVEAFKNFKPEDPTRKTKALLQMVQQFAVDFEKRIEGSGDQVDTLELSGGAKINR 364
Query: 382 IFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQISRLLDPS 441
IF + + ++ E +I A++N G ++ LF P++ FE V++QI +L PS
Sbjct: 365 IFHERFPFEIVKMEFNEKELRREISYAIKNIHGIRTGLFTPDMAFEAIVKKQIVKLKGPS 424
Query: 442 LQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIGNFLRDGLEPSETMITHIIEMEM 501
L+ + EL+ +C L FP L + + ++ N +R+ ++ + +I++++
Sbjct: 425 LKSVDLVMQELINTVKKCT-KRLANFPRLCEETERIVANHIREREGKTKDQVLLLIDIQV 483
Query: 502 DYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVSLDALESDKGLASERNGKSRG---IIA 558
YINT+H +FIG + A + + Q K S + V + + + G S+G ++
Sbjct: 484 SYINTNHEDFIGFANAQQRSSQVHKKSTIGNQVIRKGWLTVSNIGIMKGG-SKGYWFVLT 542
Query: 559 RQANGVVADHGVRGAS-----DVEKVAPSGNAGGSSWRISSIFGGRDNRVYVKENTSTKP 613
++ D + D KV SS + ++F VY K+ S +
Sbjct: 543 AESLSWYKDDEEKEKKYMLPLDNLKVRDVEKGFMSSKHVFALFNTEQRNVY-KDYRSLEL 601
Query: 614 HAEPVHNVQSSSTIHLRE---PPPVLRPSESNSEMEGV--------EITVTKLLLRSYYD 662
+ +V S LR P ++ N + E ++ + L+ SY
Sbjct: 602 ACDSQEDVDSWKASLLRAGVYPDKSFTENDENGQAENFSMDPQLERQVETIRNLVDSYMS 661
Query: 663 IARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAAKRKRCREL 722
I K I DL+PK IMH ++NN K +++ + +LY + +++E E A +R E+
Sbjct: 662 IINKCIRDLIPKTIMHLMINNVKDFINSELLAQLYSSEDQNTLMEESVEQAQRRD---EM 718
Query: 723 LRAYQ 727
LR YQ
Sbjct: 719 LRMYQ 723
>DYN1_RAT (P21575) Dynamin-1 (EC 3.6.5.5) (D100) (Dynamin, brain)
(B-dynamin)
Length = 851
Score = 483 bits (1242), Expect = e-135
Identities = 302/784 (38%), Positives = 442/784 (55%), Gaps = 55/784 (7%)
Query: 23 VISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTRR 82
+I LVNRLQD FS +G Q+ +DLPQ+AVVG QS+GKSSVLE+ VGRDFLPRG+ I TRR
Sbjct: 9 LIPLVNRLQDAFSAIG-QNADLDLPQIAVVGGQSAGKSSVLENFVGRDFLPRGSGIVTRR 67
Query: 83 PLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENKGVSEKQIRLKI 142
PLVLQLV+ S E+ EFLH G+KF DF ++R EI+AETDR G NKG+S I L++
Sbjct: 68 PLVLQLVN---STTEYAEFLHCKGKKFTDFEEVRLEIEAETDRVTGTNKGISPVPINLRV 124
Query: 143 FSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAVTPANSDLANS 202
+SP+VL++TLVDLPG+TKVPVGDQP DIE +IR M+M ++ CLILAV+PANSDLANS
Sbjct: 125 YSPHVLNLTLVDLPGMTKVPVGDQPPDIEFQIRDMLMQFVTKENCLILAVSPANSDLANS 184
Query: 203 DALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVGVVNRSQEDIQM 262
DAL++A+ DP G RTIGVITKLD+MD GTDAR++L K++PLR GY+GVVNRSQ+DI
Sbjct: 185 DALKIAKEVDPQGQRTIGVITKLDLMDEGTDARDVLENKLLPLRRGYIGVVNRSQKDIDG 244
Query: 263 NRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLPGLKARISTSL 322
+ I AL AE +FF S P Y LAD G P L K LN+ L HI+ LPGL+ ++ + L
Sbjct: 245 KKDITAALAAERKFFLSHPSYRHLADRMGTPYLQKVLNQQLTNHIRDTLPGLRNKLQSQL 304
Query: 323 VALAKEHASYGEI-TESKAGQGALVLNILSKYCDAFSSMLEGKNKEMSTCELSGGARIHY 381
+++ KE Y + A + +L ++ ++ F +EG ++ T ELSGGARI+
Sbjct: 305 LSIEKEVDEYKNFRPDDPARKTKALLQMVQQFAVDFEKRIEGSGDQIDTYELSGGARINR 364
Query: 382 IFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQISRLLDPS 441
IF + L ++ E +I A++N G ++ LF P++ FE V++Q+ +L +PS
Sbjct: 365 IFHERFPFELVKMEFDEKELRREISYAIKNIHGIRTGLFTPDLAFEATVKKQVQKLKEPS 424
Query: 442 LQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIGNFLRDGLEPSETMITHIIEMEM 501
++C + EL +C +LQ++P LR+ M+ ++ +R+ ++ + +I++E+
Sbjct: 425 IKCVDMVVSELTSTIRKCS-EKLQQYPRLREEMERIVTTHIREREGRTKEQVMLLIDIEL 483
Query: 502 DYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVSLDALESDKGLASERN-GKSRGIIARQ 560
Y+NT+H +FIG + A + + Q K + + + L KG + N G +G
Sbjct: 484 AYMNTNHEDFIGFANAQQRSNQMNK--KKTSGNQDEILVIRKGWLTINNIGIMKGGSKEY 541
Query: 561 ANGVVADHGVRGASDVE------------KVAPSGNAGGSSWRISSIFGGRDNRVYVKEN 608
+ A++ D E K+ SS I ++F VY K+
Sbjct: 542 WFVLTAENLSWYKDDEEKEKKYMLSVDNLKLRDVEKGFMSSKHIFALFNTEQRNVY-KDY 600
Query: 609 TSTKPHAEPVHNVQSSSTIHLREP--PPVLRPSESNSEME---------------GVEIT 651
+ E V S LR P + E SE E ++
Sbjct: 601 RQLELACETQEEVDSWKASFLRAGVYPERVGDKEKASETEENGSDSFMHSMDPQLERQVE 660
Query: 652 VTKLLLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAE 711
+ L+ SY I K + DL+PK IMH ++NNTK + + + LY +++E AE
Sbjct: 661 TIRNLVDSYMAIVNKTVRDLMPKTIMHLMINNTKEFIFSELLANLYSCGDQNTLMEESAE 720
Query: 712 VAAKRKRCRELLRAYQQAFR----------------DLDELPLEAETVERGYGSPEATGL 755
A +R + A ++A +D+ L+ ++V G SP ++
Sbjct: 721 QAQRRDEMLRMYHALKEALSIIGDINTTTVSTPMPPPVDDSWLQVQSVPAGRRSPTSSPT 780
Query: 756 PRIR 759
P+ R
Sbjct: 781 PQRR 784
>DYN3_HUMAN (Q9UQ16) Dynamin 3 (EC 3.6.5.5) (Dynamin, testicular)
(T-dynamin)
Length = 859
Score = 482 bits (1241), Expect = e-135
Identities = 290/725 (40%), Positives = 427/725 (58%), Gaps = 30/725 (4%)
Query: 23 VISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTRR 82
+I LVNRLQD FS +G QS +++LPQ+AVVG QS+GKSSVLE+ VGRDFLPRG+ I TRR
Sbjct: 9 LIPLVNRLQDAFSALG-QSCLLELPQIAVVGGQSAGKSSVLENFVGRDFLPRGSGIVTRR 67
