

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
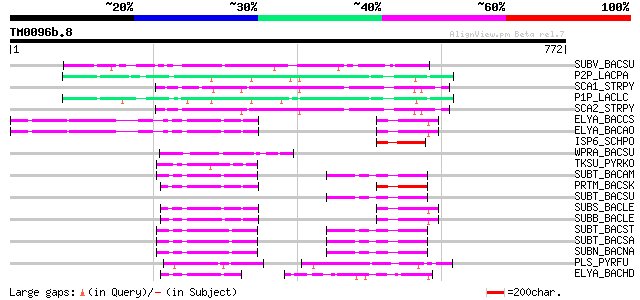
Query= TM0096b.8
(772 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SUBV_BACSU (P29141) Minor extracellular protease vpr precursor (... 123 2e-27
P2P_LACPA (Q02470) PII-type proteinase precursor (EC 3.4.21.96) ... 74 1e-12
SCA1_STRPY (P15926) C5a peptidase precursor (EC 3.4.21.-) (SCP) 73 2e-12
P1P_LACLC (P16271) PI-type proteinase precursor (EC 3.4.21.-) (W... 72 4e-12
SCA2_STRPY (P58099) C5a peptidase precursor (EC 3.4.21.-) (SCP) 70 3e-11
ELYA_BACCS (P41362) Alkaline protease precursor (EC 3.4.21.-) 56 4e-07
ELYA_BACAO (P27693) Alkaline protease precursor (EC 3.4.21.-) 56 4e-07
ISP6_SCHPO (P40903) Sexual differentiation process putative subt... 55 7e-07
WPRA_BACSU (P54423) Cell wall-associated protease precursor (EC ... 55 9e-07
TKSU_PYRKO (P58502) Tk-subtilisin precursor (EC 3.4.21.-) 52 6e-06
SUBT_BACAM (P00782) Subtilisin BPN' precursor (EC 3.4.21.62) (Su... 52 6e-06
PRTM_BACSK (Q99405) M-protease (EC 3.4.21.-) 52 6e-06
SUBT_BACSU (P04189) Subtilisin E precursor (EC 3.4.21.62) 52 8e-06
SUBS_BACLE (P29600) Subtilisin Savinase (EC 3.4.21.62) (Alkaline... 52 8e-06
SUBB_BACLE (P29599) Subtilisin BL (EC 3.4.21.62) (Alkaline prote... 52 8e-06
SUBT_BACST (P29142) Subtilisin J precursor (EC 3.4.21.62) 51 1e-05
SUBT_BACSA (P00783) Subtilisin amylosacchariticus precursor (EC ... 51 1e-05
SUBN_BACNA (P35835) Subtilisin NAT precursor (EC 3.4.21.62) 51 1e-05
PLS_PYRFU (P72186) Pyrolysin precursor (EC 3.4.21.-) 51 1e-05
ELYA_BACHD (P41363) Thermostable alkaline protease precursor (EC... 51 1e-05
>SUBV_BACSU (P29141) Minor extracellular protease vpr precursor (EC
3.4.21.-)
Length = 806
Score = 123 bits (309), Expect = 2e-27
Identities = 136/529 (25%), Positives = 223/529 (41%), Gaps = 85/529 (16%)
Query: 75 YKHVAHGFSTRLTVQEAETLAEQPGVLSVSPEVRYELHTTRTPEFLGLLKKTTTLSPGSD 134
Y+ V GFS +L E L V +V P V Y+ + + + +SP D
Sbjct: 106 YEQVFSGFSMKLPANEIPKLLAVKDVKAVYPNVTYKTDNMKDKD---VTISEDAVSPQMD 162
Query: 135 KQSQVV-------IGVLDTGVWPELKSLDDTGLSPVPSTWKGQCEAGNNMNSSSCNRKLI 187
+ + +G G+ ++ DTG+ N+ + +
Sbjct: 163 DSAPYIGANDAWDLGYTGKGI---KVAIIDTGVEY------------NHPDLKKNFGQYK 207
Query: 188 GARFFSKGYEATLGPIDVSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGM 247
