

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0098b.4
(670 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
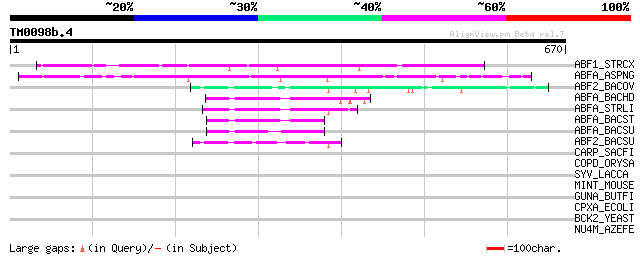
Score E
Sequences producing significant alignments: (bits) Value
ABF1_STRCX (P82593) Alpha-N-arabinofuranosidase I precursor (EC ... 294 4e-79
ABFA_ASPNG (P42254) Alpha-N-arabinofuranosidase A precursor (EC ... 258 4e-68
ABF2_BACOV (Q59219) Alpha-N-arabinofuranosidase 2 (EC 3.2.1.55) ... 84 1e-15
ABFA_BACHD (Q9KBR4) Alpha-N-arabinofuranosidase (EC 3.2.1.55) (A... 71 1e-11
ABFA_STRLI (P53627) Alpha-N-arabinofuranosidase (EC 3.2.1.55) (A... 67 2e-10
ABFA_BACST (Q9XBQ3) Alpha-N-arabinofuranosidase (EC 3.2.1.55) (A... 65 6e-10
ABFA_BACSU (P94531) Alpha-N-arabinofuranosidase 1 (EC 3.2.1.55) ... 62 6e-09
ABF2_BACSU (P94552) Alpha-N-arabinofuranosidase 2 (EC 3.2.1.55) ... 55 5e-07
CARP_SACFI (P22929) Acid protease precursor (EC 3.4.23.-) 40 0.020
COPD_ORYSA (P49661) Coatomer delta subunit (Delta-coat protein) ... 34 1.1
SYV_LACCA (P36420) Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--t... 34 1.4
MINT_MOUSE (Q62504) Msx2-interacting protein (SMART/HDAC1 associ... 33 1.9
GUNA_BUTFI (P22541) Endoglucanase A precursor (EC 3.2.1.4) (Endo... 33 3.2
CPXA_ECOLI (P08336) Sensor protein cpxA (EC 2.7.3.-) 32 5.4
BCK2_YEAST (P33306) BCK2 protein (Bypass of kinase C protein) 32 5.4
NU4M_AZEFE (P92494) NADH-ubiquinone oxidoreductase chain 4 (EC 1... 31 9.2
>ABF1_STRCX (P82593) Alpha-N-arabinofuranosidase I precursor (EC
3.2.1.55) (Arabinosidase I) (Alpha-N-AFase I)
Length = 825
Score = 294 bits (753), Expect = 4e-79
Identities = 194/572 (33%), Positives = 290/572 (49%), Gaps = 59/572 (10%)
Query: 33 TLTVDAHETSGKPISETLFGLFFEEINHAGAGGLWAELVSNRGFEAGGPNIPSN--IDPW 90
++TVD G I +T++G+FFE+IN A GGL+AELV NR FE + S + W
Sbjct: 36 SITVDP-AAKGAAIDDTMYGVFFEDINRAADGGLYAELVQNRSFEYSTDDNRSYTPLTSW 94
Query: 91 SIIGNESFINVETDRTSCFDRNKIALRLEVLCDSKGDNICPADGVGVFNPGF-WGMNIEQ 149
+ G +N + R + +RN ++L G V N G+ G+ +EQ
Sbjct: 95 IVDGTGEVVN-DAGRLNERNRNYLSL---------------GAGSSVTNAGYNTGIRVEQ 138
Query: 150 GKKYKVVFYVRSTGSLDLTVSLTGSNGVLASNVITGSASDFSNWKKVETLLEAKATNHNS 209
GK+Y + R+ + LTV+L + G LA+ + W K A T++
