

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0098e.6
(402 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
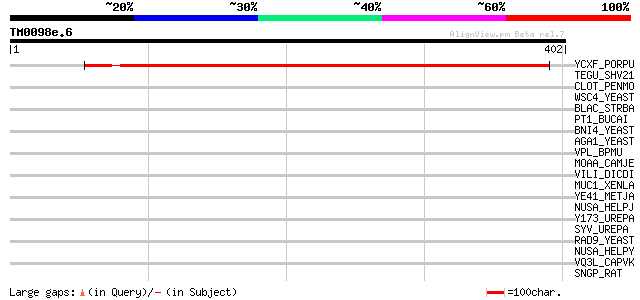
YCXF_PORPU (P51277) Hypothetical 41.5 kDa protein in YCF6-CHLB i... 446 e-125
TEGU_SHV21 (Q01056) Probable large tegument protein 37 0.069
CLOT_PENMO (Q9U572) Hemolymph clottable protein precursor 34 0.76
WSC4_YEAST (P38739) Cell wall integrity and stress response comp... 33 1.3
BLAC_STRBA (P35391) Beta-lactamase precursor (EC 3.5.2.6) (Penic... 33 1.3
PT1_BUCAI (Q9WXI6) Phosphoenolpyruvate-protein phosphotransferas... 33 1.7
BNI4_YEAST (P53858) BNI4 protein 33 1.7
AGA1_YEAST (P32323) A-agglutinin attachment subunit precursor 33 1.7
VPL_BPMU (P79678) Tail sheath protein (gpL) 32 2.2
MOAA_CAMJE (Q9PIW6) Molybdenum cofactor biosynthesis protein A 32 2.2
VILI_DICDI (P36418) Protovillin (100 kDa actin-binding protein) 32 2.9
MUC1_XENLA (Q05049) Integumentary mucin C.1 (FIM-C.1) (Fragment) 32 2.9
YE41_METJA (Q58836) Hypothetical protein MJ1441 32 3.8
NUSA_HELPJ (Q9ZJA6) Transcription elongation protein nusA 32 3.8
Y173_UREPA (Q9PQX0) Hypothetical protein UU173 31 4.9
SYV_UREPA (Q9PQM4) Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--t... 31 4.9
RAD9_YEAST (P14737) DNA repair protein RAD9 31 4.9
NUSA_HELPY (P55977) Transcription elongation protein nusA 31 4.9
VQ3L_CAPVK (Q86917) G-protein coupled receptor homolog Q2/3L 31 6.4
SNGP_RAT (Q9QUH6) Ras GTPase-activating protein SynGAP (Synaptic... 30 8.4
>YCXF_PORPU (P51277) Hypothetical 41.5 kDa protein in YCF6-CHLB
intergenic region (ORF349)
Length = 349
Score = 446 bits (1146), Expect = e-125
Identities = 207/337 (61%), Positives = 261/337 (77%), Gaps = 5/337 (1%)
Query: 55 KETLLTPRFYTTDFDEMETLFNTEINKNLNEHEFEALLQEFKTDYNQTHFVRNKEFKEAA 114
+ETLLTPRFYTTDF+EM ++ N + NE +F A+L+EF+ DYN HF+R +EF ++
Sbjct: 13 RETLLTPRFYTTDFEEMASM-----NISENEQDFFAILEEFRADYNSQHFIRGEEFNQSW 67
Query: 115 DKLDGPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGF 174
L+ + +F+EFLERSCTAEFSGFLLYKEL R+LK +NP++AE F LMSRDEARHAGF
Sbjct: 68 SNLETKTKSLFIEFLERSCTAEFSGFLLYKELSRKLKDSNPIIAECFLLMSRDEARHAGF 127
Query: 175 LNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKANPEYQ 234
LNK + DFNL+LDLGFLTK RKYTFF PKFIFYATYLSEKIGYWRYITIYRH++ PE++
Sbjct: 128 LNKAIGDFNLSLDLGFLTKTRKYTFFSPKFIFYATYLSEKIGYWRYITIYRHMEQYPEHR 187
Query: 235 CYPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQR 294
YPIFK+FENWCQDENRHGDFF+AL+K+QP FLNDWKAK+W RFF LSV+ TMYLND QR
Sbjct: 188 IYPIFKFFENWCQDENRHGDFFAALLKSQPHFLNDWKAKMWCRFFLLSVFATMYLNDFQR 247
Query: 295 TDFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKRRLDRMVEINEKILAVGE 354
