

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0098a.6
(626 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
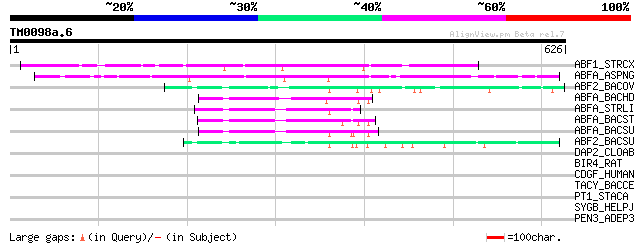
Score E
Sequences producing significant alignments: (bits) Value
ABF1_STRCX (P82593) Alpha-N-arabinofuranosidase I precursor (EC ... 272 2e-72
ABFA_ASPNG (P42254) Alpha-N-arabinofuranosidase A precursor (EC ... 228 3e-59
ABF2_BACOV (Q59219) Alpha-N-arabinofuranosidase 2 (EC 3.2.1.55) ... 94 1e-18
ABFA_BACHD (Q9KBR4) Alpha-N-arabinofuranosidase (EC 3.2.1.55) (A... 82 6e-15
ABFA_STRLI (P53627) Alpha-N-arabinofuranosidase (EC 3.2.1.55) (A... 76 2e-13
ABFA_BACST (Q9XBQ3) Alpha-N-arabinofuranosidase (EC 3.2.1.55) (A... 75 5e-13
ABFA_BACSU (P94531) Alpha-N-arabinofuranosidase 1 (EC 3.2.1.55) ... 75 7e-13
ABF2_BACSU (P94552) Alpha-N-arabinofuranosidase 2 (EC 3.2.1.55) ... 64 1e-09
DAP2_CLOAB (Q97D80) Dihydrodipicolinate synthase 2 (EC 4.2.1.52)... 33 2.2
BIR4_RAT (Q9R0I6) Baculoviral IAP repeat-containing protein 4 (I... 33 2.2
CDGF_HUMAN (Q9Y5G1) Protocadherin gamma B3 precursor (PCDH-gamma... 33 2.9
TACY_BACCE (Q45105) Hemolysin precursor (Fragment) 32 5.0
PT1_STACA (P23533) Phosphoenolpyruvate-protein phosphotransferas... 32 5.0
SYGB_HELPJ (Q9ZKM9) Glycyl-tRNA synthetase beta chain (EC 6.1.1.... 32 6.5
PEN3_ADEP3 (Q84176) Penton protein (Virion component III) (Pento... 32 6.5
>ABF1_STRCX (P82593) Alpha-N-arabinofuranosidase I precursor (EC
3.2.1.55) (Arabinosidase I) (Alpha-N-AFase I)
Length = 825
Score = 272 bits (696), Expect = 2e-72
Identities = 182/547 (33%), Positives = 271/547 (49%), Gaps = 57/547 (10%)
Query: 13 TLTVDLKSAGRPIPETLFGVFYEEINHAGTGGLWAELVNNRGFE--AGGTQTPSNIAPWT 70
++TVD + G I +T++GVF+E+IN A GGL+AELV NR FE ++ + + W
Sbjct: 36 SITVDPAAKGAAIDDTMYGVFFEDINRAADGGLYAELVQNRSFEYSTDDNRSYTPLTSWI 95
Query: 71 IVGKESLVLLQTELSSCFERNKVALRMDVLCDQCPSDGVGVSNPGY-WGMNVVQKKEYKV 129
+ G +V + ERN+ L + G V+N GY G+ V Q K Y
Sbjct: 96 VDGTGEVV---NDAGRLNERNRNYLSLGA--------GSSVTNAGYNTGIRVEQGKRYDF 144
Query: 130 VFFVKSTGSLDMTISFKKAEDGGILASQNVKASASEVANWKRMELKLVPTASNPKATLHL 189
+ ++ + +T++ K A G LA+ A W + T ++ + L +