Sbjct: 139 GKRYDFSVWARAGSASTLTVALKDAAGTLAT---ARQVAVEGGWAKYRATFTATRTSNRG 195
Query: 210 RLQLTTTAKGVIWLDQVSAMPLDTYKGR--GFRSELVGMLADLKPRFIRFPGGCFVE--- 264
RL + A LD VS P DTY+ + G R +L +A L P F+RFPGGC V
Sbjct: 196 RLAVA--ANDAAALDMVSLFPRDTYRNQQNGLRKDLAEKIAALHPGFVRFPGGCLVNTGS 253
Query: 265 --------GEWLRNAFRWKESIGPWEERPGHFGDVWKYWTDDGLGYFEFLQLAEDLGALP 316
G + +++WK+++GP EER + + W Y GLGY+E+ + +ED+GA+P
Sbjct: 254 MEDYSAASGWQRKRSYQWKDTVGPVEERATN-ANFWGYNQSYGLGYYEYFRFSEDIGAMP 312
Query: 317 IWVFN---NGVSHNDEVDTSAVLP-FVQEALDGIEFARGDPTSKWGSIRASMGHPEPFNL 372
+ V G N VD A+L +Q+ LD IEFA G TSKWG +RA MGHP PF L
Sbjct: 313 LPVVPALVTGCGQNKAVDDEALLKRHIQDTLDLIEFANGPATSKWGKVRAEMGHPRPFRL 372
Query: 373 KYVAVGNEDCGKKNYRGNYLKFYSAIRSAYPDIQIISNC--DGSSRPLDHP--------A 422
++ VGNE+ + + +F +AI + YPDI ++SN D + D
Sbjct: 373 THLEVGNEENLPDEFFDRFKQFRAAIEAEYPDITVVSNSGPDDAGTTFDTAWKLNREANV 432
Query: 423 DIYDYHIYTNANDMFSRSSTFNTALRSGPKAFVSEYAVTGNDAGTGSLLAAIAEAGFLLG 482
++ D H Y + N + +++ R GPK F+ EYA GN G ++EA F+ G
Sbjct: 433 EMVDEHYYNSPNWFLQNNDRYDSYDRGGPKVFLGEYASQGNAWKNG-----LSEAAFMTG 487
Query: 483 LEKNSDIVKMVSYAPLFVNANDRRWNPDAIVFNSFQSYGTPSYWVQLFFSESSGATLLNS 542
LE+N+D+VK+ SYAPL N + +W PD + FN+ S+ + +Y VQ F + G ++ S
Sbjct: 488 LERNADVVKLASYAPLLANEDYVQWRPDLVWFNNRASWNSANYEVQKLFMNNVGDRVVPS 547
Query: 543 SLQTSSS-SSLIASAINWQSSADKKNYIRIKV 573
T+ S I A+ + A Y +KV
Sbjct: 548 KATTTPDVSGPITGAVGLSTWATGTAYDDVKV 579
>ABFA_ASPNG (P42254) Alpha-N-arabinofuranosidase A precursor (EC
3.2.1.55) (Arabinosidase A) (ABF A)
Length = 628
Score = 258 bits (659), Expect = 4e-68
Identities = 198/641 (30%), Positives = 322/641 (49%), Gaps = 59/641 (9%)
Query: 11 AALQLCIAVSLCAIKVNADLNATLTVDAHETSGKPISETLFGLFFEEINHAGAGGLWAEL 70
+AL AVSL V +L V G S L+G FE+INH+G GG++ ++
Sbjct: 5 SALSGVSAVSLLLSLVQNAHGISLKVSTQ--GGNSSSPILYGFMFEDINHSGDGGIYGQM 62
Query: 71 VSNRGFEAGGPNIPSNIDPWSIIGNESF-INVETDRTSCFDRNKIALRLEVLCDSKGDNI 129
+ N G + PN+ + W+ +G+ + I+ ++ TS ++L + D+ G
Sbjct: 63 LQNPGLQGTAPNLTA----WAAVGDATIAIDGDSPLTSAIPST---IKLNIADDATG--- 112
Query: 130 CPADGVGVFNPGFWGMNIEQGKKYKVVFYVRSTGSLDLTVSLTGS--NGVLASNVITGSA 187
VG+ N G+WG+ ++ G ++ F+++ S D+TV L G+ S IT ++
Sbjct: 113 ----AVGLTNEGYWGIPVD-GSEFHSSFWIKGDYSGDITVRLVGNYTGTEYGSTTITHTS 167