Query: 83 PLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENKGVSEKQIRLKI 142
PLVLQLV +K E+ EFLH G+KF DF ++R EI+AETDR G NKG+S I L++
Sbjct: 68 PLVLQLVTSKA---EYAEFLHCKGKKFTDFDEVRLEIEAETDRVTGMNKGISSIPINLRV 124
Query: 143 FSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAVTPANSDLANS 202
+SP+VL++TL+DLPGITKVPVGDQP DIE +IR MIM +I CLILAVTPAN+DLANS
Sbjct: 125 YSPHVLNLTLIDLPGITKVPVGDQPPDIEYQIREMIMQFITRENCLILAVTPANTDLANS 184
Query: 203 DALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVGVVNRSQEDIQM 262
DAL++A+ DP G RTIGVITKLD+MD GTDAR++L K++PLR GYVGVVNRSQ+DI
Sbjct: 185 DALKLAKEVDPQGLRTIGVITKLDLMDEGTDARDVLENKLLPLRRGYVGVVNRSQKDIDG 244
Query: 263 NRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLPGLKARISTSL 322
+ IK A++AE +FF S P Y +AD G P L K LN+ L HI+ LP + ++ L
Sbjct: 245 KKDIKAAMLAERKFFLSHPAYRHIADRMGTPHLQKVLNQQLTNHIRDTLPNFRNKLQGQL 304
Query: 323 VALAKEHASYGEIT-ESKAGQGALVLNILSKYCDAFSSMLEGKNKEMSTCELSGGARIHY 381
+++ E +Y E + +L ++ ++ F +EG ++ T ELSGGA+I+
Sbjct: 305 LSIEHEVEAYKNFKPEDPTRKTKALLQMVQQFAVDFEKRIEGSGDQVDTLELSGGAKINR 364
Query: 382 IFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQISRLLDPS 441
IF + + ++ E +I A++N G ++ LF P++ FE V++QI +L PS
Sbjct: 365 IFHERFPFEIVKMEFNEKELRREISYAIKNIHGIRTGLFTPDMAFEAIVKKQIVKLKGPS 424
Query: 442 LQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIGNFLRDGLEPSETMITHIIEMEM 501
L+ + EL+ +C +L FP L + + ++ N +R+ ++ + +I++++
Sbjct: 425 LKSVDLVIQELINTVKKCT-KKLANFPRLCEETERIVANHIREREGKTKDQVLLLIDIQV 483
Query: 502 DYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVSLDALESDKGLASERNGKSRG---IIA 558
YINT+H +FIG + A + + Q K + V V + + + G S+G ++
Sbjct: 484 SYINTNHEDFIGFANAQQRSSQVHKKTTVGNQVIRKGWLTISNIGIMKGG-SKGYWFVLT 542
Query: 559 RQANGVVADHGVRGAS-----DVEKVAPSGNAGGSSWRISSIFGGRDNRVYVKENTSTKP 613
++ D + D KV + SS I ++F VY K+ +
Sbjct: 543 AESLSWYKDDEEKEKKYMLPLDNLKVRDVEKSFMSSKHIFALFNTEQRNVY-KDYRFLEL 601
Query: 614 HAEPVHNVQSSSTIHLRE---PPPVLRPSESNSEMEGV--------EITVTKLLLRSYYD 662
+ +V S LR P + ++ N + E ++ + L+ SY
Sbjct: 602 ACDSQEDVDSWKASLLRAGVYPDKSVAENDENGQAENFSMDPQLERQVETIRNLVDSYMS 661
Query: 663 IARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAAKRKRCREL 722
I K I DL+PK IMH ++NN K +++ + +LY + +++E AE A +R E+
Sbjct: 662 IINKCIRDLIPKTIMHLMINNVKDFINSELLAQLYSSEDQNTLMEESAEQAQRRD---EM 718
Query: 723 LRAYQ 727
LR YQ
Sbjct: 719 LRMYQ 723
>DYN1_MOUSE (P39053) Dynamin-1 (EC 3.6.5.5)
Length = 867
Score = 482 bits (1240), Expect = e-135
Identities = 301/784 (38%), Positives = 442/784 (55%), Gaps = 55/784 (7%)
Query: 23 VISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTRR 82
+I LVNRLQD FS +G Q+ +DLPQ+AVVG QS+GKSSVLE+ VGRDFLPRG+ I TRR
Sbjct: 9 LIPLVNRLQDAFSAIG-QNADLDLPQIAVVGGQSAGKSSVLENFVGRDFLPRGSGIVTRR 67
Query: 83 PLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENKGVSEKQIRLKI 142
PLVLQLV+ S E+ EFLH G+KF DF ++R EI+AETDR G NKG+S I L++
Sbjct: 68 PLVLQLVN---STTEYAEFLHCKGKKFTDFEEVRLEIEAETDRVTGTNKGISPVPINLRV 124
Query: 143 FSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAVTPANSDLANS 202
+SP+VL++TLVDLPG+TKVPVGDQP DIE +IR M+M ++ CLILAV+PANSDLANS
Sbjct: 125 YSPHVLNLTLVDLPGMTKVPVGDQPPDIEFQIRDMLMQFVTKENCLILAVSPANSDLANS 184
Query: 203 DALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVGVVNRSQEDIQM 262
DAL++A+ DP G RTIGVITKLD+MD GTDAR++L K++PLR GY+GVVNRSQ+DI
Sbjct: 185 DALKIAKEVDPQGQRTIGVITKLDLMDEGTDARDVLENKLLPLRRGYIGVVNRSQKDIDG 244
Query: 263 NRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLPGLKARISTSL 322
+ I AL AE +FF S P Y LAD G P L K LN+ L HI+ LPGL+ ++ + L
Sbjct: 245 KKDITAALAAERKFFLSHPSYRHLADRMGTPYLQKVLNQQLTNHIRDTLPGLRNKLQSQL 304
Query: 323 VALAKEHASYGEI-TESKAGQGALVLNILSKYCDAFSSMLEGKNKEMSTCELSGGARIHY 381
+++ KE Y + A + +L ++ ++ F +EG ++ T ELSGGARI+
Sbjct: 305 LSIEKEVDEYKNFRPDDPARKTKALLQMVQQFAVDFEKRIEGSGDQIDTYELSGGARINR 364
Query: 382 IFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQISRLLDPS 441
IF + L ++ E +I A++N G ++ LF P++ FE V++Q+ ++ +P
Sbjct: 365 IFHERFPFELVKMEFDEKELRREISYAIKNIHGIRTGLFTPDMAFETIVKKQVKKIREPC 424
Query: 442 LQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIGNFLRDGLEPSETMITHIIEMEM 501
L+C + EL+ +C +LQ++P LR+ M+ ++ +R+ ++ + +I++E+
Sbjct: 425 LKCVDMVISELISTVRQCT-KKLQQYPRLREEMERIVTTHIREREGRTKEQVMLLIDIEL 483
Query: 502 DYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVSLDALESDKGLASERN-GKSRGIIARQ 560
Y+NT+H +FIG + A + + Q K + + + L KG + N G +G
Sbjct: 484 AYMNTNHEDFIGFANAQQRSNQMNK--KKTSGNQDEILVIRKGWLTINNIGIMKGGSKEY 541
Query: 561 ANGVVADHGVRGASDVE------------KVAPSGNAGGSSWRISSIFGGRDNRVYVKEN 608
+ A++ D E K+ SS I ++F VY K+
Sbjct: 542 WFVLTAENLSWYKDDEEKEKKYMLSVDNLKLRDVEKGFMSSKHIFALFNTEQRNVY-KDY 600
Query: 609 TSTKPHAEPVHNVQSSSTIHLREP--PPVLRPSESNSEME---------------GVEIT 651
+ E V S LR P + E SE E ++