DFY IGL+++++DM VI +TN + AR+FP LDV+NP+F + LD N ++ + +
Sbjct: 248 IDFYNAIGLDSRQYDMQVIRKTNESAARVFPVALDVDNPKFFKYLDSCACNNRALIDIDK 307
Query: 355 SDDIPFVKNLKRIPLIAALASELLATYLMPPIESGSV 391
P VK + PL +L L+ YL+ PI+S +V
Sbjct: 308 KGSQPLVKTFMKAPLYMSLILNLIKIYLIKPIDSQAV 344
>TEGU_SHV21 (Q01056) Probable large tegument protein
Length = 2469
Score = 37.4 bits (85), Expect = 0.069
Identities = 28/83 (33%), Positives = 36/83 (42%), Gaps = 14/83 (16%)
Query: 11 TNAAPKFSNPRTISHSKFSTVRMSATTTPPSSTKPSKKANKTSIKETLLTPRFYT----- 65
T APK PR K ST+ + PP K SKK+ K K+T LTP+ T
Sbjct: 270 TPKAPK--TPRKPKTPKESTIPYDKSKKPPKIPKTSKKSKKVLTKDTALTPQHKTIEEHL 327
Query: 66 -------TDFDEMETLFNTEINK 81
T+ E TLFN + +
Sbjct: 328 RELLPPITETVEDNTLFNHPVER 350
>CLOT_PENMO (Q9U572) Hemolymph clottable protein precursor
Length = 1670
Score = 33.9 bits (76), Expect = 0.76
Identities = 23/75 (30%), Positives = 38/75 (50%), Gaps = 4/75 (5%)
Query: 8 SKFTNAAPKFSNPRTISHSKFSTVRMSATTTPPSSTKPSKKANKTSIKETLLTPRFYTTD 67
S +T ++ +T S + T S+ T+ T P + AN I++TL + R TTD
Sbjct: 198 SSYTTKTKSKTSSKTSSKTSSKT---SSKTSKTGKTSPGQLANVPHIEDTLWSVR-RTTD 253
Query: 68 FDEMETLFNTEINKN 82
FD E L + +I+ +
Sbjct: 254 FDMCENLVSLQIHSS 268
>WSC4_YEAST (P38739) Cell wall integrity and stress response
component 4 precursor
Length = 605
Score = 33.1 bits (74), Expect = 1.3
Identities = 18/49 (36%), Positives = 25/49 (50%)
Query: 18 SNPRTISHSKFSTVRMSATTTPPSSTKPSKKANKTSIKETLLTPRFYTT 66
S P T S + ST S+T+T S+T P+ + TS T T +TT
Sbjct: 241 STPTTTSSAPISTSTTSSTSTSTSTTSPTSSSAPTSSSNTTPTSTTFTT 289
>BLAC_STRBA (P35391) Beta-lactamase precursor (EC 3.5.2.6)
(Penicillinase)
Length = 313
Score = 33.1 bits (74), Expect = 1.3
Identities = 18/46 (39%), Positives = 26/46 (56%), Gaps = 5/46 (10%)
Query: 34 SATTTPPSSTKPSKKANKTSIKETLLTPRFYTTDFDEMETLFNTEI 79
S +T PPSS KP+ TS +L P+ YT DF ++E F+ +
Sbjct: 28 SDSTAPPSSAKPA-----TSASASLPRPKPYTGDFKKLEREFDARL 68
>PT1_BUCAI (Q9WXI6) Phosphoenolpyruvate-protein phosphotransferase
(EC 2.7.3.9) (Phosphotransferase system, enzyme I)
Length = 571
Score = 32.7 bits (73), Expect = 1.7
Identities = 22/77 (28%), Positives = 41/77 (52%), Gaps = 9/77 (11%)
Query: 323 IFPAVLDVENPEFKRRLDRMVEINEKILAVGESDDIPFVKNLKRIPLIAALASELLATYL 382
+FP ++ VE + R ++I K +++IPF KN+K +I AS ++A YL
Sbjct: 389 LFPMIISVEEIRILKSEVRKLQIQLK------NNNIPFDKNIKIGIMIETPASAIIAEYL 442
Query: 383 MPPIE---SGSVDLAEF 396
+ ++ G+ DL ++
Sbjct: 443 IKEVDFFSIGTNDLTQY 459
>BNI4_YEAST (P53858) BNI4 protein
Length = 892
Score = 32.7 bits (73), Expect = 1.7
Identities = 54/233 (23%), Positives = 97/233 (41%), Gaps = 59/233 (25%)
Query: 15 PKFSNPRTISHSKFSTVRMSATTTPPSSTKPSKKANKTSIKETLLTPRFYTTDFDEMETL 74
PKF + +H S V + T + S+ + S + + S+ +TL + E +