Sbjct: 145 SVWARAGSASTLTVALKDA--AGTLATARQVAVEG---GWAKYRATFTATRTSNRGRLAV 199
Query: 190 TTTQKGVIWLDQVSAMPVDTFKG--HGFRTDLVNMVLELKPAFIRFPGGCFIE------- 240
LD VS P DT++ +G R DL + L P F+RFPGGC +
Sbjct: 200 AANDAAA--LDMVSLFPRDTYRNQQNGLRKDLAEKIAALHPGFVRFPGGCLVNTGSMEDY 257
Query: 241 ----GQTLRNAFRWKDSVGPWEERPGHFGDVWGSWTDEGLGYFEGLQLAEDIGAKPIWVF 296
G + +++WKD+VGP EER + + WG GLGY+E + +EDIGA P+ V
Sbjct: 258 SAASGWQRKRSYQWKDTVGPVEERATN-ANFWGYNQSYGLGYYEYFRFSEDIGAMPLPVV 316
Query: 297 NNGISHTDQI----DTSVITPFVQEALDGIEFARGPATSKWGSLRKSMGHPEPFDLKYVA 352
++ Q D +++ +Q+ LD IEFA GPATSKWG +R MGHP PF L ++
Sbjct: 317 PALVTGCGQNKAVDDEALLKRHIQDTLDLIEFANGPATSKWGKVRAEMGHPRPFRLTHLE 376
Query: 353 IGNEDCGKKNYLGNYLAFYNAIRKAYPDIQMISNC--DASSKPLDHP--------ADLYD 402
+GNE+ + + F AI YPDI ++SN D + D ++ D
Sbjct: 377 VGNEENLPDEFFDRFKQFRAAIEAEYPDITVVSNSGPDDAGTTFDTAWKLNREANVEMVD 436
Query: 403 YHTYPNNPSNMFNNAHVFDKTPRKGPKAFVSEYALVGDQQAKLGTLIGGVSEAGFLIGLE 462
H Y N+P+ N +D R GPK F+ EYA G+ G+SEA F+ GLE
Sbjct: 437 EHYY-NSPNWFLQNNDRYDSYDRGGPKVFLGEYASQGNAWK------NGLSEAAFMTGLE 489
Query: 463 KNSDHVAMAAYAPLFVNADDRKWNPDAIVFNSNQVYGTPSYWVTHMFKESNGATFLAS-T 521
+N+D V +A+YAPL N D +W PD + FN+ + + +Y V +F + G + S
Sbjct: 490 RNADVVKLASYAPLLANEDYVQWRPDLVWFNNRASWNSANYEVQKLFMNNVGDRVVPSKA 549
Query: 522 LQTPDPS 528
TPD S
Sbjct: 550 TTTPDVS 556
>ABFA_ASPNG (P42254) Alpha-N-arabinofuranosidase A precursor (EC
3.2.1.55) (Arabinosidase A) (ABF A)
Length = 628
Score = 228 bits (582), Expect = 3e-59
Identities = 170/611 (27%), Positives = 293/611 (47%), Gaps = 49/611 (8%)
Query: 29 LFGVFYEEINHAGTGGLWAELVNNRGFEAGGTQTPSNIAPWTIVGKESLVLL-QTELSSC 87
L+G +E+INH+G GG++ +++ N G + T N+ W VG ++ + + L+S
Sbjct: 42 LYGFMFEDINHSGDGGIYGQMLQNPGLQG----TAPNLTAWAAVGDATIAIDGDSPLTSA 97
Query: 88 FERNKVALRMDVLCDQCPSDGVGVSNPGYWGMNVVQKKEYKVVFFVKSTGSLDMTISFKK 147
+++++ D + VG++N GYWG+ V E+ F++K S D+T+
Sbjct: 98 IPST---IKLNIADDA--TGAVGLTNEGYWGIPV-DGSEFHSSFWIKGDYSGDITVRLVG 151
Query: 148 AEDGGILASQNVKASASEVANWKRMELKLVPTASNPKATLHLTTTQKGVIWLDQ-----V 202
G S + S N+ + +K PT P + T G +
Sbjct: 152 NYTGTEYGSTTI-THTSTADNFTQASVKF-PTTKAPDGNVLYELTVDGSVAAGSSLNFGY 209
Query: 203 SAMPVDTFKG--HGFRTDLVNMVLELKPAFIRFPGGCFIEGQTLRNAFRWKDSVGPWEER 260