Sbjct: 601 RQLELACETQEEVDSWKASFLRAGVYPERVGDKEKASETEENGSDSFMHSMDPQLERQVE 660
Query: 652 VTKLLLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAE 711
+ L+ SY I K + DL+PK IMH ++NNTK + + + LY +++E AE
Sbjct: 661 TIRNLVDSYMAIVNKTVRDLMPKTIMHLMINNTKEFIFSELLANLYSCGDQNTLMEESAE 720
Query: 712 VAAKRKRCRELLRAYQQAFR----------------DLDELPLEAETVERGYGSPEATGL 755
A +R + A ++A +D+ L+ ++V G SP ++
Sbjct: 721 QAQRRDEMLRMYHALKEALSIIGDINTTTVSTPMPPPVDDSWLQVQSVPAGRRSPTSSPT 780
Query: 756 PRIR 759
P+ R
Sbjct: 781 PQRR 784
>DYN1_HUMAN (Q05193) Dynamin-1 (EC 3.6.5.5)
Length = 864
Score = 481 bits (1238), Expect = e-135
Identities = 300/784 (38%), Positives = 441/784 (55%), Gaps = 55/784 (7%)
Query: 23 VISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTRR 82
+I LVNRLQD FS +G Q+ +DLPQ+AVVG QS+GKSSVLE+ VGRDFLPRG+ I TRR
Sbjct: 9 LIPLVNRLQDAFSAIG-QNADLDLPQIAVVGGQSAGKSSVLENFVGRDFLPRGSGIVTRR 67
Query: 83 PLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENKGVSEKQIRLKI 142
PLVLQLV+ E+ EFLH G+KF DF ++R EI+AETDR G NKG+S I L++
Sbjct: 68 PLVLQLVNATT---EYAEFLHCKGKKFTDFEEVRLEIEAETDRVTGTNKGISPVPINLRV 124
Query: 143 FSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAVTPANSDLANS 202
+SP+VL++TLVDLPG+TKVPVGDQP DIE +IR M+M ++ CLILAV+PANSDLANS
Sbjct: 125 YSPHVLNLTLVDLPGMTKVPVGDQPPDIEFQIRDMLMQFVTKENCLILAVSPANSDLANS 184
Query: 203 DALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVGVVNRSQEDIQM 262
DAL++A+ DP G RTIGVITKLD+MD GTDAR++L K++PLR GY+GVVNRSQ+DI
Sbjct: 185 DALKVAKEVDPQGQRTIGVITKLDLMDEGTDARDVLENKLLPLRRGYIGVVNRSQKDIDG 244
Query: 263 NRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLPGLKARISTSL 322
+ I AL AE +FF S P Y LAD G P L K LN+ L HI+ LPGL+ ++ + L
Sbjct: 245 KKDITAALAAERKFFLSHPSYRHLADRMGTPYLQKVLNQQLTNHIRDTLPGLRNKLQSQL 304
Query: 323 VALAKEHASYGEI-TESKAGQGALVLNILSKYCDAFSSMLEGKNKEMSTCELSGGARIHY 381
+++ KE Y + A + +L ++ ++ F +EG ++ T ELSGGARI+
Sbjct: 305 LSIEKEVEEYKNFRPDDPARKTKALLQMVQQFAVDFEKRIEGSGDQIDTYELSGGARINR 364
Query: 382 IFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQISRLLDPS 441
IF + L ++ E +I A++N G ++ LF P++ FE V++Q+ ++ +P
Sbjct: 365 IFHERFPFELVKMEFDEKELRREISYAIKNIHGIRTGLFTPDMAFETIVKKQVKKIREPC 424
Query: 442 LQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIGNFLRDGLEPSETMITHIIEMEM 501
L+C + EL+ +C +LQ++P LR+ M+ ++ +R+ ++ + +I++E+
Sbjct: 425 LKCVDMVISELISTVRQCT-KKLQQYPRLREEMERIVTTHIREREGRTKEQVMLLIDIEL 483
Query: 502 DYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVSLDALESDKGLASERN-GKSRGIIARQ 560
Y+NT+H +FIG + A + + Q K + + + L KG + N G +G
Sbjct: 484 AYMNTNHEDFIGFANAQQRSNQMNK--KKTSGNQDEILVIRKGWLTINNIGIMKGGSKEY 541
Query: 561 ANGVVADHGVRGASDVE------------KVAPSGNAGGSSWRISSIFGGRDNRVYVKEN 608
+ A++ D E K+ SS I ++F VY K+
Sbjct: 542 WFVLTAENLSWYKDDEEKEKKYMLSVDNLKLRDVEKGFMSSKHIFALFNTEQRNVY-KDY 600
Query: 609 TSTKPHAEPVHNVQSSSTIHLREP--PPVLRPSESNSEME---------------GVEIT 651
+ E V S LR P + E SE E ++
Sbjct: 601 RQLELACETQEEVDSWKASFLRAGVYPERVGDKEKASETEENGSDSFMHSMDPQLERQVE 660
Query: 652 VTKLLLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAE 711
+ L+ SY I K + DL+PK IMH ++NNTK + + + LY +++E AE
Sbjct: 661 TIRNLVDSYMAIVNKTVRDLMPKTIMHLMINNTKEFIFSELLANLYSCGDQNTLMEESAE 720
Query: 712 VAAKRKRCRELLRAYQQAFR----------------DLDELPLEAETVERGYGSPEATGL 755
A +R + A ++A +D+ L+ ++V G SP ++
Sbjct: 721 QAQRRDEMLRMYHALKEALSIIGNINTTTVSTPMPPPVDDSWLQVQSVPAGRRSPTSSPT 780
Query: 756 PRIR 759
P+ R
Sbjct: 781 PQRR 784
>DYN2_RAT (P39052) Dynamin 2 (EC 3.6.5.5)
Length = 870
Score = 478 bits (1229), Expect = e-134
Identities = 284/742 (38%), Positives = 428/742 (57%), Gaps = 39/742 (5%)
Query: 23 VISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTRR 82
+I LVN+LQD FS +G QS +DLPQ+AVVG QS+GKSSVLE+ VGRDFLPRG+ I TRR
Sbjct: 9 LIPLVNKLQDAFSSIG-QSCHLDLPQIAVVGGQSAGKSSVLENFVGRDFLPRGSGIVTRR 67
Query: 83 PLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENKGVSEKQIRLKI 142
PL+LQL+ +K E+ EFLH +KF DF ++R EI+AETDR G NKG+S I L++
Sbjct: 68 PLILQLIFSKT---EYAEFLHCKSKKFTDFDEVRQEIEAETDRVTGTNKGISPVPINLRV 124
Query: 143 FSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAVTPANSDLANS 202
+SP+VL++TL+DLPGITKVPVGDQP DIE +I+ MI+ +I + LILAVTPAN DLANS
Sbjct: 125 YSPHVLNLTLIDLPGITKVPVGDQPPDIEYQIKDMILQFISRESSLILAVTPANMDLANS 184
Query: 203 DALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVGVVNRSQEDIQM 262
DAL++A+ DP G RTIGVITKLD+MD GTDAR++L K++PLR GY+GVVNRSQ+DI+
Sbjct: 185 DALKLAKEVDPQGLRTIGVITKLDLMDEGTDARDVLENKLLPLRRGYIGVVNRSQKDIEG 244
Query: 263 NRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLPGLKARISTSL 322
+ I+ AL AE +FF S P Y +AD G P L K LN+ L HI+ LP L++++ + L
Sbjct: 245 RKDIRAALAAERKFFLSHPAYRHMADRMGTPHLQKTLNQQLTNHIRESLPTLRSKLQSQL 304
Query: 323 VALAKEHASYGEI-TESKAGQGALVLNILSKYCDAFSSMLEGKNKEMSTCELSGGARIHY 381