Sbjct: 490 PKFLD----NHEVDSIVSLERTRSTKSNKRSSMNSQRRSLTDTLSIKA-------QSEGM 538
Query: 75 FNTEINK------NLNEHEFEALLQ----EFKTDYNQTHFVR---NKEFKEAADKLDGPL 121
F TE + +L + ++L+ E+ ++++ H N++F + D GPL
Sbjct: 539 FITEASSVVLSTPDLTKSPASSILKNGRFEYSDNFSREHSYEGTTNEDFLDIKDD-SGPL 597
Query: 122 RQ--IFVEFLERSCTAEFSGFLLYKELGRR-------------LKKTNP---VVAEIFSL 163
++ IF+E +E+ +F ++ + + LKK + V A+ +
Sbjct: 598 KKDDIFLESIEQ----KFDQLVMASDEEKTEVERDVPKPREEPLKKDSERQSVFADDDNE 653
Query: 164 MSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIG 216
+ D A F+N G D NL LDLG T + YAT E +G
Sbjct: 654 LISDIMEFASFINFGDDDLNLDLDLGDTTAS------------YATETPEPVG 694
>AGA1_YEAST (P32323) A-agglutinin attachment subunit precursor
Length = 725
Score = 32.7 bits (73), Expect = 1.7
Identities = 21/56 (37%), Positives = 31/56 (54%), Gaps = 2/56 (3%)
Query: 2 ALVKPISKFTNAAPKFSNPRTISHSKFSTVRMSATTTPPSSTKPSKKANKTSIKET 57
+++ P++ T ++ SNP T S S ST S+T+T PSST S + TS T
Sbjct: 164 SIISPVTS-TLSSTTSSNPTTTSLSSTST-SPSSTSTSPSSTSTSSSSTSTSSSST 217
>VPL_BPMU (P79678) Tail sheath protein (gpL)
Length = 495
Score = 32.3 bits (72), Expect = 2.2
Identities = 29/113 (25%), Positives = 48/113 (41%), Gaps = 10/113 (8%)
Query: 8 SKFTNAAPKFSNPRTISHSKFSTVRMSATTTPPSSTKPSKKANKTSIKETLLTPRFYTTD 67
+K+ ++ P + N TI+ S +R S T K A+ + RF T
Sbjct: 376 NKYGDSDPSYLNVNTIA--TLSYLRYSLRTRITQKFPNYKLASDGT--------RFATGQ 425
Query: 68 FDEMETLFNTEINKNLNEHEFEALLQEFKTDYNQTHFVRNKEFKEAADKLDGP 120
++ TE+ E E L+++F T + + RNK+ K+ D L GP
Sbjct: 426 AVVTPSVIKTELLALFEEWENAGLVEDFDTFKEELYVARNKDDKDRLDVLCGP 478
>MOAA_CAMJE (Q9PIW6) Molybdenum cofactor biosynthesis protein A
Length = 320
Score = 32.3 bits (72), Expect = 2.2
Identities = 19/62 (30%), Positives = 30/62 (47%)
Query: 70 EMETLFNTEINKNLNEHEFEALLQEFKTDYNQTHFVRNKEFKEAADKLDGPLRQIFVEFL 129
+++ NT KNLN+ E +LL+ K+ Q F+ E A KL G R ++ L
Sbjct: 151 DLKVKLNTVALKNLNDDELISLLEFAKSKKAQIRFIEFMENTHAYGKLQGLKRDEIIQIL 210
Query: 130 ER 131
+
Sbjct: 211 SQ 212
>VILI_DICDI (P36418) Protovillin (100 kDa actin-binding protein)
Length = 959
Score = 32.0 bits (71), Expect = 2.9
Identities = 31/117 (26%), Positives = 50/117 (42%), Gaps = 9/117 (7%)
Query: 2 ALVKPISKFTNAAPKFSNPRTISHSKFSTVRMSATTTPPSSTKPSKKA--NKTSIKETLL 59
A KPI+ T PK T++ K TV + T P + T PSK A TS
Sbjct: 839 ATPKPITTPTVTTPKPITTPTVATLK--TVTPAVTLKPTTVTTPSKVATTTNTSTPSPTT 896
Query: 60 TPRFYTTDFDEMETLFNTEINKN-----LNEHEFEALLQEFKTDYNQTHFVRNKEFK 111
FY + +T +I+K+ L++ EF + + K + +T + K+ +
Sbjct: 897 ITTFYPLSVLKQKTNLPNDIDKSCLHLYLSDEEFLSTFKMTKEIFQKTPAWKTKQLR 953
>MUC1_XENLA (Q05049) Integumentary mucin C.1 (FIM-C.1) (Fragment)
Length = 662
Score = 32.0 bits (71), Expect = 2.9
Identities = 20/77 (25%), Positives = 32/77 (40%)
