

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0100b.1
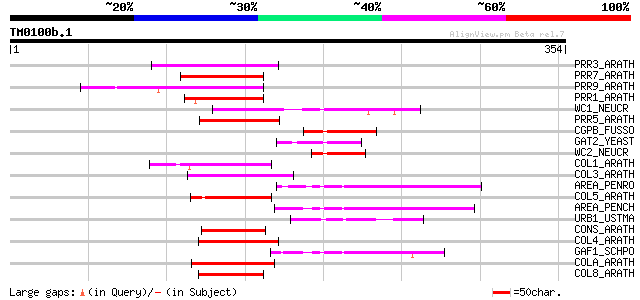
(354 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
PRR3_ARATH (Q9LVG4) Two-component response regulator-like APRR3 ... 61 4e-09
PRR7_ARATH (Q93WK5) Two-component response regulator-like APRR7 ... 57 7e-08
PRR9_ARATH (Q8L500) Two-component response regulator-like APRR9 ... 54 5e-07
PRR1_ARATH (Q9LKL2) Two-component response regulator-like APRR1 ... 54 6e-07
WC1_NEUCR (Q01371) White collar 1 protein (WC1) 52 2e-06
PRR5_ARATH (Q6LA42) Two-component response regulator-like APRR5 ... 52 2e-06
CGPB_FUSSO (Q00858) Cutinase gene palindrome-binding protein (PBP) 52 3e-06
GAT2_YEAST (P40209) GAT2 protein 51 5e-06
WC2_NEUCR (P78714) White collar 2 protein (WC2) 50 9e-06
COL1_ARATH (O50055) Zinc finger protein CONSTANS-LIKE 1 50 1e-05
COL3_ARATH (Q9SK53) Zinc finger protein CONSTANS-LIKE 3 47 6e-05
AREA_PENRO (O13508) Nitrogen regulatory protein areA (Nitrogen r... 47 6e-05
COL5_ARATH (Q9FHH8) Zinc finger protein CONSTANS-LIKE 5 46 2e-04
AREA_PENCH (Q01582) Nitrogen regulatory protein areA (Nitrogen r... 45 2e-04
URB1_USTMA (P40349) Siderophore biosynthesis regulatory protein ... 45 3e-04
CONS_ARATH (Q39057) Zinc finger protein CONSTANS 45 3e-04
COL4_ARATH (Q940T9) Zinc finger protein CONSTANS-LIKE 4 44 5e-04
GAF1_SCHPO (Q10280) Transcription factor gaf1 (Gaf-1) 44 6e-04
COLA_ARATH (Q9LUA9) Zinc finger protein CONSTANS-LIKE 10 44 6e-04
COL8_ARATH (Q9M9B3) Putative zinc finger protein CONSTANS-LIKE 8 44 8e-04
>PRR3_ARATH (Q9LVG4) Two-component response regulator-like APRR3
(Pseudo-response regulator 3)
Length = 495
Score = 61.2 bits (147), Expect = 4e-09
Identities = 32/81 (39%), Positives = 49/81 (59%)
Query: 91 PNSTPNSGLLQQKNFPQIKGINDPSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKE 150
P+ +P + LL + S S ++R A+L++FR KRKERCFEKK+RY RK+
Sbjct: 410 PHDSPIAKLLGSSSSSDNPLKQQSSGSDRWAQREAALMKFRLKRKERCFEKKVRYHSRKK 469
Query: 151 VAQRMHRKNGQFASLKEDYKS 171
+A++ GQF ++D+KS
Sbjct: 470 LAEQRPHVKGQFIRKRDDHKS 490
>PRR7_ARATH (Q93WK5) Two-component response regulator-like APRR7
(Pseudo-response regulator 7)
Length = 727
Score = 57.0 bits (136), Expect = 7e-08
Identities = 25/53 (47%), Positives = 38/53 (71%)
Query: 110 GINDPSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQF 162
G + + + +S+R A+L +FR+KRKERCF KK+RY RK++A++ R GQF
Sbjct: 656 GSGNLADENKISQREAALTKFRQKRKERCFRKKVRYQSRKKLAEQRPRVRGQF 708
>PRR9_ARATH (Q8L500) Two-component response regulator-like APRR9
(Pseudo-response regulator 9)
Length = 468
Score = 54.3 bits (129), Expect = 5e-07
Identities = 37/126 (29%), Positives = 66/126 (52%), Gaps = 10/126 (7%)
Query: 46 ASNASAPHPRAGELTISFEGEVYVFPAVTPEKVQAVLLLLGQKDIPNS--------TPNS 97
++ S+ G ++SF +V + VT +K + + + ++ S + N
Sbjct: 332 SATTSSNQENIGSSSVSFRNQV-LQSTVTNQKQDSPIPVESNREKAASKEVEAGSQSTNE 390
Query: 98 GLL-QQKNFPQIKGINDPSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMH 156
G+ Q + + K Q + S+R A+L++FR KRK+RCF+KK+RY RK++A++
