

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0097b.12
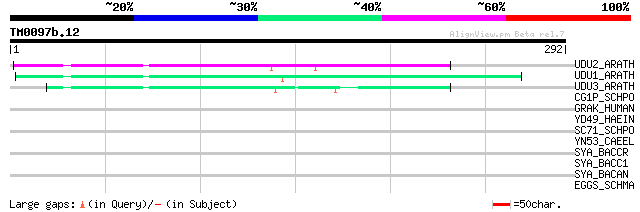
(292 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UDU2_ARATH (Q9FHD4) DUF26 domain containing protein 2 precursor 100 7e-21
UDU1_ARATH (Q9FHD5) DUF26 domain containing protein 1 precursor 95 2e-19
UDU3_ARATH (Q9FHD3) Hypothetical DUF26 domain containing protein... 80 8e-15
CG1P_SCHPO (P25009) Cyclin puc1 31 3.1
GRAK_HUMAN (P49863) Granzyme K precursor (EC 3.4.21.-) (Granzyme... 31 4.1
YD49_HAEIN (P45173) Protein HI1349 30 7.0
SC71_SCHPO (Q9UT02) Protein transport protein sec71 30 7.0
YN53_CAEEL (P34587) Hypothetical protein ZC21.3 in chromosome III 30 9.2
SYA_BACCR (Q817Z0) Alanyl-tRNA synthetase (EC 6.1.1.7) (Alanine-... 30 9.2
SYA_BACC1 (P61697) Alanyl-tRNA synthetase (EC 6.1.1.7) (Alanine-... 30 9.2
SYA_BACAN (Q81LK0) Alanyl-tRNA synthetase (EC 6.1.1.7) (Alanine-... 30 9.2
EGGS_SCHMA (P08016) Putative eggshell protein (Fragment) 30 9.2
>UDU2_ARATH (Q9FHD4) DUF26 domain containing protein 2 precursor
Length = 287
Score = 99.8 bits (247), Expect = 7e-21
Identities = 65/242 (26%), Positives = 102/242 (41%), Gaps = 19/242 (7%)
Query: 3 LLLRLLFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSAS 62
L L +T++ + S ++TF C + S+ Y +N+LL++L N +S
Sbjct: 7 LFALLCLFVTMNQAISVSDPDDMETF----CMKSSRNTTSNTTYNKNLNTLLSTLSNQSS 62
Query: 63 FTNYNNFTVPATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLE 122
F NY N T T + DT++G++ C GD+N C CV A ++ C+ R +
Sbjct: 63 FANYYNLT---TGLASDTVHGMFLCTGDVNRTTCNACVKNATIEIAKNCTNHREAIIYNV 119
Query: 123 GCFVKYDNVKFLGV---------EDKTMVVKKCGPSIGLTSDALTR---RDAVLAYLQTS 170
C V+Y + FL ++ K G SD + R ++L+ T
Sbjct: 120 DCMVRYSDKFFLTTLETNPSYWWSSNDLIPKSFGKFGQRLSDKMGEVIVRSSLLSSSFTP 179
Query: 171 DGVYRTWRISAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYA 230
+ T R D + QCT DL P C CL ++Q L CG W +Y C
Sbjct: 180 YYLMDTTRFDNLYDLESIVQCTPDLDPRNCTTCLKLALQELTECCGNQVWAFIYTPNCMV 239
Query: 231 RY 232
+
Sbjct: 240 SF 241
Score = 30.8 bits (68), Expect = 4.1
Identities = 13/48 (27%), Positives = 21/48 (43%)
Query: 187 GAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSE 234
G CTGD++ + C C+ + + C +Y C RYS+
Sbjct: 80 GMFLCTGDVNRTTCNACVKNATIEIAKNCTNHREAIIYNVDCMVRYSD 127
Score = 30.4 bits (67), Expect = 5.4
Identities = 28/92 (30%), Positives = 39/92 (41%), Gaps = 11/92 (11%)
Query: 41 VSDPVYETGVNSLLTSLVNSASFTNYNNFTVPATAGSGDTLYGL---YQCRGDLNNDQCY 97
+SD + E V S L S +SFT Y + + T D LY L QC DL+ C