Query: 11 TNAAPKFSNPRTISHSKFSTVRMSATTTPPSSTKPSKKANKTSIKETLLTPRFYTTDFDE 70
T A P + + + +T + + TTT P++T + KA T T TP TT
Sbjct: 239 TKATPTTTTTTKATPTTTTTTKATTTTTTPTTTTTTTKATTTPTTTTTTTPTTTTTKATT 298
Query: 71 METLFNTEINKNLNEHE 87
T + E ++ E
Sbjct: 299 TTTTTSGECKMEPSKRE 315
Score = 30.8 bits (68), Expect = 6.4
Identities = 14/38 (36%), Positives = 23/38 (59%)
Query: 29 STVRMSATTTPPSSTKPSKKANKTSIKETLLTPRFYTT 66
+T++++ TTT P++T + KA T+ T TP TT
Sbjct: 221 TTIQIATTTTTPTTTTTTTKATPTTTTTTKATPTTTTT 258
>YE41_METJA (Q58836) Hypothetical protein MJ1441
Length = 1226
Score = 31.6 bits (70), Expect = 3.8
Identities = 42/174 (24%), Positives = 75/174 (42%), Gaps = 45/174 (25%)
Query: 39 PPSSTKPSKKANKTSIK--ETLLTPRFYTTDFDEMETLFNTEINKNLNE------HEFEA 90
PP T +++ T I +T++ FY +E+ETL ++ I + L E H+ E
Sbjct: 568 PPEGTIAKRRSYATIIDHMQTVMVDAFY----EELETL-DSYIEEYLKEMDASRRHQLEH 622
Query: 91 LLQEFKTDYNQTHFVRNKEFKEAADKLDGPLRQIFVEFLER-----------SCTAEFSG 139
L+ E + +T+ ++ KE E +K +G + + F E + C G
Sbjct: 623 LIVE---EVKKTNLLKIKEKIEKIEK-EGKIHENFKEIFDELRDILEMIKNSKCN---DG 675
Query: 140 FLLYKELG---RRLKKTNPVVAEIF-----------SLMSRDEARHAGFLNKGL 179
++ EL +R++ ++ IF + R EA H G+ N+GL
Sbjct: 676 MHIFGELPSGEKRVEFIKSILEAIFIQNNTMNSKRRGIAERSEAMHPGYPNRGL 729
>NUSA_HELPJ (Q9ZJA6) Transcription elongation protein nusA
Length = 395
Score = 31.6 bits (70), Expect = 3.8
Identities = 21/77 (27%), Positives = 36/77 (46%), Gaps = 3/77 (3%)
Query: 296 DFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKRRLDRMVEINEKILAVGES 355
D + + K +++E +RTT ++ A+L++E PE K D+ +EI G
Sbjct: 180 DSIKAVLTQVKRTKKGLLLELSRTTPKMLEALLELEVPEIK---DKEIEIIHCARIPGNR 236
Query: 356 DDIPFVKNLKRIPLIAA 372
+ F + RI I A
Sbjct: 237 AKVSFFSHNSRIDPIGA 253
>Y173_UREPA (Q9PQX0) Hypothetical protein UU173
Length = 690
Score = 31.2 bits (69), Expect = 4.9
Identities = 40/135 (29%), Positives = 57/135 (41%), Gaps = 32/135 (23%)
Query: 81 KNLNEHEFEALLQEFKTDY---------NQTHFVRNKEFKEAADKLDGPLRQIFVEFLER 131
KN++ ++FE L E + DY QT F+ E + D +D P
Sbjct: 39 KNVSSYDFEDLDDEEEHDYEIDAYEIYKEQTDFLSQLEINKNID-VDNPK---------- 87
Query: 132 SCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFLNKGLSDF-NLALDLGF 190
AE G LL K +KKT + +I + + F NK L + NL L+L
Sbjct: 88 --IAE--GILLDKASQEDIKKTYHYIKKIINFET-----DPLFKNKSLEELANLTLEL-- 136
Query: 191 LTKARKYTFFKPKFI 205
+ K K FF+P FI
Sbjct: 137 IDKNDKIIFFQPVFI 151
>SYV_UREPA (Q9PQM4) Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA
ligase) (ValRS)
Length = 874
Score = 31.2 bits (69), Expect = 4.9
Identities = 23/78 (29%), Positives = 36/78 (45%), Gaps = 4/78 (5%)
Query: 211 LSEKIGYWRYITIYRHLKANPEYQCYPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDW 270