Query: 188 SDFSNWKKVETLLEAKATNHNSRLQLT---TTAKGVIWLDQVSAMPLDTYKGR--GFRSE 242
+ + + KA + N +LT + A G + +TYK R G + +
Sbjct: 168 TADNFTQASVKFPTTKAPDGNVLYELTVDGSVAAGSSLNFGYLTLFGETYKSRENGLKPQ 227
Query: 243 LVGMLADLKPRFIRFPGGCFVEGEWLRNAFRWKESIGPWEERPGHFGDVWKYWTDDGLGY 302
L +L D+K F+RFPGG +EG N ++W E+IG +RPG G W Y+ DGLG
Sbjct: 228 LANVLDDMKGSFLRFPGGNNLEGNSAENRWKWNETIGDLCDRPGREG-TWTYYNTDGLGL 286
Query: 303 FEFLQLAEDLGALPIWVFNNGVS----HNDEVDTSAVLPFVQEALDGIEFARGDPTSKWG 358
E+ EDLG +P+ +G + N + A+ P++ + L+ +E+ GD ++ +G
Sbjct: 287 HEYFYWCEDLGLVPVLGVWDGFALESGGNTPLTGDALTPYIDDVLNELEYILGDTSTTYG 346
Query: 359 SIRASMGHPEPFNLKYVAVGNEDC---GKKNYRGNYLKFYSAIRSAYPD-IQIISNCDGS 414
+ RA+ G EP+NL V +GNED G ++Y + FY AI +AYPD I I S +
Sbjct: 347 AWRAANGQEEPWNLTMVEIGNEDMLGGGCESYAERFTAFYDAIHAAYPDLILIASTSEAD 406
Query: 415 SRPLDHPADIY-DYHIYTNANDMFSRSSTFNTALRSGPKAFVSEYAVTGNDAGTGSLLAA 473
P P + DYH Y+ + + + + F+ RS P F+ EY+ D ++ +
Sbjct: 407 CLPESMPEGSWVDYHDYSTPDGLVGQFNYFDNLNRSVP-YFIGEYSRWEID--WPNMKGS 463
Query: 474 IAEAGFLLGLEKNSDIVKMVSYAPLFVNANDRRWNPDAIVFNSFQSYG----TPSYWVQL 529
+AEA F++G E+NSD+VKM +YAPL N +W PD I + QS G + SY+VQ
Sbjct: 464 VAEAVFMIGFERNSDVVKMAAYAPLLQLINSTQWTPDLIGYT--QSPGDIFLSTSYYVQE 521
Query: 530 FFSESSGATLLNSSLQTSSSSSLIASAINWQSSADKKNYIRIKVVNFGANTVNLKISLNG 589
FS + G T+ + +S S + W +S+ +Y +K+ N+G+ T +L +S+ G
Sbjct: 522 MFSRNRGDTI----KEVTSDSDF--GPLYWVASSAGDSYY-VKLANYGSETQDLTVSIPG 574
Query: 590 LDPNSLQPSGSTRTVLTSANVMDENSFSQPKKVVPIQSLLQ 630
L TVL ++ NS +Q V P +S +Q
Sbjct: 575 TSTGKL-------TVLADSDPDAYNSDTQ-TLVTPSESTVQ 607
>ABF2_BACOV (Q59219) Alpha-N-arabinofuranosidase 2 (EC 3.2.1.55)
(Arabinosidase 2)
Length = 514
Score = 84.0 bits (206), Expect = 1e-15
Identities = 116/492 (23%), Positives = 188/492 (37%), Gaps = 93/492 (18%)
Query: 219 GVIWLDQVSAMPLDTYKGRGFRSELVGMLADLKPRFIRFPGGCFVEGEWLRNAFRWKESI 278
G +W+ + S +P +G+R+++ L DL +R+PGGCF + + W + I
Sbjct: 55 GGLWVGENSDIP----NIKGYRTDVFNALKDLSVPVLRWPGGCFAD------EYHWMDGI 104
Query: 279 GPWEERPGHFGDVWKYWTDDG-LGYFEFLQLAEDLGALPIWVFNNGVSHNDEVDTSAVLP 337
GP E RP + W +D G EFL L E LG P VS N T
Sbjct: 105 GPKENRPKMVNNNWGGTIEDNSFGTHEFLNLCEMLGCEPY------VSGNVGSGT----- 153
Query: 338 FVQEALDGIEFARGDPTSKWGSIRASMGHPEPFNLKYVAVGNEDCG-----KKNYRGNYL 392
V+E +E+ D S ++R G + + LKY+ VGNE G + Y +