Sbjct: 159 LSDKMGEVIVRSSLLS----SSFTPY--YLMDTTRF--DNLYDLESIVQCTPDLDPRNCT 210
Query: 98 RCVARAVSQLGTLCSAVRGGALQLEGCFVKYD 129
C+ A+ +L C + C V +D
Sbjct: 211 TCLKLALQELTECCGNQVWAFIYTPNCMVSFD 242
>UDU1_ARATH (Q9FHD5) DUF26 domain containing protein 1 precursor
Length = 286
Score = 94.7 bits (234), Expect = 2e-19
Identities = 71/279 (25%), Positives = 111/279 (39%), Gaps = 20/279 (7%)
Query: 4 LLRLLFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASF 63
+L LF AI+ + + TF C+ + Y T +N+LL++L N +SF
Sbjct: 9 VLLCLFFTMNQAISESDSDEHMATF----CNDSSGNFTRNTTYNTNLNTLLSTLSNQSSF 64
Query: 64 TNYNNFTVPATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEG 123
NY N T T DT++G++ C GD+N C CV A ++ C+ R +
Sbjct: 65 ANYYNLT---TGLGSDTVHGMFLCIGDVNRTTCNACVKNATIEIAKNCTNHREAIIYYFS 121
Query: 124 CFVKYDNVKFLG-VEDKTMV-----------VKKCGPSIGLTSDALTRRDAVLAYLQTSD 171
C V+Y + FL +E K K G + + R ++L+ T
Sbjct: 122 CMVRYSDKFFLSTLETKPNTYWSSDDPIPKSYDKFGQRLSDKMGEVIIRSSLLSSSFTPY 181
Query: 172 GVYRTWRISAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYAR 231
+ T D + QC+ L P C CL ++Q L CG W ++ KC
Sbjct: 182 YLMDTTTFDNLYDLESVVQCSPHLDPKNCTTCLKLALQELTQCCGDQLWAFIFTPKCLVS 241
Query: 232 YSEGGDHSRHS-DDDSNHNDDEIEKTLAILIGLIAGVAL 269
+ S S I IL+G+I V++
Sbjct: 242 FDTSNSSSLPPLPPPSRSGSFSIRGNNKILVGMILAVSV 280
>UDU3_ARATH (Q9FHD3) Hypothetical DUF26 domain containing protein 3
precursor
Length = 287
Score = 79.7 bits (195), Expect = 8e-15
Identities = 58/235 (24%), Positives = 92/235 (38%), Gaps = 39/235 (16%)
Query: 20 STSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNYNNFTVPATAGSGD 79
S + +DTF C + Y +N++L++ N +S N N T T + D
Sbjct: 28 SETDHMDTF----CIDSSRNTTGNTTYNKNLNTMLSTFRNQSSIVNNYNLT---TGLASD 80
Query: 80 TLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKYDNVKFLGVED- 138
T+YG++ C GD+N C CV A ++ C+ R + C V+Y + FL +
Sbjct: 81 TVYGMFLCTGDVNITTCNNCVKNATIEIVKNCTNHREAIIYYIDCMVRYSDKFFLSTFEK 140
Query: 139 -------------------KTMVVKKCGPSIGLTSDALTRRDAVLAYLQTS--DGVYRTW 177
K + KK G +I + S L+ YL + DG Y
Sbjct: 141 KPNSIWSGDDPIPKSLGPFKKRLYKKMGEAI-VRSSTLSSALTPYYYLDVTRFDGSY--- 196
Query: 178 RISAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARY 232
D QC+ L+P C CL ++Q + C W ++ C+ Y
Sbjct: 197 ------DLDSLVQCSPHLNPENCTICLEYALQEIIDCCSDKFWAMIFTPNCFVNY 245
Score = 31.6 bits (70), Expect = 2.4
Identities = 13/48 (27%), Positives = 22/48 (45%)
Query: 187 GAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSE 234
G CTGD++ + C +C+ + + C +Y C RYS+