+ +T+K +G + L N++ ++K +F+RFPGG +EG + N ++W +++G +R
Sbjct: 210 LTLFGETYKSRENGLKPQLANVLDDMKGSFLRFPGGNNLEGNSAENRWKWNETIGDLCDR 269
Query: 261 PGHFGDVWGSWTDEGLGYFEGLQLAEDIGAKPIWVFNNGISHTDQIDT----SVITPFVQ 316
PG G W + +GLG E ED+G P+ +G + +T +TP++
Sbjct: 270 PGREG-TWTYYNTDGLGLHEYFYWCEDLGLVPVLGVWDGFALESGGNTPLTGDALTPYID 328
Query: 317 EALDGIEFARGPATSKWGSLRKSMGHPEPFDLKYVAIGNEDC---GKKNYLGNYLAFYNA 373
+ L+ +E+ G ++ +G+ R + G EP++L V IGNED G ++Y + AFY+A
Sbjct: 329 DVLNELEYILGDTSTTYGAWRAANGQEEPWNLTMVEIGNEDMLGGGCESYAERFTAFYDA 388
Query: 374 IRKAYPDIQMI-SNCDASSKPLDHPADLY-DYHTYPNNPSNMFNNAHVFDKTPRKGPKAF 431
I AYPD+ +I S +A P P + DYH Y + P + + FD R P F
Sbjct: 389 IHAAYPDLILIASTSEADCLPESMPEGSWVDYHDY-STPDGLVGQFNYFDNLNRSVP-YF 446
Query: 432 VSEYALVGDQQAKLGTLIGGVSEAGFLIGLEKNSDHVAMAAYAPLFVNADDRKWNPDAIV 491
+ EY+ + + G V+EA F+IG E+NSD V MAAYAPL + +W PD I
Sbjct: 447 IGEYS---RWEIDWPNMKGSVAEAVFMIGFERNSDVVKMAAYAPLLQLINSTQWTPDLIG 503
Query: 492 FNSN--QVYGTPSYWVTHMFKESNGATFLASTLQTPDPSSFDASTILWQNPQDKKTYLKI 549
+ + ++ + SY+V MF + G T T S D + W +Y +
Sbjct: 504 YTQSPGDIFLSTSYYVQEMFSRNRGDTIKEVT------SDSDFGPLYWVASSAGDSYY-V 556
Query: 550 KVANLGNNQVKLGIVVHGLESSKIIGTKTVLTSKNALDENTFLEPRKIVPQQTPLEEASA 609
K+AN G+ L + + G + K+ TVL + N+ + + P ++ ++ ++
Sbjct: 557 KLANYGSETQDLTVSIPGTSTGKL----TVLADSDPDAYNSDTQ-TLVTPSESTVQASNG 611
Query: 610 NMNVELPPLSV 620
LP +V
Sbjct: 612 TFTFSLPAWAV 622
>ABF2_BACOV (Q59219) Alpha-N-arabinofuranosidase 2 (EC 3.2.1.55)
(Arabinosidase 2)
Length = 514
Score = 94.0 bits (232), Expect = 1e-18
Identities = 117/513 (22%), Positives = 205/513 (39%), Gaps = 96/513 (18%)
Query: 175 KLVPTASNPKATLHLTTTQKGVIWLDQVSAMPVDTFKGHGFRTDLVNMVLELKPAFIRFP 234
+++P + HL + G +W+ + S +P G+RTD+ N + +L +R+P
Sbjct: 35 EIIPKEIYGQFAEHLGSCIYGGLWVGENSDIP----NIKGYRTDVFNALKDLSVPVLRWP 90
Query: 235 GGCFIEGQTLRNAFRWKDSVGPWEERPGHFGDVW-GSWTDEGLGYFEGLQLAEDIGAKPI 293
GGCF + + W D +GP E RP + W G+ D G E L L E +G +P
Sbjct: 91 GGCFAD------EYHWMDGIGPKENRPKMVNNNWGGTIEDNSFGTHEFLNLCEMLGCEP- 143
Query: 294 WVFNNGISHTDQIDTSVITPFVQEALDGIEFARGPATSKWGSLRKSMGHPEPFDLKYVAI 353
+V N S T V+E +E+ S +LR+ G + + LKY+ +