+S ++ + I ++ H K N Y K ENW QDE+ +FS+ M P W
Sbjct: 407 ISRQLWWGHQIPVWYHKKTNQIYCDTIPPKDLENWIQDEDVLDTWFSSGM--WPLLTTKW 464
Query: 271 --KAKLWSRFFCLSVYVT 286
+ + R+F S+ VT
Sbjct: 465 NYNSHFFDRYFPTSLIVT 482
>RAD9_YEAST (P14737) DNA repair protein RAD9
Length = 1309
Score = 31.2 bits (69), Expect = 4.9
Identities = 20/100 (20%), Positives = 44/100 (44%), Gaps = 1/100 (1%)
Query: 17 FSNPRTISHSKFSTVRMSATTTPPSSTKPSKKANKTSIKETLLTPRFYTTDFDEMETLFN 76
F P T+S F + T+ PS SK +N + I + T + + E +F+
Sbjct: 472 FETPVTLSRINFEPILEVPETSSPSKNTMSKPSNSSPIPKEKDTFNIHEREV-ETNNVFS 530
Query: 77 TEINKNLNEHEFEALLQEFKTDYNQTHFVRNKEFKEAADK 116
+I + N + ++ +D+N+ + ++ + + + K
Sbjct: 531 NDIQNSSNAATRDDIIIAGSSDFNEQKEITDRIYLQLSGK 570
>NUSA_HELPY (P55977) Transcription elongation protein nusA
Length = 395
Score = 31.2 bits (69), Expect = 4.9
Identities = 21/77 (27%), Positives = 36/77 (46%), Gaps = 3/77 (3%)
Query: 296 DFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKRRLDRMVEINEKILAVGES 355
D + + K +++E +RTT ++ A+L++E PE K D+ +EI G
Sbjct: 180 DSIKAVLTQVKRTKKGLLLELSRTTPKMLEALLELEVPEIK---DKEIEIIHCARIPGNR 236
Query: 356 DDIPFVKNLKRIPLIAA 372
+ F + RI I A
Sbjct: 237 AKVSFFSHNARIDPIGA 253
>VQ3L_CAPVK (Q86917) G-protein coupled receptor homolog Q2/3L
Length = 381
Score = 30.8 bits (68), Expect = 6.4
Identities = 18/57 (31%), Positives = 29/57 (50%), Gaps = 2/57 (3%)
Query: 18 SNPRTISHSKFSTVRMSATTTPPSSTKPSKKANKTSIKE--TLLTPRFYTTDFDEME 72
SN TI+ + ST+ + +T + T PS N T+I T +Y+ D+D+ E
Sbjct: 18 SNITTIATTIISTILSTISTNQNNVTTPSTYENTTTISNYTTAYNTTYYSDDYDDYE 74
>SNGP_RAT (Q9QUH6) Ras GTPase-activating protein SynGAP (Synaptic
Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1)
(Neuronal RasGAP) (p135 SynGAP)
Length = 1293
Score = 30.4 bits (67), Expect = 8.4
Identities = 19/68 (27%), Positives = 31/68 (44%)
Query: 119 GPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFLNKG 178
G ++Q E S F Y +LGR L + ++ E+ +S++ G L +
Sbjct: 643 GSMQQFLYEISNLDTLTNSSSFEGYIDLGRELSTLHALLWEVLPQLSKEALLKLGPLPRL 702
Query: 179 LSDFNLAL 186
LSD + AL
Sbjct: 703 LSDISTAL 710
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.137 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 46,308,033
Number of Sequences: 164201
Number of extensions: 1953367
Number of successful extensions: 5375
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 5329
Number of HSP's gapped (non-prelim): 45
length of query: 402
length of database: 59,974,054
effective HSP length: 112
effective length of query: 290
effective length of database: 41,583,542
effective search space: 12059227180
effective search space used: 12059227180
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0098e.6