Sbjct: 154 -VEELAKWVEYMTSDGDSPMANLRRKNGRDKAWKLKYLGVGNESWGCGGSMRPEYYADLY 212
Query: 393 KFYSAIRSAYPDIQIISNCDGSS-----------RPLDHPADIYDYHIYT---------- 431
+ YS Y ++ G+S + H D H YT
Sbjct: 213 RRYSTYCRNYDGNRLFKIASGASDYDYKWTDVLMNRVGHRMDGLSLHYYTVTGWSGSKGS 272
Query: 432 -----------------NANDMFSRSSTFNTALRSGPK-AFVSEYAVTGNDAGTGSLLAA 473
D+ + T K A + + T D G++
Sbjct: 273 ATQFNKDDYYWTMGKCLEVEDVLKKHCTIMDKYDKDKKIALLLDEWGTWWDEEPGTIKGH 332
Query: 474 IAEAGFL-------LGLE---KNSDIVKMVSYAPLFVNANDRRWNPDAIVFNSFQSYGTP 523
+ + L L L+ K +D +KM + A + VN I+ + TP
Sbjct: 333 LYQQNTLRDAFVASLSLDVFHKYTDRLKMANIAQI-VNVLQ-----SMILTKDKEMVLTP 386
Query: 524 SYWVQLFFSESSGATLLNSSL---QTSSSSSLIASAINWQSSADKKNYIRIKVVNFGANT 580
+Y+V + AT L L + S + ++ +S +K I I + N A+
Sbjct: 387 TYYVFKMYKVHQDATYLPIDLTCEKMSVRDNRTVPMVSATASKNKDGVIHISLSNVDADE 446
Query: 581 VNLKISLNGLDPNSLQPSGSTRTVLTSANVMDENSFSQPK--KVVPIQSLLQNVGKEMNV 638
+I++N D + + G +LT++ + D NSF +P K P + + N G M V
Sbjct: 447 AQ-EITINLGDTKAKKAIGE---ILTASKLTDYNSFEKPNIVKPAPFKEVKINKG-TMKV 501
Query: 639 VVPPHSFSSFDL 650
+P S + +L
Sbjct: 502 KLPAKSIVTLEL 513
Score = 32.3 bits (72), Expect = 4.1
Identities = 24/64 (37%), Positives = 33/64 (51%), Gaps = 8/64 (12%)
Query: 5 KASFIVAALQLCIAVSLCAIKVNADLNATLTVDAHETSGKPI-SETLFGLFFEEINHAGA 63
KA +V+ L +VSL A K +AT+TV H GK I + ++G F E +
Sbjct: 2 KAKLLVSTAFLAASVSLSAQK-----SATITV--HADQGKEIIPKEIYGQFAEHLGSCIY 54
Query: 64 GGLW 67
GGLW
Sbjct: 55 GGLW 58
>ABFA_BACHD (Q9KBR4) Alpha-N-arabinofuranosidase (EC 3.2.1.55)
(Arabinosidase)
Length = 500
Score = 70.9 bits (172), Expect = 1e-11
Identities = 56/220 (25%), Positives = 91/220 (40%), Gaps = 39/220 (17%)
Query: 237 RGFRSELVGMLADLKPRFIRFPGGCFVEGEWLRNAFRWKESIGPWEERPGHFGDVWKYWT 296
+GFR +++ ++ +L+ +R+PGG FV G + W++ +GP ERP W+
Sbjct: 49 QGFRKDVIRLVQELQVPLVRYPGGNFVSG------YNWEDGVGPVSERPKRLDLAWRTTE 102
Query: 297 DDGLGYFEFLQLAEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGIEFARGDPTSK 356
+ +G EF+ A+ +GA N G V A + +E+ S
Sbjct: 103 TNEIGTNEFVDWAKKVGAEVNMAVNLGSRG------------VDAARNLVEYCNHPSGSY 150
Query: 357 WGSIRASMGHPEPFNLKYVAVGNEDCGKKNYRGNYLKFYSAI--------RSAYPDIQII 408
W +R S G+ +P N+K +GNE G + Y + + P I+++
Sbjct: 151 WSDLRISHGYKDPHNIKTWCLGNEMDGPWQIGQKTAEEYGRVAAEAGKVMKLVDPSIELV 210