Sbjct: 144 YVSGNVGSGT-----------VEELAKWVEYMTSDGDSPMANLRRKNGRDKAWKLKYLGV 192
Query: 354 GNEDCG-----KKNYLGNYLAFYNAIRKAYPDIQMISNCDASS-----------KPLDHP 397
GNE G + Y + Y+ + Y ++ +S + H
Sbjct: 193 GNESWGCGGSMRPEYYADLYRRYSTYCRNYDGNRLFKIASGASDYDYKWTDVLMNRVGHR 252
Query: 398 ADLYDYHTY-------PNNPSNMFN--------------------NAHVFDKTPR-KGPK 429
D H Y + FN + + DK + K
Sbjct: 253 MDGLSLHYYTVTGWSGSKGSATQFNKDDYYWTMGKCLEVEDVLKKHCTIMDKYDKDKKIA 312
Query: 430 AFVSEYALVGDQQAKLGTLIGGVSEA-----GFLIGL-----EKNSDHVAMAAYAPLFVN 479
+ E+ D++ GT+ G + + F+ L K +D + MA A + VN
Sbjct: 313 LLLDEWGTWWDEEP--GTIKGHLYQQNTLRDAFVASLSLDVFHKYTDRLKMANIAQI-VN 369
Query: 480 ADDRKWNPDAIVFNSNQVYGTPSYWVTHMFKESNGATFLASTLQTPDPSSFDASTILWQN 539
I+ ++ TP+Y+V M+K AT+L L S D T+ +
Sbjct: 370 V-----LQSMILTKDKEMVLTPTYYVFKMYKVHQDATYLPIDLTCEKMSVRDNRTVPMVS 424
Query: 540 ---PQDKKTYLKIKVANLGNNQV-KLGIVVHGLESSKIIGTKTVLTSKNALDENTFLEPR 595
++K + I ++N+ ++ ++ I + ++ K IG +LT+ D N+F +P
Sbjct: 425 ATASKNKDGVIHISLSNVDADEAQEITINLGDTKAKKAIG--EILTASKLTDYNSFEKPN 482
Query: 596 KIVPQQTPLEEASAN---MNVELPPLSVTSFDI 625
+ P P +E N M V+LP S+ + ++
Sbjct: 483 IVKP--APFKEVKINKGTMKVKLPAKSIVTLEL 513
>ABFA_BACHD (Q9KBR4) Alpha-N-arabinofuranosidase (EC 3.2.1.55)
(Arabinosidase)
Length = 500
Score = 81.6 bits (200), Expect = 6e-15
Identities = 63/217 (29%), Positives = 95/217 (43%), Gaps = 39/217 (17%)
Query: 214 GFRTDLVNMVLELKPAFIRFPGGCFIEGQTLRNAFRWKDSVGPWEERPGHFGDVWGSWTD 273
GFR D++ +V EL+ +R+PGG F+ G + W+D VGP ERP W +
Sbjct: 50 GFRKDVIRLVQELQVPLVRYPGGNFVSG------YNWEDGVGPVSERPKRLDLAWRTTET 103
Query: 274 EGLGYFEGLQLAEDIGAKPIWVFNNGISHTDQIDTSVITPFVQEALDGIEFARGPATSKW 333
+G E + A+ +GA+ N G D A + +E+ P+ S W
Sbjct: 104 NEIGTNEFVDWAKKVGAEVNMAVNLGSRGVD------------AARNLVEYCNHPSGSYW 151
Query: 334 GSLRKSMGHPEPFDLKYVAIGNE-----DCGKK--NYLGNYLAFYNAIRKAY-PDIQMIS 385
LR S G+ +P ++K +GNE G+K G A + K P I++++
Sbjct: 152 SDLRISHGYKDPHNIKTWCLGNEMDGPWQIGQKTAEEYGRVAAEAGKVMKLVDPSIELVA 211
Query: 386 NCDASSK----------PLDHPADLYDY---HTYPNN 409
++SK LDH D DY HTY N
Sbjct: 212 CGSSNSKMATFADWEATVLDHTYDYVDYISLHTYYGN 248
>ABFA_STRLI (P53627) Alpha-N-arabinofuranosidase (EC 3.2.1.55)
(Arabinosidase) (ABF) (Alpha-L-AF)
Length = 662