++L KE Y + + +L ++ ++ F +EG ++ T ELSGGARI+
Sbjct: 305 LSLEKEVEEYKNFRPDDPTRKTKALLQMVQQFGVDFEKRIEGSGDQVDTLELSGGARINR 364
Query: 382 IFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQISRLLDPS 441
IF + L ++ E +I A++N G ++ LF P++ FE V++Q+ +L +P
Sbjct: 365 IFHERFPFELVKMEFDEKDLRREISYAIKNIHGVRTGLFTPDLAFEAIVKKQVVKLKEPC 424
Query: 442 LQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIGNFLRDGLEPSETMITHIIEMEM 501
L+C + EL+ +C ++L +P LR+ + ++ ++R+ ++ I +I++E
Sbjct: 425 LKCVDLVIQELISTVRQCT-SKLSSYPRLREETERIVTTYIREREGRTKDQILLLIDIEQ 483
Query: 502 DYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVSLDALESDKGLASERNGKSRGIIARQA 561
YINT+H +FIG + A + + Q K + P + L +G + N +++
Sbjct: 484 SYINTNHEDFIGFANAQQRSTQLNKKRAI--PNQGEILVIRRGWLTINNISLMKGGSKEY 541
Query: 562 NGVVADHGVRGASDVE-------------KVAPSGNAGGSSWRISSIFGGRDNRVYVKEN 608
V+ + D E K+ S+ + +IF VY K+
Sbjct: 542 WFVLTAESLSWYKDEEEKEKKYMLPLDNLKIRDVEKGFMSNKHVFAIFNTEQRNVY-KDL 600
Query: 609 TSTKPHAEPVHNVQSSSTIHLREPPPVLRPSESNSEMEG--------------VEITVTK 654
+ + +V S LR + P + +E E ++ +
Sbjct: 601 RQIELACDSQEDVDSWKASFLRAG---VYPEKDQAENEDGAQENTFSMDPQLERQVETIR 657
Query: 655 LLLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAA 714
L+ SY I K+I DL+PK IMH ++NNTK +H+ + LY +++E AE A
Sbjct: 658 NLVDSYVAIINKSIRDLMPKTIMHLMINNTKAFIHHELLAYLYSSADQSSLMEESAEQAQ 717
Query: 715 KRKRCRELLRAYQQAFRDLDEL 736
+R + A ++A + ++
Sbjct: 718 RRDDMLRMYHALKEALNIIGDI 739
>DYN2_MOUSE (P39054) Dynamin 2 (EC 3.6.5.5) (Dynamin UDNM)
Length = 870
Score = 478 bits (1229), Expect = e-134
Identities = 284/742 (38%), Positives = 428/742 (57%), Gaps = 39/742 (5%)
Query: 23 VISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTRR 82
+I LVN+LQD FS +G QS +DLPQ+AVVG QS+GKSSVLE+ VGRDFLPRG+ I TRR
Sbjct: 9 LIPLVNKLQDAFSSIG-QSCHLDLPQIAVVGGQSAGKSSVLENFVGRDFLPRGSGIVTRR 67
Query: 83 PLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENKGVSEKQIRLKI 142
PL+LQL+ +K E+ EFLH +KF DF ++R EI+AETDR G NKG+S I L++
Sbjct: 68 PLILQLIFSKT---EYAEFLHCKSKKFTDFDEVRQEIEAETDRVTGTNKGISPVPINLRV 124
Query: 143 FSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAVTPANSDLANS 202
+SP+VL++TL+DLPGITKVPVGDQP DIE +I+ MI+ +I + LILAVTPAN DLANS
Sbjct: 125 YSPHVLNLTLIDLPGITKVPVGDQPPDIEYQIKDMILQFISRESSLILAVTPANMDLANS 184
Query: 203 DALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVGVVNRSQEDIQM 262
DAL++A+ DP G RTIGVITKLD+MD GTDAR++L K++PLR GY+GVVNRSQ+DI+
Sbjct: 185 DALKLAKEVDPQGLRTIGVITKLDLMDEGTDARDVLENKLLPLRRGYIGVVNRSQKDIEG 244
Query: 263 NRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLPGLKARISTSL 322
+ I+ AL AE +FF S P Y +AD G P L K LN+ L HI+ LP L++++ + L
Sbjct: 245 KKDIRAALAAERKFFLSHPAYRHMADRMGTPHLQKTLNQQLTNHIRESLPTLRSKLQSQL 304
Query: 323 VALAKEHASYGEI-TESKAGQGALVLNILSKYCDAFSSMLEGKNKEMSTCELSGGARIHY 381
++L KE Y + + +L ++ ++ F +EG ++ T ELSGGARI+
Sbjct: 305 LSLEKEVEEYKNFRPDDPTRKTKALLQMVQQFGVDFEKRIEGSGDQVDTLELSGGARINR 364
Query: 382 IFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQISRLLDPS 441
IF + L ++ E +I A++N G ++ LF P++ FE V++Q+ +L +P
Sbjct: 365 IFHERFPFELVKMEFDEKDLRREISYAIKNIHGVRTGLFTPDLAFEAIVKKQVVKLKEPC 424
Query: 442 LQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIGNFLRDGLEPSETMITHIIEMEM 501
L+C + EL+ +C ++L +P LR+ + ++ ++R+ ++ I +I++E
Sbjct: 425 LKCVDLVIQELISTVRQCT-SKLSSYPRLREETERIVTTYIREREGRTKDQILLLIDIEQ 483
Query: 502 DYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVSLDALESDKGLASERNGKSRGIIARQA 561
YINT+H +FIG + A + + Q K + P + L +G + N +++
Sbjct: 484 SYINTNHEDFIGFANAQQRSTQLNKKRAI--PNQGEILVIRRGWLTINNISLMKGGSKEY 541
Query: 562 NGVVADHGVRGASDVE-------------KVAPSGNAGGSSWRISSIFGGRDNRVYVKEN 608
V+ + D E K+ S+ + +IF VY K+
Sbjct: 542 WFVLTAESLSWYKDEEEKEKKYMLPLDNLKIRDVEKGFMSNKHVFAIFNTEQRNVY-KDL 600
Query: 609 TSTKPHAEPVHNVQSSSTIHLREPPPVLRPSESNSEMEG--------------VEITVTK 654
+ + +V S LR + P + +E E ++ +
Sbjct: 601 RQIELACDSQEDVDSWKASFLRAG---VYPEKDQAENEDGAQENTFSMDPQLERQVETIR 657
Query: 655 LLLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAA 714
L+ SY I K+I DL+PK IMH ++NNTK +H+ + LY +++E AE A
Sbjct: 658 NLVDSYVAIINKSIRDLMPKTIMHLMINNTKAFIHHELLAYLYSSADQSSLMEESAEQAQ 717
Query: 715 KRKRCRELLRAYQQAFRDLDEL 736
+R + A ++A + ++
Sbjct: 718 RRDDMLRMYHALKEALNIIGDI 739
>DYN2_HUMAN (P50570) Dynamin 2 (EC 3.6.5.5)
Length = 870
Score = 477 bits (1228), Expect = e-134
Identities = 283/742 (38%), Positives = 427/742 (57%), Gaps = 39/742 (5%)
Query: 23 VISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTRR 82
+I LVN+LQD FS +G QS +DLPQ+AVVG QS+GKSSVLE+ VGRDFLPRG+ I TRR
Sbjct: 9 LIPLVNKLQDAFSSIG-QSCHLDLPQIAVVGGQSAGKSSVLENFVGRDFLPRGSGIVTRR 67
Query: 83 PLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENKGVSEKQIRLKI 142
PL+LQL+ +K H EFLH +KF DF ++R EI+AETDR G NKG+S I L++