Query: 409 ----SN------CDGSSRPLDHPADIYDY---HIYTNAND 435
SN D + LDH D DY H Y D
Sbjct: 211 ACGSSNSKMATFADWEATVLDHTYDYVDYISLHTYYGNRD 250
>ABFA_STRLI (P53627) Alpha-N-arabinofuranosidase (EC 3.2.1.55)
(Arabinosidase) (ABF) (Alpha-L-AF)
Length = 662
Score = 66.6 bits (161), Expect = 2e-10
Identities = 48/197 (24%), Positives = 88/197 (44%), Gaps = 29/197 (14%)
Query: 233 TYKGRGFRSELVGMLADLKPRFIRFPGGCFVEGEWLRNAFRWKESIGPWEERPGHFGDVW 292
T G R +++ ++ +L +R+PGG FV G ++W++S+GP E+RP W
Sbjct: 44 TADAEGLRQDVLELVRELGVTAVRYPGGNFVSG------YKWEDSVGPVEDRPRRLDLAW 97
Query: 293 KYWTDDGLGYFEFLQLAEDLG--ALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGIEFAR 350
+ + G E++ + +G A P+ N G V EAL+ E+A
Sbjct: 98 RSTETNRFGLSEYIAFLKKIGPQAEPMMAVNLGTRG------------VAEALELQEYAN 145
Query: 351 GDPTSKWGSIRASMGHPEPFNLKYVAVGNEDCG--------KKNYRGNYLKFYSAIRSAY 402
+ +RA G +PF ++ +GNE G + Y + A+R
Sbjct: 146 HPSGTALSDLRAEHGDKDPFGIRLWCLGNEMDGPWQTGHKTAEEYGRVAAETARAMRQID 205
Query: 403 PDIQIISNCDGSSRPLD 419
PD+++++ C S + ++
Sbjct: 206 PDVELVA-CGSSGQSME 221
>ABFA_BACST (Q9XBQ3) Alpha-N-arabinofuranosidase (EC 3.2.1.55)
(Arabinosidase)
Length = 501
Score = 65.1 bits (157), Expect = 6e-10
Identities = 39/143 (27%), Positives = 67/143 (46%), Gaps = 18/143 (12%)
Query: 238 GFRSELVGMLADLKPRFIRFPGGCFVEGEWLRNAFRWKESIGPWEERPGHFGDVWKYWTD 297
GFR +++ ++ +L+ IR+PGG FV G + W++ +GP E+RP WK
Sbjct: 50 GFRQDVIELVKELQVPIIRYPGGNFVSG------YNWEDGVGPKEQRPRRLDLAWKSVET 103
Query: 298 DGLGYFEFLQLAEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGIEFARGDPTSKW 357
+ +G EF+ A+ +GA N G + A + +E+ S +
Sbjct: 104 NEIGLNEFMDWAKMVGAEVNMAVNLGTRG------------IDAARNLVEYCNHPSGSYY 151
Query: 358 GSIRASMGHPEPFNLKYVAVGNE 380
+R + G+ EP +K +GNE
Sbjct: 152 SDLRIAHGYKEPHKIKTWCLGNE 174
>ABFA_BACSU (P94531) Alpha-N-arabinofuranosidase 1 (EC 3.2.1.55)
(Arabinosidase 1)
Length = 500
Score = 61.6 bits (148), Expect = 6e-09
Identities = 38/145 (26%), Positives = 67/145 (46%), Gaps = 22/145 (15%)
Query: 238 GFRSELVGMLADLKPRFIRFPGGCFVEGEWLRNAFRWKESIGPWEERPGHFGDVWKYWTD 297
GFR ++ ++ +L+ IR+PGG F+ G + W++ +GP E RP W+
Sbjct: 49 GFRKDVQSLIKELQVPIIRYPGGNFLSG------YNWEDGVGPVENRPRRLDLAWQTTET 102