Score = 76.3 bits (186), Expect = 2e-13
Identities = 56/197 (28%), Positives = 90/197 (45%), Gaps = 29/197 (14%)
Query: 209 TFKGHGFRTDLVNMVLELKPAFIRFPGGCFIEGQTLRNAFRWKDSVGPWEERPGHFGDVW 268
T G R D++ +V EL +R+PGG F+ G ++W+DSVGP E+RP W
Sbjct: 44 TADAEGLRQDVLELVRELGVTAVRYPGGNFVSG------YKWEDSVGPVEDRPRRLDLAW 97
Query: 269 GSWTDEGLGYFEGLQLAEDIG--AKPIWVFNNGISHTDQIDTSVITPFVQEALDGIEFAR 326
S G E + + IG A+P+ N G T V EAL+ E+A
Sbjct: 98 RSTETNRFGLSEYIAFLKKIGPQAEPMMAVNLG------------TRGVAEALELQEYAN 145
Query: 327 GPATSKWGSLRKSMGHPEPFDLKYVAIGNEDCG--------KKNYLGNYLAFYNAIRKAY 378
P+ + LR G +PF ++ +GNE G + Y A+R+
Sbjct: 146 HPSGTALSDLRAEHGDKDPFGIRLWCLGNEMDGPWQTGHKTAEEYGRVAAETARAMRQID 205
Query: 379 PDIQMISNCDASSKPLD 395
PD+++++ C +S + ++
Sbjct: 206 PDVELVA-CGSSGQSME 221
>ABFA_BACST (Q9XBQ3) Alpha-N-arabinofuranosidase (EC 3.2.1.55)
(Arabinosidase)
Length = 501
Score = 75.1 bits (183), Expect = 5e-13
Identities = 62/222 (27%), Positives = 94/222 (41%), Gaps = 41/222 (18%)
Query: 213 HGFRTDLVNMVLELKPAFIRFPGGCFIEGQTLRNAFRWKDSVGPWEERPGHFGDVWGSWT 272
+GFR D++ +V EL+ IR+PGG F+ G + W+D VGP E+RP W S
Sbjct: 49 NGFRQDVIELVKELQVPIIRYPGGNFVSG------YNWEDGVGPKEQRPRRLDLAWKSVE 102
Query: 273 DEGLGYFEGLQLAEDIGAKPIWVFNNGISHTDQIDTSVITPFVQEALDGIEFARGPATSK 332
+G E + A+ +GA+ N G T + A + +E+ P+ S
Sbjct: 103 TNEIGLNEFMDWAKMVGAEVNMAVNLG------------TRGIDAARNLVEYCNHPSGSY 150
Query: 333 WGSLRKSMGHPEPFDLKYVAIGNEDCGKKNYLGNYLAFYNAI--------RKAYPDIQMI 384
+ LR + G+ EP +K +GNE G Y I + P I+++
Sbjct: 151 YSDLRIAHGYKEPHKIKTWCLGNEMDGPWQIGHKTAVEYGRIACEAAKVMKWVDPTIELV 210
Query: 385 SNCDASSK-----------PLDHPADLYDY---HTYPNNPSN 412
C +S++ LDH D DY H Y N N
Sbjct: 211 V-CGSSNRNMPTFAEWEATVLDHTYDHVDYISLHQYYGNRDN 251
>ABFA_BACSU (P94531) Alpha-N-arabinofuranosidase 1 (EC 3.2.1.55)
(Arabinosidase 1)
Length = 500
Score = 74.7 bits (182), Expect = 7e-13
Identities = 62/224 (27%), Positives = 94/224 (41%), Gaps = 39/224 (17%)
Query: 214 GFRTDLVNMVLELKPAFIRFPGGCFIEGQTLRNAFRWKDSVGPWEERPGHFGDVWGSWTD 273
GFR D+ +++ EL+ IR+PGG F+ G + W+D VGP E RP W +
Sbjct: 49 GFRKDVQSLIKELQVPIIRYPGGNFLSG------YNWEDGVGPVENRPRRLDLAWQTTET 102
Query: 274 EGLGYFEGLQLAEDIGAKPIWVFNNGISHTDQIDTSVITPFVQEALDGIEFARGPATSKW 333