Sbjct: 68 PLILQLIFSKTEH---AEFLHCKSKKFTDFDEVRQEIEAETDRVTGTNKGISPVPINLRV 124
Query: 143 FSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAVTPANSDLANS 202
+SP+VL++TL+DLPGITKVPVGDQP DIE +I+ MI+ +I + LILAVTPAN DLANS
Sbjct: 125 YSPHVLNLTLIDLPGITKVPVGDQPPDIEYQIKDMILQFISRESSLILAVTPANMDLANS 184
Query: 203 DALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVGVVNRSQEDIQM 262
DAL++A+ DP G RTIGVITKLD+MD GTDAR++L K++PLR GY+GVVNRSQ+DI+
Sbjct: 185 DALKLAKEVDPQGLRTIGVITKLDLMDEGTDARDVLENKLLPLRRGYIGVVNRSQKDIEG 244
Query: 263 NRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLPGLKARISTSL 322
+ I+ AL AE +FF S P Y +AD G P L K LN+ L HI+ LP L++++ + L
Sbjct: 245 KKDIRAALAAERKFFLSHPAYRHMADRMGTPHLQKTLNQQLTNHIRESLPALRSKLQSQL 304
Query: 323 VALAKEHASYGEI-TESKAGQGALVLNILSKYCDAFSSMLEGKNKEMSTCELSGGARIHY 381
++L KE Y + + +L ++ ++ F +EG ++ T ELSGGARI+
Sbjct: 305 LSLEKEVEEYKNFRPDDPTRKTKALLQMVQQFGVDFEKRIEGSGDQVDTLELSGGARINR 364
Query: 382 IFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQISRLLDPS 441
IF + L ++ E +I A++N G ++ LF P++ FE V++Q+ +L +P
Sbjct: 365 IFHERFPFELVKMEFDEKDLRREISYAIKNIHGVRTGLFTPDLAFEAIVKKQVVKLKEPC 424
Query: 442 LQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIGNFLRDGLEPSETMITHIIEMEM 501
L+C + EL+ +C ++L +P LR+ + ++ ++R+ ++ I +I++E
Sbjct: 425 LKCVDLVIQELINTVRQCT-SKLSSYPRLREETERIVTTYIREREGRTKDQILLLIDIEQ 483
Query: 502 DYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVSLDALESDKGLASERNGKSRGIIARQA 561
YINT+H +FIG + A + + Q K + P + L +G + N +++
Sbjct: 484 SYINTNHEDFIGFANAQQRSTQLNKKRAI--PNQGEILVIRRGWLTINNISLMKGGSKEY 541
Query: 562 NGVVADHGVRGASDVE-------------KVAPSGNAGGSSWRISSIFGGRDNRVYVKEN 608
V+ + D E K+ S+ + +IF VY K+
Sbjct: 542 WFVLTAESLSWYKDEEEKEKKYMLPLDNLKIRDVEKGFMSNKHVFAIFNTEQRNVY-KDL 600
Query: 609 TSTKPHAEPVHNVQSSSTIHLREPPPVLRPSESNSEMEG--------------VEITVTK 654
+ + +V S LR + P + +E E ++ +
Sbjct: 601 RQIELACDSQEDVDSWKASFLRAG---VYPEKDQAENEDGAQENTFSMDPQLERQVETIR 657
Query: 655 LLLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAA 714
L+ SY I K+I DL+PK IMH ++NNTK +H+ + LY +++E A+ A
Sbjct: 658 NLVDSYVAIINKSIRDLMPKTIMHLMINNTKAFIHHELLAYLYSSADQSSLMEESADQAQ 717
Query: 715 KRKRCRELLRAYQQAFRDLDEL 736
+R + A ++A + ++
Sbjct: 718 RRDDMLRMYHALKEALNIIGDI 739
>DYN1_CAEEL (P39055) Dynamin (EC 3.6.5.5)
Length = 830
Score = 451 bits (1160), Expect = e-126
Identities = 278/742 (37%), Positives = 418/742 (55%), Gaps = 39/742 (5%)
Query: 22 SVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTR 81
++I ++NR+QD FS++G S +LPQ+AVVG QS+GKSSVLE+ VG+DFLPRG+ I TR
Sbjct: 10 ALIPVINRVQDAFSQLGT-SVSFELPQIAVVGGQSAGKSSVLENFVGKDFLPRGSGIVTR 68
Query: 82 RPLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENKGVSEKQIRLK 141
RPL+LQL+ + E+ EFLH G +F DF +R EI+ ETDR G+NKG+S I L+
Sbjct: 69 RPLILQLIQDRN---EYAEFLHKKGHRFVDFDAVRKEIEDETDRVTGQNKGISPHPINLR 125
Query: 142 IFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAVTPANSDLAN 201
+FSPNVL++TL+DLPG+TKVPVGDQP+DIE +IR MI+++I TCLILAVTPANSDLA
Sbjct: 126 VFSPNVLNLTLIDLPGLTKVPVGDQPADIEQQIRDMILTFINRETCLILAVTPANSDLAT 185
Query: 202 SDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVGVVNRSQEDIQ 261
SDAL++A+ DP G RTIGV+TKLD+MD GTDAR +L K+ LR GYVGVVNR Q+DI
Sbjct: 186 SDALKLAKEVDPQGLRTIGVLTKLDLMDEGTDAREILENKLFTLRRGYVGVVNRGQKDIV 245
Query: 262 MNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLPGLKARISTS 321
+ I+ AL AE +FF S P Y +AD G L LN+ L HI+ LP L+ +
Sbjct: 246 GRKDIRAALDAERKFFISHPSYRHMADRLGTSYLQHTLNQQLTNHIRDTLPTLRDSLQKK 305
Query: 322 LVALAKEHASYGEITESKAGQGA-LVLNILSKYCDAFSSMLEGKN-KEMSTCELSGGARI 379
+ A+ K+ A Y + G+ +L +++++ +EG + K +ST ELSGGARI
Sbjct: 306 MFAMEKDVAEYKNYQPNDPGRKTKALLQMVTQFNADIERSIEGSSAKLVSTNELSGGARI 365
Query: 380 HYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQISRLLD 439
+ +F + + ++ E +I+ A++N G + LF P++ FE ++QI+RL +
Sbjct: 366 NRLFHERFPFEIVKMEIDEKEMRKEIQYAIRNIHGIRVGLFTPDMAFEAIAKKQITRLKE 425
Query: 440 PSLQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIGNFLRDGLEPSETMITHIIEM 499
PSL+C + +EL + +C T + R+P LR ++ ++ + +R+ + ++ I I++
Sbjct: 426 PSLKCVDLVVNELANVIRQCADT-MARYPRLRDELERIVVSHMREREQIAKQQIGLIVDY 484
Query: 500 EMDYINTSHPNFIGGSKALEAAVQ-QTKPSRVSPPVSLDALESDKGLASERNGKSRGIIA 558
E+ Y+NT+H +FIG S A A Q Q+ + V S ++ R K
Sbjct: 485 ELAYMNTNHEDFIGFSNAEAKASQGQSAKKNLGNQVIRKGWLSLSNVSFVRGSKDNWF-- 542
Query: 559 RQANGVVADHGVRGASDVEKVAPSGNAGGSSWRISSIFGG------------RDNRVYVK 606
V+ + D E+ ++ I GG D + K
Sbjct: 543 -----VLMSDSLSWYKDDEEKEKKYMLPLDGVKLKDIEGGFMSRNHKFALFYPDGKNIYK 597
Query: 607 ENTSTKPHAEPVHNVQSSSTIHLR----EPPPVLRPSESNSEMEGV--------EITVTK 654
+ + + + + LR + ES EME ++ +
Sbjct: 598 DYKQLELGCTNLDEIDAWKASFLRAGVYPEKQKAQEDESQQEMEDTSIDPQLERQVETIR 657
Query: 655 LLLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAA 714