G F Y+ P + + R D HG+H T A A+GT +G+
Sbjct: 208 GYDFVDNDYDPKETP---TGDPRGEATD--HGTHVAGTVA------------ANGTIKGV 250
Query: 248 ATQARVAAYKVCWLGGCFSSD-IAAGIDKAIEDGVNIISMSIGGS--SADYFRDIIAIGA 304
A A + AY+V GG +++ + AG+++A++DG +++++S+G S + D+ +
Sbjct: 251 APDATLLAYRVLGPGGSGTTENVIAGVERAVQDGADVMNLSLGNSLNNPDW---ATSTAL 307
Query: 305 FTANSHGILVSTSAGNGGPSPSSLSNTAPWITTVGAGTIDRDFPAYITLGNNITHTGASL 364
A S G++ TS GN GP+ ++ + + G Y + +
Sbjct: 308 DWAMSEGVVAVTSNGNSGPNGWTVGSPGTSREAISVGATQLPLNEYAVTFGSYSSAKVMG 367
Query: 365 YRG----KPLSDSPLPLVYAG--NASNFSVGYLCLPDSLVPSKVLGKIVICERGGNARVE 418
Y K L++ + LV AG A +F + GK+ + +RG A V+
Sbjct: 368 YNKEDDVKALNNKEVELVEAGIGEAKDFE-----------GKDLTGKVAVVKRGSIAFVD 416
Query: 419 KGLVVKRAGGIGMILANNEEFGEELVADSHLLPAAAL----GERSSKALKDYVFSSRNPT 474
K K+AG IGM++ NN E +P L GE+ ALK T
Sbjct: 417 KADNAKKAGAIGMVVYNNLSGEIEANVPGMSVPTIKLSLEDGEKLVSALKA---GETKTT 473
Query: 475 AKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPV 534
KL + VA FSSRGP + ++KPD+ APGVNI++ PT P
Sbjct: 474 FKLT-----VSKALGEQVADFSSRGPV-MDTWMIKPDISAPGVNIVSTI-----PTHDP- 521
Query: 535 DTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTTSYT 583
+ GTSM+ PH++G A++K + P+WS I++A+M T+ T
Sbjct: 522 -DHPYGYGSKQGTSMASPHIAGAVAVIKQAKPKWSVEQIKAAIMNTAVT 569
>P2P_LACPA (Q02470) PII-type proteinase precursor (EC 3.4.21.96)
(Lactocepin) (Cell wall-associated serine proteinase)
(LP151)
Length = 1902
Score = 74.3 bits (181), Expect = 1e-12
Identities = 136/586 (23%), Positives = 223/586 (37%), Gaps = 78/586 (13%)
Query: 74 TYKHVAHGFSTRLTVQEAETLAEQPGVLSVSPEVRYELHTTRTPEFLGLLKKTTTLSPGS 133
+Y +V +GFST++ V + L + GV +V+ Y + + S
Sbjct: 149 SYGYVVNGFSTKVRVVDIPKLKQIAGVKTVTLAKVYYPTDAKANSMANV---QAVWSNYK 205
Query: 134 DKQSQVVIGVLDTGVWPELKSLDDTGLSPVPSTWKGQCEAGNNMNSSSCNRKLIGARFFS 193
K V+ V+DTG+ P K + + V T K E + R+F+
Sbjct: 206 YKGEGTVVSVIDTGIDPTHKDMRLSDDKDVKLT-KYDVEKFTDTAKHG--------RYFT 256
Query: 194 KGYEATLGPIDVSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGL-ASGTARGMATQAR 252
D + D+ HG H + + GA+ G + + G+A +A+
Sbjct: 257 SKVPYGFNYADNNDTITDDTVDEQHGMHV------AGIIGANGTGDDPTKSVVGVAPEAQ 310
Query: 253 VAAYKVCWLGGCFSSDIAAGIDKAIED----GVNIISMSIGGSSADYFRDIIAIGAF-TA 307
+ A KV ++ +A + AIED G ++++MS+G S + + I A A
Sbjct: 311 LLAMKVFTNSDTSATTGSATLVSAIEDSAKIGADVLNMSLGSDSGNQTLEDPEIAAVQNA 370