+G E L + + + N G T + A + +E+ P S W
Sbjct: 103 NEVGTNEFLSWPKKVNTEVNMAVNLG------------TRGIDAARNLVEYCNHPKGSYW 150
Query: 334 GSLRKSMGHPEPFDLKYVAIGNEDCG-------KKNYLGNYLAFYNAIRK-AYPDIQMI- 384
LR+S G+ +P+ +K +GNE G + G A + K P I+++
Sbjct: 151 SDLRRSHGYEQPYGIKTWCLGNEMDGPWQIGHKTADEYGRLAAETAKVMKWVDPSIELVA 210
Query: 385 ---SN------CDASSKPLDHPADLYDY---HTYPNNPSNMFNN 416
SN D +K L+H + DY HTY N N N
Sbjct: 211 CGSSNSGMPTFIDWEAKVLEHTYEHVDYISLHTYYGNRDNNLPN 254
>ABF2_BACSU (P94552) Alpha-N-arabinofuranosidase 2 (EC 3.2.1.55)
(Arabinosidase 2)
Length = 495
Score = 63.9 bits (154), Expect = 1e-09
Identities = 109/477 (22%), Positives = 190/477 (38%), Gaps = 80/477 (16%)
Query: 197 IWLDQVSAMPVDTFKGHGFRTDLVNMVLELKPAFIRFPGGCFIEGQTLRNAFRWKDSVGP 256
IW+ S +P +G R D++ + +L +R+PGGCF + + W + VG
Sbjct: 37 IWVGTDSDIP----NINGIRKDVLEALKQLHIPVLRWPGGCFAD------EYHWANGVG- 85
Query: 257 WEERPGHFGDVWGSWTDEG-LGYFEGLQLAEDIGAKPIWVFNNGISHTDQIDTSVITPFV 315
+R WG + G E + L E + +P N G ++
Sbjct: 86 --DRKTMLNTHWGGTIESNEFGTHEFMMLCELLECEPYICGNVGSGTVQEMS-------- 135
Query: 316 QEALDGIEFARGPATSKWGSLRKSMGHPEPFDLKYVAIGNED--CG----KKNYLGNYLA 369
E ++ + F G S W RK G EP+ LKY +GNE+ CG + Y Y
Sbjct: 136 -EWIEYMTFEEGTPMSDW---RKQNGREEPWKLKYFGVGNENWGCGGNMHPEYYADLYRR 191
Query: 370 FYNAIRK-AYPDIQMIS---NCDA---SSKPLDHPADLYD-----YHTYPNNPSNMFNNA 417
F +R + DI I+ N D + + A L D Y+T P + +A
Sbjct: 192 FQTYVRNYSGNDIYKIAGGANVDDFNWTDVLMKKAAGLMDGLSLHYYTIPGDFWKGKGSA 251
Query: 418 HVFDK-----TPRKGP--KAFVSEYALVGDQ---QAKLGTLIGGV---------SEAGFL 458
F + T +K + ++ + D+ + ++G +I + GFL
Sbjct: 252 TEFTEDEWFITMKKAKYIDELIQKHGTIMDRYDPEQRVGLIIDEWGTWFDPEPGTNPGFL 311
Query: 459 IGLEKNSDHVAMAAYAPLFVNADDRKWNPDA----------IVFNSNQVYGTPSYWVTHM 508
D + A++ +F R + I+ ++ TP+Y V +M
Sbjct: 312 YQQNTIRDALVAASHFHIFHQHCRRVQMANIAQTVNVLQAMILTEGERMLLTPTYHVFNM 371
Query: 509 FKESNGATFLASTLQTPDPSSFDAST---ILWQNPQDKKTYLKIKVANLGN-NQVKLGIV 564
FK A+ LA+ + D ++ T I + + + I + N+ + N+ + I
Sbjct: 372 FKVHQDASLLATETMSAD-YEWNGETFPQISISASKQAEGDINITICNIDHQNKAEAEIE 430
Query: 565 VHGLESSKIIGTKTVLTSKNALDENTFLEPRKIVPQQTPLEEASAN-MNVELPPLSV 620
+ GL + + +LT++ NTF +P + P+ S N + V+LPP+SV