L+ SY I K I+DLVPKA+MH +VN T + + + LY+ + +++E A
Sbjct: 658 NLVDSYMRIITKTIKDLVPKAVMHLIVNQTGEFMKDELLAHLYQCGDTDALMEESQIEAQ 717
Query: 715 KRKRCRELLRAYQQAFRDLDEL 736
KR+ + A ++A R + E+
Sbjct: 718 KREEMLRMYHACKEALRIISEV 739
>DYN_DROME (P27619) Dynamin (EC 3.6.5.5) (dDyn) (Shibire protein)
Length = 877
Score = 450 bits (1158), Expect = e-126
Identities = 278/747 (37%), Positives = 419/747 (55%), Gaps = 41/747 (5%)
Query: 22 SVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTR 81
S+I++VN+LQD F+ +G + DLPQ+AVVG QS+GKSSVLE+ VG+DFLPRG+ I TR
Sbjct: 3 SLITIVNKLQDAFTSLGVHMQL-DLPQIAVVGGQSAGKSSVLENFVGKDFLPRGSGIVTR 61
Query: 82 RPLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENKGVSEKQIRLK 141
RPL+LQL++ E+GEFLH+ G+KF F +IR EI+ ETDR G NKG+S I L+
Sbjct: 62 RPLILQLIN---GVTEYGEFLHIKGKKFSSFDEIRKEIEDETDRVTGSNKGISNIPINLR 118
Query: 142 IFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAVTPANSDLAN 201
++SP+VL++TL+DLPG+TKV +GDQP DIE +I+ MI +I+ TCLILAVTPAN+DLAN
Sbjct: 119 VYSPHVLNLTLIDLPGLTKVAIGDQPVDIEQQIKQMIFQFIRKETCLILAVTPANTDLAN 178
Query: 202 SDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVGVVNRSQEDIQ 261
SDAL++A+ DP G RTIGVITKLD+MD GTDAR++L K++PLR GY+GVVNRSQ+DI+
Sbjct: 179 SDALKLAKEVDPQGVRTIGVITKLDLMDEGTDARDILENKLLPLRRGYIGVVNRSQKDIE 238
Query: 262 MNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLPGLKARISTS 321
+ I AL AE +FF S P Y +AD G P L + LN+ L HI+ LPGL+ ++
Sbjct: 239 GRKDIHQALAAERKFFLSHPSYRHMADRLGTPYLQRVLNQQLTNHIRDTLPGLRDKLQKQ 298
Query: 322 LVALAKEHASYGEITESKAG-QGALVLNILSKYCDAFSSMLEGKNKEM-STCELSGGARI 379
++ L KE + A + +L ++ + F +EG + +T ELSGGA+I
Sbjct: 299 MLTLEKEVEEFKHFQPGDASIKTKAMLQMIQQLQSDFERTIEGSGSALVNTNELSGGAKI 358
Query: 380 HYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQISRLLD 439
+ IF + + E +I A++N G + LF P++ FE V+RQI+ L +
Sbjct: 359 NRIFHERLRFEIVKMACDEKELRREISFAIRNIHGIRVGLFTPDMAFEAIVKRQIALLKE 418
Query: 440 PSLQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIGNFLRDGLEPSETMITHIIEM 499
P ++C + EL + C ++ R+P LR+ + +I +R + I +I+
Sbjct: 419 PVIKCVDLVVQELSVVVRMCT-AKMSRYPRLREETERIITTHVRQREHSCKEQILLLIDF 477
Query: 500 EMDYINTSHPNFIGGSKALEAAVQQTKPSR--------VSPPVSLDALESDKG------- 544
E+ Y+NT+H +FIG + A + K + + L KG
Sbjct: 478 ELAYMNTNHEDFIGFANAQNKSENANKTGTRQLGNQVIRKGHMVIQNLGIMKGGSRPYWF 537
Query: 545 -LASERNGKSRGIIARQANGVVADHGVRGASDVEKVAPSGNAGGSSWRISSIFGGRDNRV 603
L SE + ++ ++ G++ D+E+ S S R++ D R
Sbjct: 538 VLTSESISWYKDEDEKEKKFMLPLDGLK-LRDIEQGFMS-----MSRRVTFALFSPDGRN 591
Query: 604 YVKENTSTKPHAEPVHNVQSSSTIHLREPP-PVLRPSESNSEMEGVE-----------IT 651
K+ + E V +V+S LR P + ++ N + E +
Sbjct: 592 VYKDYKQLELSCETVEDVESWKASFLRAGVYPEKQETQENGDESASEESSSDPQLERQVE 651
Query: 652 VTKLLLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAE 711
+ L+ SY I K D+VPKAIM ++NN K ++ + LY +M++E AE
Sbjct: 652 TIRNLVDSYMKIVTKTTRDMVPKAIMMLIINNAKDFINGELLAHLYASGDQAQMMEESAE 711
Query: 712 VAAKRKRCRELLRAYQQAFRDLDELPL 738
A +R+ + RA + A + + ++ +
Sbjct: 712 SATRREEMLRMYRACKDALQIIGDVSM 738
>DYNL_ARATH (P42697) Dynamin-like protein
Length = 610
Score = 322 bits (826), Expect = 2e-87
Identities = 191/493 (38%), Positives = 286/493 (57%), Gaps = 18/493 (3%)
Query: 22 SVISLVNRLQDIFSRVGNQS------TIID-LPQVAVVGSQSSGKSSVLESLVGRDFLPR 74
++ISLVN++Q + +G+ T+ D LP +AVVG QSSGKSSVLES+VG+DFLPR
Sbjct: 3 NLISLVNKIQRACTALGDHGDSSALPTLWDSLPAIAVVGGQSSGKSSVLESIVGKDFLPR 62
Query: 75 GTDICTRRPLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENKGVS 134
G+ I TRRPLVLQL E+ EFLHLP +KF DF +R EIQ ETDRE G +K +S
Sbjct: 63 GSGIVTRRPLVLQLQKIDDGTREYAEFLHLPRKKFTDFAAVRKEIQDETDRETGRSKAIS 122
Query: 135 EKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAVTP 194
I L I+SPNV+++TL+DLPG+TKV V Q I I M+ SYI+ P C+ILA++P
Sbjct: 123 SVPIHLSIYSPNVVNLTLIDLPGLTKVAVDGQSDSIVKDIENMVRSYIEKPNCIILAISP 182
Query: 195 ANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVGVVN 254
AN DLA SDA+++++ DP G+RT GV+TK+D+MD+GTDA +L G+ L+ +VGVVN
Sbjct: 183 ANQDLATSDAIKISREVDPSGDRTFGVLTKIDLMDKGTDAVEILEGRSFKLKYPWVGVVN 242
Query: 255 RSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLPGL 314
RSQ DI N + A E E+F + Y LA+ G LAK L+K L + IK+ +PG+
Sbjct: 243 RSQADINKNVDMIAARKREREYFSNTTEYRHLANKMGSEHLAKMLSKHLERVIKSRIPGI 302
Query: 315 KARISTSLVALAKEHASYGE-ITESKAGQGALVLNILSKYCDAFSSMLEGKNKEMSTCEL 373
++ I+ +++ L E + G+ I G+ ++ I + F L+G
Sbjct: 303 QSLINKTVLELETELSRLGKPIAADAGGKLYSIMEICRLFDQIFKEHLDGVR-------- 354
Query: 374 SGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQ 433
+GG +++ +F + +L+ + + L D+IR + A G + L PE + +
Sbjct: 355 AGGEKVYNVFDNQLPAALKRLQFDKQLAMDNIRKLVTEADGYQPHLIAPEQGYRRLIESS 414
Query: 434 ISRLLDPSLQCARFIYDELMKISHRCM--FTELQRFPFLRKHMDEVIGNFLRDGLEPSET 491
I + P+ ++ L + H+ + EL+++P LR + L E S+