Sbjct: 391 GIAGQSSSTEKPKEEESAKQRWSRSQREAALMKFRLKRKDRCFDKKVRYQSRKKLAEQRP 450
Query: 157 RKNGQF 162
R GQF
Sbjct: 451 RVKGQF 456
>PRR1_ARATH (Q9LKL2) Two-component response regulator-like APRR1
(Pseudo-response regulator 1) (Timing of CAB expression
1) (ABI3-interacting protein 1)
Length = 618
Score = 53.9 bits (128), Expect = 6e-07
Identities = 27/54 (50%), Positives = 35/54 (64%), Gaps = 3/54 (5%)
Query: 112 NDPSQS---SNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQF 162
N PS + L RR +L++FR KR +RCF+KKIRY RK +A+R R GQF
Sbjct: 519 NPPSNEVRVNKLDRREEALLKFRRKRNQRCFDKKIRYVNRKRLAERRPRVKGQF 572
>WC1_NEUCR (Q01371) White collar 1 protein (WC1)
Length = 1167
Score = 52.4 bits (124), Expect = 2e-06
Identities = 40/147 (27%), Positives = 63/147 (42%), Gaps = 27/147 (18%)
Query: 130 FREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLKEDYKSPAGNLDSSNGTPPCPESI 189
F E + RC + ++V + + + Q S K+ K G ++
Sbjct: 882 FDELKTTRCTSWQYELRQMEKVNRMLAEELAQLLSNKKKRKRRKGG-----------GNM 930
Query: 190 ERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANK------------GTLRDLSKAA 237
R C +C T ++TP RRGP+G R LCN+CGL WA + G +SK +
Sbjct: 931 VRDCANCHT--RNTPEWRRGPSGNRDLCNSCGLRWAKQTGRVSPRTSSRGGNGDSMSKKS 988
Query: 238 KTPYEQN--ERDTSADMKPSTTEPENS 262
+P + R+ D +TT +NS
Sbjct: 989 NSPSHSSPLHREVGNDSPSTTTATKNS 1015
>PRR5_ARATH (Q6LA42) Two-component response regulator-like APRR5
(Pseudo-response regulator 5)
Length = 558
Score = 52.4 bits (124), Expect = 2e-06
Identities = 23/51 (45%), Positives = 36/51 (70%)
Query: 122 RRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLKEDYKSP 172
+R A+L +FR KRK+RC+EKK+RY RK++A++ R GQF + ++P
Sbjct: 508 QREAALTKFRMKRKDRCYEKKVRYESRKKLAEQRPRIKGQFVRQVQSTQAP 558
>CGPB_FUSSO (Q00858) Cutinase gene palindrome-binding protein (PBP)
Length = 457
Score = 51.6 bits (122), Expect = 3e-06
Identities = 24/47 (51%), Positives = 31/47 (65%), Gaps = 2/47 (4%)
Query: 188 SIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLS 234
S E C CGT + +P R+GP+GP++LCNACGL WA K R+ S
Sbjct: 397 SEEYVCTDCGTLD--SPEWRKGPSGPKTLCNACGLRWAKKEKKRNSS 441
>GAT2_YEAST (P40209) GAT2 protein
Length = 560
Score = 50.8 bits (120), Expect = 5e-06
Identities = 26/54 (48%), Positives = 32/54 (59%), Gaps = 5/54 (9%)
Query: 171 SPAGNLDSSNGTPPCPESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMW 224
+PA N D N P + I C HCG +E TP R+GP G R+LCNACGL +
Sbjct: 453 TPAANSDEKN---PNAKKIIEFCFHCGETE--TPEWRKGPYGTRTLCNACGLFY 501
>WC2_NEUCR (P78714) White collar 2 protein (WC2)
Length = 530
Score = 50.1 bits (118), Expect = 9e-06
Identities = 20/35 (57%), Positives = 26/35 (74%), Gaps = 2/35 (5%)
Query: 193 CQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANK 227
C CGT + +P R+GP+GP++LCNACGL WA K
Sbjct: 468 CTDCGTLD--SPEWRKGPSGPKTLCNACGLRWAKK 500
>COL1_ARATH (O50055) Zinc finger protein CONSTANS-LIKE 1
Length = 355
Score = 49.7 bits (117), Expect = 1e-05
Identities = 30/83 (36%), Positives = 44/83 (52%), Gaps = 7/83 (8%)