Query: 308 NSHGILVSTSAGNGGPSPSSLSNTAPWI------TTVGAGTIDRDFPAYITLGN-NITHT 360
N G SAGN G S S+ VG R + N ++
Sbjct: 371 NESGTAAVISAGNSGTSGSATQGVNKDYYGLQDNEMVGTPGTSRGATTVASAENTDVISQ 430
Query: 361 GASLYRGKPLSDSPLPLVYAGNASNFSVG----YLCLPDSLVPSK---------VLGKIV 407
++ GK L P + + N S Y+ S SK GKI
Sbjct: 431 AVTITDGKDLQLGPETIQLSSNDFTGSFDQKKFYVVKDASGDLSKGAAADYTADAKGKIA 490
Query: 408 ICERGGNARVEKGLVVKRAGGIGMILANNEEFGEEL--VADSHLLPAAALGERSSKALKD 465
I +RG +K + AG G+I+ NN+ L + + P L ++ + L D
Sbjct: 491 IVKRGELNFADKQKYAQAAGAAGLIIVNNDGTATPLTSIRLTTTFPTFGLSSKTGQKLVD 550
Query: 466 YVFSSRNPTAKLVFGGTHL--QVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGW 523
+V + + + + T L Q ++ F+S GP ++ KPD+ APG NI W
Sbjct: 551 WVTAHPDDSLGVKIALTLLPNQKYTEDKMSDFTSYGP--VSNLSFKPDITAPGGNI---W 605
Query: 524 TGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAILK-----------GSHPEWSPAA 572
+ + + +SGTSM+ P ++G A+LK + + A
Sbjct: 606 S----------TQNNNGYTNMSGTSMASPFIAGSQALLKQALNNKNNPFYADYKQLKGTA 655
Query: 573 IRSALMTTSYTAYKNGQTIQDVATGKP-ATPLDFGAGHVDPVASLD 617
+ L T Q I D+ +P GAG VD A++D
Sbjct: 656 LTDFLKTVE---MNTAQPINDINYNNVIVSPRRQGAGLVDVKAAID 698
>SCA1_STRPY (P15926) C5a peptidase precursor (EC 3.4.21.-) (SCP)
Length = 1167
Score = 73.2 bits (178), Expect = 2e-12
Identities = 104/432 (24%), Positives = 178/432 (41%), Gaps = 59/432 (13%)
Query: 204 DVSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCWLGG 263
D S + ++A D + HG+H +G+A + G +A++ +V + G
Sbjct: 180 DYSKDGKTAVDQE-HGTHVSGILSGNAPSETK----EPYRLEGAMPEAQLLLMRVEIVNG 234
Query: 264 CFSSDIAAGIDKAIEDGVN----IISMSIGGSSADYFR--DIIAIGAFTANSHGILVSTS 317
+D A +AI D VN +I+MS G ++ Y D A S G+ + TS
Sbjct: 235 L--ADYARNYAQAIRDAVNLGAKVINMSFGNAALAYANLPDETKKAFDYAKSKGVSIVTS 292
Query: 318 AGN----GGPSPSSLSNTAPWITTVGAGTIDRDFPAYITLGNNITHTGASLYRGKPLSDS 373
AGN GG + L++ P VG + + T ++ + D
Sbjct: 293 AGNDSSFGGKTRLPLADH-PDYGVVGTPAAADSTLTVASYSPDKQLTETAMVKTDDQQDK 351
Query: 374 PLPLVYAGNAS-NFSVGYLCLPDSLVPSK---VLGKIVICERGGNARVEKGLVVKRAGGI 429
+P++ N + Y + V GKI + ERG +K K+AG +
Sbjct: 352 EMPVLSTNRFEPNKAYDYAYANRGMKEDDFKDVKGKIALIERGDIDFKDKVANAKKAGAV 411
Query: 430 GMILANNEEFGEEL-VADSHLLPAAALGERSSKALKDYVFSSRNPTAKLVFGGTHLQVKP 488
G+++ +N++ G + + + +PAA + + LKD NP + F T +V P