Sbjct: 415 IVSIRGPAEASVDTVHAILKDLVHKSVNETVELKQYPALRVEVTNAAIESLDKMREGSKK 474
Query: 492 MITHIIEMEMDYI 504
+++ME Y+
Sbjct: 475 ATLQLVDMECSYL 487
>MX3_RAT (P18590) Interferon-induced GTP-binding protein Mx3
Length = 659
Score = 211 bits (536), Expect = 8e-54
Identities = 143/405 (35%), Positives = 223/405 (54%), Gaps = 10/405 (2%)
Query: 46 LPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTRRPLVLQLVHTKPSHPEFGEFLHLP 105
LP +AV+G QSSGKSSVLE+L G LPRG+ I TR PLVL+L G+ +
Sbjct: 68 LPAIAVIGDQSSGKSSVLEALSGVA-LPRGSGIVTRCPLVLKLKKLNQGEEWKGKVTYDD 126
Query: 106 GR-KFHDFTQIRHEIQAETDREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVG 164
+ D +++ I + AG G+S+K I L + SP+V D+TL+DLPGIT+V VG
Sbjct: 127 IEVELSDPSEVEEAINTGQNHIAGVGLGISDKLISLDVSSPHVPDLTLIDLPGITRVAVG 186
Query: 165 DQPSDIEARIRTMIMSYIKIPTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITK 224
+QPSDI +I+ +I +YI+ + L V P+N D+A ++AL MAQ DPDG+RTIG++TK
Sbjct: 187 NQPSDIGRQIKRLITNYIQKQETINLVVVPSNVDIATTEALSMAQEVDPDGDRTIGILTK 246
Query: 225 LDIMDRGTDAR--NLLLGKVIPLRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPV 282
D++DRGT+ + +++ V L+ GY+ V R Q+DIQ S+ +AL E+ FF+ P
Sbjct: 247 PDLVDRGTEDKVVDVVRNLVCHLKKGYMIVKCRGQQDIQEQLSLAEALQKEQVFFKEHPQ 306
Query: 283 YSGLAD--SCGVPQLAKKLNKILAQHIKAVLPGLKARISTSLVALAKEHASYG-EITESK 339
+ L + VP LA++L L HI LP L+ +I S + ++E YG +I E +
Sbjct: 307 FRALLEDGKATVPCLAERLTMELISHICKSLPLLENQIKESHQSTSEELQKYGADIPEDE 366
Query: 340 AGQGALVLNILSKYCDAFSSMLEGKN-KEMSTCELSGGARIHYIFQSIYVRSLEAVDPCE 398
+ ++ ++ + ++++EG+ C L R + S +
Sbjct: 367 NEKTLFLIEKINAFNQDITAIVEGEEIVREKECRLFTKLRKEFFLWSEEIE--RNFQKGS 424
Query: 399 DLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQISRLLDPSLQ 443
D ++ T G + F+ FE +RRQI L +P+++
Sbjct: 425 DALYKEVYTFEMQYRGRELPGFVNYKTFENIIRRQIKTLEEPAME 469
>MX1_PIG (P27594) Interferon-induced GTP-binding protein Mx1
Length = 663
Score = 210 bits (534), Expect = 1e-53
Identities = 154/472 (32%), Positives = 248/472 (51%), Gaps = 29/472 (6%)
Query: 46 LPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTRRPLVLQLVHTKPSHPEF-GEFLHL 104
LP +AV+G QSSGKSSVLE+L G LPRG+ I TR PLVL+L E+ G+ +
Sbjct: 70 LPAIAVIGDQSSGKSSVLEALSGVA-LPRGSGIVTRCPLVLKLKKLVNEEDEWKGKVSYR 128
Query: 105 PGR-KFHDFTQIRHEIQAETDREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPV 163
+ D +Q+ E+ A AGE G+S + I L++ SP+V D+TL+DLPGIT+V V
Sbjct: 129 DSEIELSDASQVEKEVSAAQIAIAGEGVGISHELISLEVSSPHVPDLTLIDLPGITRVAV 188
Query: 164 GDQPSDIEARIRTMIMSYIKIPTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVIT 223
G+QP DIE +I+++I YI + L V P N D+A ++AL+MAQ DP+G+RTIG++T
Sbjct: 189 GNQPYDIEYQIKSLIKKYICKQETINLVVVPCNVDIATTEALRMAQEVDPEGDRTIGILT 248
Query: 224 KLDIMDRGTD------ARNLLLGKVIPLRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFF 277
K D++D+GT+ ARNL V L+ GY+ V R Q+DIQ S+ AL E+ FF
Sbjct: 249 KPDLVDKGTEDKIVDVARNL----VFHLKKGYMIVKCRGQQDIQEQLSLAKALQKEQAFF 304
Query: 278 RSRPVYSGLADS--CGVPQLAKKLNKILAQHIKAVLPGLKARISTSLVALAKEHASYG-E 334
+ + L + +P LA++L L HI LP L+ +I S + +E YG +
Sbjct: 305 ENHAHFRDLLEEGRATIPCLAERLTSELIMHICKTLPLLENQIKESHQKITEELQKYGSD 364
Query: 335 ITESKAGQGALVLNILSKYCDAFSSMLEGKNKEMS-TCELSGGARIHY-----IFQSIYV 388
I E ++G+ +++ + + +++++G+ + C L R + + + +
Sbjct: 365 IPEDESGKMFFLIDKIDAFNSDITALIQGEELVVEYECRLFTKMRNEFCRWSAVVEKNFK 424
Query: 389 RSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQISRLLDPSLQCARFI 448
+A+ L ++ R G + F+ FE +++Q+S L +P++ +
Sbjct: 425 NGYDAICKQIQLFENQYR-------GRELPGFVNYKTFETIIKKQVSVLEEPAVDMLHTV 477
Query: 449 YDELMKISHRCMFTELQRFPFLRKHMDEVIGNFLRDGLEPSETMITHIIEME 500
D + T F L + I + + + +ET I +ME
Sbjct: 478 TDLVRLAFTDVSETNFNEFFNLHRTAKSKIEDIKLEQEKEAETSIRLHFQME 529
>MX2_RAT (P18589) Interferon-induced GTP-binding protein Mx2
Length = 659
Score = 209 bits (533), Expect = 2e-53
Identities = 142/405 (35%), Positives = 223/405 (55%), Gaps = 10/405 (2%)
Query: 46 LPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTRRPLVLQLVHTKPSHPEFGEFLHLP 105
LP +AV+G QSSGKSSVLE+L G LPRG+ I TR PLVL+L G+ +
Sbjct: 68 LPAIAVIGDQSSGKSSVLEALSGVA-LPRGSGIVTRCPLVLKLKKLNQGEEWKGKVTYDD 126
Query: 106 GR-KFHDFTQIRHEIQAETDREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVG 164
+ D +++ I + AG G+S+K I L + SP+V D+TL+DLPGIT+V VG
Sbjct: 127 IEVELSDPSEVEEAINTGQNHIAGVGLGISDKLISLDVSSPHVPDLTLIDLPGITRVAVG 186
Query: 165 DQPSDIEARIRTMIMSYIKIPTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITK 224
+QP+DI +I+ +I +YI+ + L V P+N D+A ++AL MAQ DPDG+RTIG++TK
Sbjct: 187 NQPADIGRQIKRLITNYIQKQETINLVVVPSNVDIATTEALSMAQKVDPDGDRTIGILTK 246
Query: 225 LDIMDRGTDAR--NLLLGKVIPLRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPV 282
D++DRGT+ + +++ V L+ GY+ V R Q+DIQ S+ +AL E+ FF+ P
Sbjct: 247 PDLVDRGTEDKVVDVVRNLVCHLKKGYMIVKCRGQQDIQEQLSLAEALQKEQVFFKEHPQ 306