Query: 90 IPNSTPNSGLLQQKNFPQIKGIND-----PSQSSNLSRRFASLVRFREKRKERCFEKKIR 144
+P ST + + P K + D P+Q + R A ++R+REK+K R FEK IR
Sbjct: 250 VPESTTSDATVSNPRSP--KAVTDQPPYPPAQMLSPRDREARVLRYREKKKMRKFEKTIR 307
Query: 145 YTCRKEVAQRMHRKNGQFASLKE 167
Y RK A++ R G+FA K+
Sbjct: 308 YASRKAYAEKRPRIKGRFAKKKD 330
>COL3_ARATH (Q9SK53) Zinc finger protein CONSTANS-LIKE 3
Length = 294
Score = 47.4 bits (111), Expect = 6e-05
Identities = 25/68 (36%), Positives = 38/68 (55%)
Query: 114 PSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLKEDYKSPA 173
P+ + + R A ++R+REKRK R FEK IRY RK A+ R G+FA + ++
Sbjct: 220 PAVQLSPAEREARVLRYREKRKNRKFEKTIRYASRKAYAEMRPRIKGRFAKRTDSRENDG 279
Query: 174 GNLDSSNG 181
G++ G
Sbjct: 280 GDVGVYGG 287
>AREA_PENRO (O13508) Nitrogen regulatory protein areA (Nitrogen
regulator nmc)
Length = 860
Score = 47.4 bits (111), Expect = 6e-05
Identities = 38/131 (29%), Positives = 55/131 (41%), Gaps = 9/131 (6%)
Query: 171 SPAGNLDSSNGTPPCPESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTL 230
SP G S NG P + C +C T ++TP RR P G + LCNACGL G +
Sbjct: 644 SPGG---SKNGDPNAGPTT---CTNCFT--QTTPLWRRNPEG-QPLCNACGLFLKLHGVV 694
Query: 231 RDLSKAAKTPYEQNERDTSADMKPSTTEPENSCADQAEEGTPEENKPVPMDSRETPEKKN 290
R LS ++N S ++ + S + + P + M++ E+P N
Sbjct: 695 RPLSLKTDVIKKRNRSSASTLAVGTSRSSKKSSRKNSIQHAPSTSISSRMNTSESPPSMN 754
Query: 291 EQDILGTAQTV 301
LG V
Sbjct: 755 GSSSLGKTGVV 765
>COL5_ARATH (Q9FHH8) Zinc finger protein CONSTANS-LIKE 5
Length = 355
Score = 45.8 bits (107), Expect = 2e-04
Identities = 25/52 (48%), Positives = 33/52 (63%), Gaps = 1/52 (1%)
Query: 116 QSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLKE 167
Q+S++ R A ++R+REKRK R FEK IRY RK A+ R G+FA E
Sbjct: 279 QASSMDRE-ARVLRYREKRKNRKFEKTIRYASRKAYAESRPRIKGRFAKRTE 329
>AREA_PENCH (Q01582) Nitrogen regulatory protein areA (Nitrogen
regulator nre)
Length = 725
Score = 45.4 bits (106), Expect = 2e-04
Identities = 34/127 (26%), Positives = 53/127 (40%), Gaps = 8/127 (6%)
Query: 170 KSPAGNLDSSNGTPPCPESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGT 229
+ P G + S P P C +C T ++TP RR P G + LCNACGL G
Sbjct: 507 RQPGGLKNGSTNAGPEPA-----CTNCFT--QTTPLWRRNPEG-QPLCNACGLFLKLHGV 558
Query: 230 LRDLSKAAKTPYEQNERDTSADMKPSTTEPENSCADQAEEGTPEENKPVPMDSRETPEKK 289
+R LS ++N + ++ + S + + P + M++ E+P
Sbjct: 559 VRPLSLKTDVIKKRNRSSANTLTVGTSRSSKKSSRKNSIQHAPSTSISSRMNTSESPPSI 618
Query: 290 NEQDILG 296
N LG
Sbjct: 619 NGSSTLG 625
>URB1_USTMA (P40349) Siderophore biosynthesis regulatory protein
URBS1
Length = 950
Score = 45.1 bits (105), Expect = 3e-04
Identities = 29/85 (34%), Positives = 40/85 (46%), Gaps = 13/85 (15%)
Query: 180 NGTPPCPESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAAKT 239
+G+PP RC +CG + STP RR P G ++CNACGL + T R S
Sbjct: 325 SGSPPAAHHAGMRCSNCGVT--STPLWRRAPDGS-TICNACGLYIKSHSTHRSAS----- 376
Query: 240 PYEQNERDTSADMKPSTTEPENSCA 264
R + +D P T E + + A
Sbjct: 377 -----NRLSGSDASPPTHEAKLAAA 396
Score = 32.7 bits (73), Expect = 1.4
Identities = 20/64 (31%), Positives = 30/64 (46%), Gaps = 3/64 (4%)