Query: 298 DGLGYFEFLQLAEDLGALPIWVFNNGVSHNDEVDTSAVLPF--VQEALDGIEFARGDPTS 355
+ +G EFL + + N EV+ + L + A + +E+ S
Sbjct: 103 NEVGTNEFLSWPKKV--------------NTEVNMAVNLGTRGIDAARNLVEYCNHPKGS 148
Query: 356 KWGSIRASMGHPEPFNLKYVAVGNE 380
W +R S G+ +P+ +K +GNE
Sbjct: 149 YWSDLRRSHGYEQPYGIKTWCLGNE 173
>ABF2_BACSU (P94552) Alpha-N-arabinofuranosidase 2 (EC 3.2.1.55)
(Arabinosidase 2)
Length = 495
Score = 55.5 bits (132), Expect = 5e-07
Identities = 50/187 (26%), Positives = 77/187 (40%), Gaps = 32/187 (17%)
Query: 221 IWLDQVSAMPLDTYKGRGFRSELVGMLADLKPRFIRFPGGCFVEGEWLRNAFRWKESIGP 280
IW+ S +P G R +++ L L +R+PGGCF + + W +G
Sbjct: 37 IWVGTDSDIP----NINGIRKDVLEALKQLHIPVLRWPGGCFAD------EYHWANGVGD 86
Query: 281 WEERPG-HFGDVWKYWTDDGLGYFEFLQLAEDLGALPIWVFNNGVSHNDEVDTSAVLPFV 339
+ H+G + + G EF+ L E L P N G S + +
Sbjct: 87 RKTMLNTHWGGTIE---SNEFGTHEFMMLCELLECEPYICGNVG---------SGTVQEM 134
Query: 340 QEALDGIEFARGDPTSKWGSIRASMGHPEPFNLKYVAVGNEDCG------KKNYRGNYLK 393
E ++ + F G P S W R G EP+ LKY VGNE+ G + Y Y +
Sbjct: 135 SEWIEYMTFEEGTPMSDW---RKQNGREEPWKLKYFGVGNENWGCGGNMHPEYYADLYRR 191
Query: 394 FYSAIRS 400
F + +R+
Sbjct: 192 FQTYVRN 198
>CARP_SACFI (P22929) Acid protease precursor (EC 3.4.23.-)
Length = 390
Score = 40.0 bits (92), Expect = 0.020
Identities = 50/227 (22%), Positives = 84/227 (36%), Gaps = 14/227 (6%)
Query: 306 LQLAEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGIEFARGDPTSK---WGSIRA 362
LQ+ D G+ +WV G S ++ G + GD +S W
Sbjct: 88 LQVDVDTGSSDLWVPGQGTSSLYGTYDHTKSTSYKKDRSGFSISYGDGSSARGDWAQETV 147
Query: 363 SMGHPEPFNLKYVAVGNEDCGKK----NYRGNYLKFYSAIRSAYPDIQIISNCDGSSRPL 418
S+G L++ ++D G+ +GN S+ Y ++ + G +
Sbjct: 148 SIGGASITGLEFGDATSQDVGQGLLGIGLKGNEASAQSSNSFTYDNLPLKLKDQGL---I 204
Query: 419 DHPADIYDYHIYTNANDMFSRSSTFNTALRSGPKAFVSEYAVTGNDAGTGSLLAAIAEAG 478
D A Y +Y N+ D S S F + S ++ + D S A+A
Sbjct: 205 DKAA----YSLYLNSEDATSGSILFGGSDSSKYSGSLATLDLVNIDDEGDSTSGAVAFFV 260
Query: 479 FLLGLEKNSDIVKMVSYAPLFVNANDRRWNPDAIVFNSFQSYGTPSY 525
L G+E S + +Y L + + P +I + + YGT SY
Sbjct: 261 ELEGIEAGSSSITKTTYPALLDSGTTLIYAPSSIASSIGREYGTYSY 307
>COPD_ORYSA (P49661) Coatomer delta subunit (Delta-coat protein)
(Delta-COP) (Archain)
Length = 518
Score = 34.3 bits (77), Expect = 1.1
Identities = 32/149 (21%), Positives = 64/149 (42%), Gaps = 7/149 (4%)