Query: 283 YSGLAD--SCGVPQLAKKLNKILAQHIKAVLPGLKARISTSLVALAKEHASYG-EITESK 339
+ L + VP LA++L L HI LP L+ +I S + ++E YG +I E +
Sbjct: 307 FRALLEDGKATVPCLAERLTMELISHICKSLPLLENQIKESHQSTSEELQKYGADIPEDE 366
Query: 340 AGQGALVLNILSKYCDAFSSMLEGKN-KEMSTCELSGGARIHYIFQSIYVRSLEAVDPCE 398
+ ++ ++ + ++++EG+ C L R + S +
Sbjct: 367 NEKTLFLIEKINAFNQDITAIVEGEEIVREKECRLFTKLRKEFFLWSEEIE--RNFQKGS 424
Query: 399 DLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQISRLLDPSLQ 443
D ++ T G + F+ FE +RRQI L +P+++
Sbjct: 425 DALYKEVYTFEMQYRGRELPGFVNYKTFENIIRRQIKTLEEPAME 469
>MX1_RAT (P18588) Interferon-induced GTP-binding protein Mx1
Length = 652
Score = 204 bits (520), Expect = 5e-52
Identities = 138/404 (34%), Positives = 222/404 (54%), Gaps = 8/404 (1%)
Query: 46 LPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTRRPLVLQLVHTKPSHPEFGEFLHLP 105
LP +AV+G QSSGKSSVLE+L G LPRG+ I TR PLVL+L K G+ ++
Sbjct: 61 LPAIAVIGDQSSGKSSVLEALSGVA-LPRGSGIVTRCPLVLKLKQLKQGEKWSGKVIYKD 119
Query: 106 GR-KFHDFTQIRHEIQAETDREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVG 164
+ + + EI + AGE +S I L++ SP+V D+TL+DLPGIT+V VG
Sbjct: 120 TEIEISHPSLVEREINKAQNLIAGEGLKISSDLISLEVSSPHVPDLTLIDLPGITRVAVG 179
Query: 165 DQPSDIEARIRTMIMSYIKIPTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITK 224
DQP+DIE +I+ +I YI+ + L V P+N D+A ++AL+MAQ DP G+RTIG++TK
Sbjct: 180 DQPADIEHKIKRLITEYIQKQETINLVVVPSNVDIATTEALKMAQEVDPQGDRTIGILTK 239
Query: 225 LDIMDRGTDAR--NLLLGKVIPLRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPV 282
D++DRGT+ + +++ V L+ GY+ V R Q+DIQ S+ +AL E+ FF+ P
Sbjct: 240 PDLVDRGTEDKVVDVVRNLVCHLKKGYMIVKCRGQQDIQEQLSLAEALQKEQVFFKEHPQ 299
Query: 283 YSGLAD--SCGVPQLAKKLNKILAQHIKAVLPGLKARISTSLVALAKEHASYG-EITESK 339
+ L + VP LAK+L L HI LP L+ +I+ + ++E YG +I E
Sbjct: 300 FRVLLEDGKATVPCLAKRLTMELTSHICKSLPILENQINVNHQIASEELQKYGADIPEDD 359
Query: 340 AGQGALVLNILSKYCDAFSSMLEG-KNKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCE 398
+ + + ++N ++ + S+++ +N L R ++ + Y+
Sbjct: 360 SKRLSFLMNKINVFNKDILSLVQAQENISWEESRLFTKLRNEFLAWNDYIEEHFKKTLGS 419
Query: 399 DLTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQISRLLDPSL 442
+ + G + F+ FE +++++ L +P+L
Sbjct: 420 SEKHSQMEKFESHYRGRELPGFVDYKAFENIIKKEVKALEEPAL 463
>MX1_MOUSE (P09922) Interferon-induced GTP-binding protein Mx1
(Influenza resistance protein)
Length = 631
Score = 203 bits (516), Expect = 2e-51
Identities = 127/314 (40%), Positives = 193/314 (61%), Gaps = 7/314 (2%)
Query: 46 LPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTRRPLVLQLVHTKPSHPEFGEFLHLP 105
LP +AV+G QSSGKSSVLE+L G LPRG+ I TR PLVL+L K G+ +
Sbjct: 36 LPAIAVIGDQSSGKSSVLEALSGVA-LPRGSGIVTRCPLVLKLRKLKEGEEWRGKVSYDD 94
Query: 106 GR-KFHDFTQIRHEIQAETDREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVG 164
+ D +++ I + AG G+S+K I L + SPNV D+TL+DLPGIT+V VG
Sbjct: 95 IEVELSDPSEVEEAINKGQNFIAGVGLGISDKLISLDVSSPNVPDLTLIDLPGITRVAVG 154
Query: 165 DQPSDIEARIRTMIMSYIKIPTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITK 224
+QP+DI +I+ +I +YI+ + L V P+N D+A ++AL MAQ DP+G+RTIGV+TK
Sbjct: 155 NQPADIGRQIKRLIKTYIQKQETINLVVVPSNVDIATTEALSMAQEVDPEGDRTIGVLTK 214
Query: 225 LDIMDRGTDAR--NLLLGKVIPLRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPV 282
D++DRG + + +++ V PL+ GY+ V R Q+DIQ S+ +A E+ FF+
Sbjct: 215 PDLVDRGAEGKVLDVMRNLVYPLKKGYMIVKCRGQQDIQEQLSLTEAFQKEQVFFKDHSY 274
Query: 283 YSGLAD--SCGVPQLAKKLNKILAQHIKAVLPGLKARISTSLVALAKEHASYG-EITESK 339
+S L + VP LA++L + L HI LP L+ +I++S + ++E YG +I E
Sbjct: 275 FSILLEDGKATVPCLAERLTEELTSHICKSLPLLEDQINSSHQSASEELQKYGADIPEDD 334
Query: 340 AGQGALVLNILSKY 353
+ + ++N +S +
Sbjct: 335 RTRMSFLVNKISAF 348
Score = 37.4 bits (85), Expect = 0.15
Identities = 22/96 (22%), Positives = 46/96 (47%), Gaps = 2/96 (2%)
Query: 640 ESNSEMEGVEITVTKLL--LRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLY 697
++NS+ +T T++ L++YY R+NI +P I +F++ E+ + ++ L
Sbjct: 531 QNNSQFPQKGLTTTEMTQHLKAYYQECRRNIGRQIPLIIQYFILKTFGEEIEKMMLQLLQ 590
Query: 698 REDLFEEMLQEPAEVAAKRKRCRELLRAYQQAFRDL 733
L+E ++ K+K + L +A + L
Sbjct: 591 DTSKCSWFLEEQSDTREKKKFLKRRLLRLDEARQKL 626
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.135 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 87,239,191
Number of Sequences: 164201
Number of extensions: 3758164
Number of successful extensions: 11162
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 45
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 10967
Number of HSP's gapped (non-prelim): 103
length of query: 773
length of database: 59,974,054
effective HSP length: 118
effective length of query: 655
effective length of database: 40,598,336
effective search space: 26591910080
effective search space used: 26591910080
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0100c.1