Query: 192 RCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAAKTPYEQNERDTSAD 251
RC +C T+ +TP RR G ++CNACGL GT R + + +A
Sbjct: 481 RCTNCQTT--TTPLWRRDEDG-NNICNACGLYHKLHGTHRPIGMKKSVIKRRKRIPANAT 537
Query: 252 MKPS 255
+P+
Sbjct: 538 AQPT 541
>CONS_ARATH (Q39057) Zinc finger protein CONSTANS
Length = 373
Score = 45.1 bits (105), Expect = 3e-04
Identities = 23/41 (56%), Positives = 28/41 (68%)
Query: 123 RFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFA 163
R A ++R+REKRK R FEK IRY RK A+ R NG+FA
Sbjct: 306 REARVLRYREKRKTRKFEKTIRYASRKAYAEIRPRVNGRFA 346
>COL4_ARATH (Q940T9) Zinc finger protein CONSTANS-LIKE 4
Length = 362
Score = 44.3 bits (103), Expect = 5e-04
Identities = 23/51 (45%), Positives = 31/51 (60%)
Query: 121 SRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLKEDYKS 171
+ R A ++R+REKRK R FEK IRY RK A+ R G+FA + +S
Sbjct: 293 AEREARVMRYREKRKNRKFEKTIRYASRKAYAEMRPRIKGRFAKRTDTNES 343
>GAF1_SCHPO (Q10280) Transcription factor gaf1 (Gaf-1)
Length = 855
Score = 43.9 bits (102), Expect = 6e-04
Identities = 39/116 (33%), Positives = 53/116 (45%), Gaps = 14/116 (12%)
Query: 167 EDYKSPAGNLDSSNGTPPCPESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWAN 226
ED K A N +N T P P C +C T ++TP RR P G + LCNACGL
Sbjct: 615 EDKKGDA-NTRRANATNPTPT-----CTNCQT--RTTPLWRRSPDG-QPLCNACGLFMKI 665
Query: 227 KGTLRDLSKAAKTPYEQNER-DTSADMKPS----TTEPENSCADQAEEGTPEENKP 277
G +R LS ++N TSA K S + ++S + + T + KP
Sbjct: 666 NGVVRPLSLKTDVIKKRNRGVGTSATPKQSGGRKGSTRKSSSKSSSAKSTAADMKP 721
>COLA_ARATH (Q9LUA9) Zinc finger protein CONSTANS-LIKE 10
Length = 373
Score = 43.9 bits (102), Expect = 6e-04
Identities = 21/53 (39%), Positives = 33/53 (61%)
Query: 117 SSNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLKEDY 169
SS+ + R +++R++EK+K R F+K++RY RKE A R G+F E Y
Sbjct: 310 SSHATTRNNAVMRYKEKKKARKFDKRVRYVSRKERADVRRRVKGRFVKSGEAY 362
>COL8_ARATH (Q9M9B3) Putative zinc finger protein CONSTANS-LIKE 8
Length = 313
Score = 43.5 bits (101), Expect = 8e-04
Identities = 22/42 (52%), Positives = 27/42 (63%)
Query: 121 SRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQF 162
S R A + R+R+KRK R FEKKIRY RK A + R G+F
Sbjct: 263 SEREARVWRYRDKRKNRLFEKKIRYEVRKVNADKRPRMKGRF 304
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.311 0.128 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 43,787,450
Number of Sequences: 164201
Number of extensions: 1945591
Number of successful extensions: 5325
Number of sequences better than 10.0: 163
Number of HSP's better than 10.0 without gapping: 37
Number of HSP's successfully gapped in prelim test: 127
Number of HSP's that attempted gapping in prelim test: 5218
Number of HSP's gapped (non-prelim): 246
length of query: 354
length of database: 59,974,054
effective HSP length: 111
effective length of query: 243
effective length of database: 41,747,743
effective search space: 10144701549
effective search space used: 10144701549
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0100b.1