Sbjct: 431 LRGLHKA-ADHSGVILTAEKMNAHNTFDDPHHVKPESFRQYTLSKNKLKVKLPPMSV 486
>DAP2_CLOAB (Q97D80) Dihydrodipicolinate synthase 2 (EC 4.2.1.52)
(DHDPS 2)
Length = 286
Score = 33.1 bits (74), Expect = 2.2
Identities = 25/88 (28%), Positives = 41/88 (46%)
Query: 518 LASTLQTPDPSSFDASTILWQNPQDKKTYLKIKVANLGNNQVKLGIVVHGLESSKIIGTK 577
LA+T +TP S ++ +IL + + + L I V GNN K+ + L+ I G
Sbjct: 42 LATTGETPTLSDYEYQSILEKTVECNELNLPIYVGFGGNNTEKMTKDIKMLDKYNIKGIL 101
Query: 578 TVLTSKNALDENTFLEPRKIVPQQTPLE 605
+V N D+ E K + + T L+
Sbjct: 102 SVCPYYNRPDQRGIYEHFKRISESTSLD 129
>BIR4_RAT (Q9R0I6) Baculoviral IAP repeat-containing protein 4
(Inhibitor of apoptosis protein 3) (X-linked inhibitor
of apoptosis protein) (X-linked IAP) (IAP homolog A)
(RIAP3) (RIAP-3)
Length = 496
Score = 33.1 bits (74), Expect = 2.2
Identities = 19/71 (26%), Positives = 30/71 (41%), Gaps = 1/71 (1%)
Query: 250 WKDSVGPWEERPGHFGDVWGSWTDEGLGYFEGLQLAEDIGAKPIWVFNNGISHTDQIDTS 309
WK S PWE+ + ++G Y + L +G + S T +ID +
Sbjct: 309 WKPSEDPWEQHAKWYPGCKYLLDEKGQEYINNIHLTHSLGESVVRTAEKTPSVTKKIDDT 368
Query: 310 VI-TPFVQEAL 319
+ P VQEA+
Sbjct: 369 IFQNPMVQEAI 379
>CDGF_HUMAN (Q9Y5G1) Protocadherin gamma B3 precursor
(PCDH-gamma-B3)
Length = 929
Score = 32.7 bits (73), Expect = 2.9
Identities = 23/81 (28%), Positives = 35/81 (42%), Gaps = 2/81 (2%)
Query: 393 PLDHPADLYDYHTYPNNPSNMFNNAHVFDKTPRKGPKAFVSEYALVGDQQAKLGTLIGGV 452
P+ H A Y H NNP + AHV P GP VS Y + D + + + V
Sbjct: 450 PVFHQAS-YTVHVAENNPPGA-SIAHVRASDPDLGPNGLVSYYIVASDLEPRELSSYVSV 507
Query: 453 SEAGFLIGLEKNSDHVAMAAY 473
S ++ ++ DH + A+
Sbjct: 508 SARSGVVFAQRAFDHEQLRAF 528
>TACY_BACCE (Q45105) Hemolysin precursor (Fragment)
Length = 485
Score = 32.0 bits (71), Expect = 5.0
Identities = 23/76 (30%), Positives = 30/76 (39%), Gaps = 5/76 (6%)
Query: 407 PNNPSNMFNNAHVFDKTPRKG-----PKAFVSEYALVGDQQAKLGTLIGGVSEAGFLIGL 461
PNNPS++F+N+ F + RKG P VS A KL T L
Sbjct: 245 PNNPSDLFDNSVTFGELTRKGVSNSAPPVMVSNVAYGRTVYVKLETTSKSKDVQAAFKAL 304
Query: 462 EKNSDHVAMAAYAPLF 477
KN+ Y +F
Sbjct: 305 LKNNSVETSGQYKDIF 320
>PT1_STACA (P23533) Phosphoenolpyruvate-protein phosphotransferase
(EC 2.7.3.9) (Phosphotransferase system, enzyme I)