Sbjct: 84 GMFLCTGDVNITTCNNCVKNATIEIVKNCTNHREAIIYYIDCMVRYSD 131
>CG1P_SCHPO (P25009) Cyclin puc1
Length = 359
Score = 31.2 bits (69), Expect = 3.1
Identities = 20/47 (42%), Positives = 25/47 (52%), Gaps = 1/47 (2%)
Query: 18 TPSTSSSIDTFIFGGCSQPKNT-PVSDPVYETGVNSLLTSLVNSASF 63
TP++SSSI+ I CS + P S V GVNS SL N +F
Sbjct: 14 TPTSSSSIEPKILAACSYSLSVGPCSLAVSPKGVNSKSPSLKNETAF 60
>GRAK_HUMAN (P49863) Granzyme K precursor (EC 3.4.21.-) (Granzyme 3)
(NK-tryptase-2) (NK-TRYP-2) (Fragmentin 3)
Length = 264
Score = 30.8 bits (68), Expect = 4.1
Identities = 22/80 (27%), Positives = 31/80 (38%)
Query: 32 GCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNYNNFTVPATAGSGDTLYGLYQCRGDL 91
G + P + SD + E V L L NS S+ N + F +GD C+GD
Sbjct: 155 GATDPDSLRPSDTLREVTVTVLSRKLCNSQSYYNGDPFITKDMVCAGDAKGQKDSCKGDS 214
Query: 92 NNDQCYRCVARAVSQLGTLC 111
+ V A+ G C
Sbjct: 215 GGPLICKGVFHAIVSGGHEC 234
>YD49_HAEIN (P45173) Protein HI1349
Length = 160
Score = 30.0 bits (66), Expect = 7.0
Identities = 13/34 (38%), Positives = 22/34 (64%)
Query: 180 SAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKT 213
+AY + ++ D++ SE Q+CLS ++Q LKT
Sbjct: 80 NAYSQYLKISRIKEDIAVSEAQECLSGTLQGLKT 113
>SC71_SCHPO (Q9UT02) Protein transport protein sec71
Length = 1811
Score = 30.0 bits (66), Expect = 7.0
Identities = 39/164 (23%), Positives = 66/164 (39%), Gaps = 25/164 (15%)
Query: 10 ILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDP--VYETGVNSLLTSLVNSASFTNYN 67
I+TLS I + STS +F+F + + P VY T +N + N
Sbjct: 549 IVTLSRIASQSTSDPPPSFVF----RDDQLVIDKPGFVYHT-LNDI----------PQLN 593
Query: 68 NFTVPATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVK 127
+ T+ + S + Y YQ R + YRC+ +S L T C+ +++ K
Sbjct: 594 SSTIGSYVHSHNPPYFDYQIR-----LKSYRCLISTLSSLFTWCNQTFAPTVEIT---AK 645
Query: 128 YDNVKFLGVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQTSD 171
D + ++ K PS G+ S ++ ++ L T D
Sbjct: 646 DDETESTSKGEEPQKSKSEPPSAGINSTSMDNLESSGQALATDD 689
>YN53_CAEEL (P34587) Hypothetical protein ZC21.3 in chromosome III
Length = 321
Score = 29.6 bits (65), Expect = 9.2
Identities = 35/157 (22%), Positives = 56/157 (35%), Gaps = 20/157 (12%)
Query: 45 VYETGVNSLLTSLVNSASFTNYNNFTVPATAGSGDTLYGLYQCRGDLNNDQCYRCVARAV 104
+Y+TG + T S+S Y+N P+T S + YG + Q Y + +
Sbjct: 59 MYQTGTTNYPTLYQPSSSSPQYSN-GAPSTTYSPTSTYGY--------SSQNYNPSSASS 109
Query: 105 SQLGTLCSAVRGGALQLEGCFVKYDN-----------VKFLGVEDKTMVVKKCGPSIGLT 153
S + S G Q +V YD V + + V P