Query: 70 LVSNRGFEAGGPNIPSNIDPW-SIIGNESFINVETDRTSCFDRNKIALRLEVLCDSKGDN 128
LV R E ++P ++ W S+ GNE+++N+E + + FD + + + + + +
Sbjct: 368 LVKWRIQELNESSLPLAVNCWPSVSGNETYVNIEYEASEMFDLHNVVISIPLPALREAPG 427
Query: 129 ICPADGVGVFNPGFWGMNIEQGKKYKVVFYVRSTGSLDLTVSLTGSNGVLASNVITGSAS 188
+ DG ++ N +V +GSL+ TV + +V +++
Sbjct: 428 VRQIDGEWRYD----SRNSVLEWSIILVDQSNRSGSLEFTVPAADPSTFFPISVGFSASN 483
Query: 189 DFSNWK--KVETLLEAKATNHNSRLQLTT 215
FS+ K + L E + R +L T
Sbjct: 484 TFSDLKVTAIRPLREGSPPKFSQRNRLVT 512
>SYV_LACCA (P36420) Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA
ligase) (ValRS)
Length = 901
Score = 33.9 bits (76), Expect = 1.4
Identities = 29/108 (26%), Positives = 42/108 (38%), Gaps = 9/108 (8%)
Query: 293 KYWTDDGLGYFEFLQLAED--LGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGIEFAR 350
KY DG GY E + +G + V + D V T +LP + IE A
Sbjct: 226 KYPFADGSGYIEIATTRPETMMGDTAVAVHPGDERYKDMVGTELILPLANRKIPIIEDAY 285
Query: 351 GDPTSKWGSIRASMGH-PEPF------NLKYVAVGNEDCGKKNYRGNY 391
DP G+++ + H P F +LK + N+D G Y
Sbjct: 286 VDPEFGTGAVKITPAHDPNDFQVGNRHDLKRINTMNDDGTMNENAGKY 333
>MINT_MOUSE (Q62504) Msx2-interacting protein (SMART/HDAC1 associated
repressor protein)
Length = 3644
Score = 33.5 bits (75), Expect = 1.9
Identities = 19/59 (32%), Positives = 30/59 (50%), Gaps = 8/59 (13%)
Query: 589 GLDPNSLQPSGSTRTVL-TSANVMDENSFSQPKKVVPIQSLLQNVGKEMNVVVPPHSFS 646
GL P PS +T T L T+A VM +P+ + ++ E +V++PPHS +
Sbjct: 3009 GLFPRPCHPSSTTSTALSTNATVMLAAG-------IPVPQFISSIHPEQSVIMPPHSIT 3060
>GUNA_BUTFI (P22541) Endoglucanase A precursor (EC 3.2.1.4)
(Endo-1,4-beta-glucanase) (Cellulase) (EGA)
Length = 429
Score = 32.7 bits (73), Expect = 3.2
Identities = 19/55 (34%), Positives = 27/55 (48%), Gaps = 2/55 (3%)
Query: 416 RPLDHPADIYDYHIYTNANDMFSRSSTFNTALRSGPKAFVSEYAVTGNDAGTGSL 470
+PLD +Y YH Y + R++ AL G F+SEY + D G G+L
Sbjct: 294 KPLDFDNIMYTYHFYAGTHHKAERNA-LRDALDEGLPVFISEYGLVDAD-GDGNL 346
>CPXA_ECOLI (P08336) Sensor protein cpxA (EC 2.7.3.-)
Length = 457
Score = 32.0 bits (71), Expect = 5.4
Identities = 30/133 (22%), Positives = 53/133 (39%), Gaps = 26/133 (19%)
Query: 23 AIKVNADLNATLTVDAHETSGKPISETLFGLFFEEINHAGAGGLWAELVSNRGFEAG--G 80
A ++++ +N L + ++ +SET+ A LW+E++ N FEA G