Sbjct: 412 GVLIYDNQDKGFPIELPNVDQMPAAFISRKDGLLLKD------NPQKTITFNATP-KVLP 464
Query: 489 SPVVAAFSSRGPNGLTPK-ILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGT 547
+ S GLT +KPD+ APG +IL+ + + +SGT
Sbjct: 465 TASGTKLSRFSSWGLTADGNIKPDIAAPGQDILS-------------SVANNKYAKLSGT 511
Query: 548 SMSCPHVSGLAAIL----KGSHPEWSPA----AIRSALMTTSYTAYKNGQTIQDVATGKP 599
SMS P V+G+ +L + +P+ +P+ + LM+++ Y +
Sbjct: 512 SMSAPLVAGIMGLLQKQYETQYPDMTPSERLDLAKKVLMSSATALYDEDEKAY------- 564
Query: 600 ATPLDFGAGHVD 611
+P GAG VD
Sbjct: 565 FSPRQQGAGAVD 576
>P1P_LACLC (P16271) PI-type proteinase precursor (EC 3.4.21.-)
(Wall-associated serine proteinase)
Length = 1902
Score = 72.4 bits (176), Expect = 4e-12
Identities = 139/595 (23%), Positives = 230/595 (38%), Gaps = 96/595 (16%)
Query: 74 TYKHVAHGFSTRLTVQEAETLAEQPGVLSVSPEVRYELHTTRTPEFLGLLKKTTTLSPGS 133
+Y +V +GFST++ V + L + GV +V+ Y + + S
Sbjct: 149 SYGYVVNGFSTKVRVVDIPKLKQIAGVKTVTLAKVYYPTDAKANSMANV---QAVWSNYK 205
Query: 134 DKQSQVVIGVLDTGVWPELKSL---DDTGLSPVPSTWKGQCEAGNNMNSSSCNRKLIGAR 190
K V+ V+D+G+ P K + DD + S + + + R
Sbjct: 206 YKGEGTVVSVIDSGIDPTHKDMRLSDDKDVKLTKSDVEKFTDTAKH------------GR 253
Query: 191 FFSKGYEATLGPIDVSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTAR---GM 247
+F+ D + D+ HG H + + GA+ G A+ G+
Sbjct: 254 YFNSKVPYGFNYADNNDTITDDTVDEQHGMHV------AGIIGAN--GTGDDPAKSVVGV 305
Query: 248 ATQARVAAYKVCWLGGCFSSDIAAGIDKAIED----GVNIISMSIGGSSADYFRDIIAIG 303
A +A++ A KV ++ ++ + AIED G ++++MS+G S + + +
Sbjct: 306 APEAQLLAMKVFTNSDTSATTGSSTLVSAIEDSAKIGADVLNMSLGSDSGNQTLEDPELA 365
Query: 304 AF-TANSHGILVSTSAGNGGPSPSSLSNTAPWI-------------TTVGAGTI-DRDFP 348
A AN G SAGN G S S+ T+ GA T+ +
Sbjct: 366 AVQNANESGTAAVISAGNSGTSGSATEGVNKDYYGLQDNEMVGTPGTSRGATTVASAENT 425
Query: 349 AYITLGNNITH-TGASLYRGK-PLSDSPLP--------LVYAGNASNFSVGYLCLPDSLV 398
IT IT TG L G LS + V + N S G L D
Sbjct: 426 DVITQAVTITDGTGLQLGPGTIQLSSNDFTGSFDQKKFYVVKDASGNLSKG--ALADYTA 483
Query: 399 PSKVLGKIVICERGGNARVEKGLVVKRAGGIGMILANNEEFGEEL--VADSHLLPAAALG 456
+K GKI I +RG + +K + AG G+I+ NN+ + +A + P L
Sbjct: 484 DAK--GKIAIVKRGELSFDDKQKYAQAAGAAGLIIVNNDGTATPVTSMALTTTFPTFGLS 541
Query: 457 ERSSKALKDYVFSSRNPT--AKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIA 514
+ + L D+V + + + K+ Q ++ F+S GP ++ KPD+ A
Sbjct: 542 SVTGQKLVDWVTAHPDDSLGVKIALTLVPNQKYTEDKMSDFTSYGP--VSNLSFKPDITA 599