Length = 574
Score = 32.0 bits (71), Expect = 5.0
Identities = 21/73 (28%), Positives = 34/73 (45%), Gaps = 7/73 (9%)
Query: 522 LQTPDPSSFDASTILWQN---PQD----KKTYLKIKVANLGNNQVKLGIVVHGLESSKII 574
++ P+PS D S ++ N P D K Y++ V N+G I+ LE ++
Sbjct: 146 VELPNPSIVDESVVIIGNDLTPSDTAQLNKEYVQGFVTNIGGRTSHSAIMSRSLEIPAVV 205
Query: 575 GTKTVLTSKNALD 587
GTK++ A D
Sbjct: 206 GTKSITEEVEAGD 218
>SYGB_HELPJ (Q9ZKM9) Glycyl-tRNA synthetase beta chain (EC 6.1.1.14)
(Glycine--tRNA ligase beta chain) (GlyRS)
Length = 701
Score = 31.6 bits (70), Expect = 6.5
Identities = 20/74 (27%), Positives = 35/74 (47%), Gaps = 8/74 (10%)
Query: 275 GLGYFEGLQLAEDIGAKPIWVFNNGIS-----HTDQIDTSVITPFVQEALDGIEFARGPA 329
GLG+++ L L + + + N + H Q +I P V E L+G+ F +
Sbjct: 94 GLGFYQKLGLKDHQHFQTAFKSNKEVLYHAKIHAKQPTKDLIMPIVLEFLEGLNFGK--- 150
Query: 330 TSKWGSLRKSMGHP 343
+ +WG++ KS P
Sbjct: 151 SMRWGNVEKSFIRP 164
>PEN3_ADEP3 (Q84176) Penton protein (Virion component III) (Penton
base protein)
Length = 484
Score = 31.6 bits (70), Expect = 6.5
Identities = 28/123 (22%), Positives = 53/123 (42%), Gaps = 11/123 (8%)
Query: 437 LVGDQQAKLGTLIGGVSEAGFLIGLEKNSDHVAMAAYAPLFVNADDR-KWNPDAIVFNSN 495
L+ ++ A + +L + + FL + +NSD M A + +N D+R +W + +
Sbjct: 63 LIDNKSADIASLNYQNNHSDFLTSVVQNSDFTPMEA-STQTINLDERSRWGGEFKSILTT 121
Query: 496 QVYGTPSYWVTHMF---------KESNGATFLASTLQTPDPSSFDASTILWQNPQDKKTY 546
+ Y ++ F KE+N T+ TL P+ + D + I N + Y
Sbjct: 122 NIPNVTQYMFSNSFRVRLMSARDKETNAPTYEWFTLTLPEGNFSDIAVIDLMNNAIVENY 181
Query: 547 LKI 549
L +
Sbjct: 182 LAV 184
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.135 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 78,302,396
Number of Sequences: 164201
Number of extensions: 3498082
Number of successful extensions: 7143
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 7104
Number of HSP's gapped (non-prelim): 21
length of query: 626
length of database: 59,974,054
effective HSP length: 116
effective length of query: 510
effective length of database: 40,926,738
effective search space: 20872636380
effective search space used: 20872636380
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0098a.6