Sbjct: 281 AQRLDSMINDLLVMSRNQQKNALVSETI-----------KANQLWSEVLDNAAFEAEQMG 329
Query: 81 PNIPSNI--DPWSIIGN----ESFINVETDRTSCFDRNKIAL-------RLEVLCDSKGD 127
++ N PW + GN ES + + KI + + + D G
Sbjct: 330 KSLTVNFPPGPWPLYGNPNALESALENIVRNALRYSHTKIEVGFAVDKDGITITVDDDGP 389
Query: 128 NICPADGVGVFNP 140
+ P D +F P
Sbjct: 390 GVSPEDREQIFRP 402
>BCK2_YEAST (P33306) BCK2 protein (Bypass of kinase C protein)
Length = 851
Score = 32.0 bits (71), Expect = 5.4
Identities = 52/218 (23%), Positives = 82/218 (36%), Gaps = 32/218 (14%)
Query: 430 YTNANDMFSRSSTFNTALRSGPKAFVSEYAVTGNDAGTGSLLAAIAEAGFLLGLEKNSDI 489
Y+N ND S FNT + + P A+ G T L E L+ DI
Sbjct: 564 YSNENDASGEFSAFNTPMENIP-------ALKGIPRST---LEENEEEDVLV-----QDI 608
Query: 490 VKMVSYAPLFVNANDRRWNPDAIVFNSFQSYG--TPSYWVQLFFSESSGATLLNSS---L 544
+ + D D++ FNS +G T S V + S T N++
Sbjct: 609 PNTAHFQRRDIMGMDTHRKDDSLDFNSLMPHGSTTSSSIVDSVMTNSISTTTSNATGNYF 668
Query: 545 QTSSSSSLIASAINWQSSADKKNYIRIKVVNFGA----NTVNLKISLNGLDPNSLQPSGS 600
Q +L+ + + S A+ ++IR + + N ++S +G PN++
Sbjct: 669 QDQDKYTLVNTGLG-LSDANLDHFIRSQWKHASRSESNNNTGNRVSYSGSTPNNVD---- 723
Query: 601 TRTVLTSANVMDENSFSQPKKVVPIQS-LLQNVGKEMN 637
T T+ V E F P+ QS LL +G N
Sbjct: 724 --TTKTNLQVYTEFDFENPESFFHEQSKLLGEMGHSNN 759
>NU4M_AZEFE (P92494) NADH-ubiquinone oxidoreductase chain 4 (EC
1.6.5.3) (Fragment)
Length = 231
Score = 31.2 bits (69), Expect = 9.2
Identities = 15/52 (28%), Positives = 25/52 (47%)
Query: 507 WNPDAIVFNSFQSYGTPSYWVQLFFSESSGATLLNSSLQTSSSSSLIASAIN 558
W P I+ T SY + +F S +G TLLN+ + + S + A++
Sbjct: 165 WCPTTIILLGLSMLITASYSLHMFLSTQAGPTLLNNQTEPTHSREHLLMALH 216
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.134 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 80,756,405
Number of Sequences: 164201
Number of extensions: 3582587
Number of successful extensions: 8263
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 8220
Number of HSP's gapped (non-prelim): 24
length of query: 670
length of database: 59,974,054
effective HSP length: 117
effective length of query: 553
effective length of database: 40,762,537
effective search space: 22541682961
effective search space used: 22541682961
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0098b.4