Sbjct: 110 SNVYRNPSTSYGNTQQSNQNYVAYDTTNQNSPYYYQRVPSTAQYNSVLGVNAGFPQSSNQ 169
Query: 154 SDALTRRDAVLAYLQTSDGVYRTWRISAYGDFQGAAQ 190
+ L + Y+ + G+Y T YG+ QG+ Q
Sbjct: 170 NQVLRDQSNNQGYMNNNQGMYNTQTTQGYGNNQGSYQ 206
>SYA_BACCR (Q817Z0) Alanyl-tRNA synthetase (EC 6.1.1.7)
(Alanine--tRNA ligase) (AlaRS)
Length = 880
Score = 29.6 bits (65), Expect = 9.2
Identities = 23/82 (28%), Positives = 38/82 (46%), Gaps = 5/82 (6%)
Query: 39 TPVSDPVYETGVN-SLLTSLVNSASFTNYNNFTVPATAGSGDTLYGLYQCRGDLNNDQCY 97
TP+ +TG+ +TS+V TN++ G+ +T+ G GDL D +
Sbjct: 218 TPLPKKNIDTGMGLERMTSIVQDVP-TNFDTDLFMPMIGATETISGEKYRNGDLEKDMAF 276
Query: 98 RCVARAVSQLGTLCSAVRGGAL 119
+ +A + T+ AV GAL
Sbjct: 277 KVIA---DHIRTVTFAVGDGAL 295
>SYA_BACC1 (P61697) Alanyl-tRNA synthetase (EC 6.1.1.7)
(Alanine--tRNA ligase) (AlaRS)
Length = 880
Score = 29.6 bits (65), Expect = 9.2
Identities = 23/82 (28%), Positives = 38/82 (46%), Gaps = 5/82 (6%)
Query: 39 TPVSDPVYETGVN-SLLTSLVNSASFTNYNNFTVPATAGSGDTLYGLYQCRGDLNNDQCY 97
TP+ +TG+ +TS+V TN++ G+ +T+ G GDL D +
Sbjct: 218 TPLPKKNIDTGMGLERMTSIVQDVP-TNFDTDLFMPMIGATETISGEKYRNGDLEKDMAF 276
Query: 98 RCVARAVSQLGTLCSAVRGGAL 119
+ +A + T+ AV GAL
Sbjct: 277 KVIA---DHIRTVTFAVGDGAL 295
>SYA_BACAN (Q81LK0) Alanyl-tRNA synthetase (EC 6.1.1.7)
(Alanine--tRNA ligase) (AlaRS)
Length = 880
Score = 29.6 bits (65), Expect = 9.2
Identities = 23/82 (28%), Positives = 38/82 (46%), Gaps = 5/82 (6%)
Query: 39 TPVSDPVYETGVN-SLLTSLVNSASFTNYNNFTVPATAGSGDTLYGLYQCRGDLNNDQCY 97
TP+ +TG+ +TS+V TN++ G+ +T+ G GDL D +
Sbjct: 218 TPLPKKNIDTGMGLERMTSIVQDVP-TNFDTDLFMPMIGATETISGEKYRNGDLEKDMAF 276
Query: 98 RCVARAVSQLGTLCSAVRGGAL 119
+ +A + T+ AV GAL
Sbjct: 277 KVIA---DHIRTVTFAVGDGAL 295
>EGGS_SCHMA (P08016) Putative eggshell protein (Fragment)
Length = 149
Score = 29.6 bits (65), Expect = 9.2
Identities = 10/23 (43%), Positives = 15/23 (64%)
Query: 229 YARYSEGGDHSRHSDDDSNHNDD 251
Y +YS DH +H D+ +H+DD
Sbjct: 100 YEKYSYRKDHDKHDHDEHDHHDD 122
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 34,146,079
Number of Sequences: 164201
Number of extensions: 1397780
Number of successful extensions: 3862
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 3845
Number of HSP's gapped (non-prelim): 15
length of query: 292
length of database: 59,974,054
effective HSP length: 109
effective length of query: 183
effective length of database: 42,076,145
effective search space: 7699934535
effective search space used: 7699934535
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0097b.12