Query: 515 PGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAILKGS---------- 564
PG NI W+ + + +SGTSM+ P ++G A+LK +
Sbjct: 600 PGGNI---WS----------TQNNNGYTNMSGTSMASPFIAGSQALLKQALNNKNNPFYA 646
Query: 565 -HPEWSPAAIRSALMTTSYTAYKNGQTIQDVATGKP-ATPLDFGAGHVDPVASLD 617
+ + A+ L T Q I D+ +P GAG VD A++D
Sbjct: 647 YYKQLKGTALTDFLKTVE---MNTAQPINDINYNNVIVSPRRQGAGLVDVKAAID 698
>SCA2_STRPY (P58099) C5a peptidase precursor (EC 3.4.21.-) (SCP)
Length = 1181
Score = 69.7 bits (169), Expect = 3e-11
Identities = 100/430 (23%), Positives = 176/430 (40%), Gaps = 55/430 (12%)
Query: 204 DVSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCWLGG 263
D S + ++A D + HG+H +G+A + G +A++ +V + G
Sbjct: 180 DYSKDGKTAVDQE-HGTHVSGILSGNAPSETK----EPYRLEGAMPEAQLLLMRVEIVNG 234
Query: 264 C--FSSDIAAGIDKAIEDGVNIISMSIGGSSADYFR--DIIAIGAFTANSHGILVSTSAG 319
++ + A I A+ G +I+MS G ++ Y D A S G+ + TSAG
Sbjct: 235 LADYARNYAQAIIDAVNLGAKVINMSFGNAALAYANLPDETKKAFDYAKSKGVSIVTSAG 294
Query: 320 N----GGPSPSSLSNTAPWITTVGAGTIDRDFPAYITLGNNITHTGASLYRGKPLSDSPL 375
N GG + L++ P VG + + T + + D +
Sbjct: 295 NDSSFGGKTRLPLADH-PDYGVVGTPAAADSTLTVASYSPDKQLTETATVKTADQQDKEM 353
Query: 376 PLVYAGNAS-NFSVGYLCLPDSLVPSK---VLGKIVICERGGNARVEKGLVVKRAGGIGM 431
P++ N + Y + V GKI + ERG +K K+AG +G+
Sbjct: 354 PVLSTNRFEPNKAYDYAYANRGMKEDDFKDVKGKIALIERGDIDFKDKIANAKKAGAVGV 413
Query: 432 ILANNEEFGEEL-VADSHLLPAAALGERSSKALKDYVFSSRNPTAKLVFGGTHLQVKPSP 490
++ +N++ G + + + +PAA + + LK+ NP + F T +V P+
Sbjct: 414 LIYDNQDKGFPIELPNVDQMPAAFISRKDGLLLKE------NPQKTITFNATP-KVLPTA 466
Query: 491 VVAAFSSRGPNGLTPK-ILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSM 549
S GLT +KPD+ APG +IL+ + + +SGTSM
Sbjct: 467 SGTKLSRFSSWGLTADGNIKPDIAAPGQDILS-------------SVANNKYAKLSGTSM 513
Query: 550 SCPHVSGLAAIL----KGSHPEWSPA----AIRSALMTTSYTAYKNGQTIQDVATGKPAT 601
S P V+G+ +L + +P+ +P+ + LM+++ Y + +
Sbjct: 514 SAPLVAGIMGLLQKQYETQYPDMTPSERLDLAKKVLMSSATALYDEDEKAY-------FS 566
Query: 602 PLDFGAGHVD 611
P GAG VD
Sbjct: 567 PRQQGAGAVD 576
>ELYA_BACCS (P41362) Alkaline protease precursor (EC 3.4.21.-)
Length = 380
Score = 55.8 bits (133), Expect = 4e-07
Identities = 80/351 (22%), Positives = 144/351 (40%), Gaps = 68/351 (19%)
Query: 1 MKMPIFQLLQS-ALLLLLIFCSSYTIAEKKTAQQAKKTYIIHMDKSTMPATFNDHQHWFD 59
MK P+ +++ S ALL+ + F SS A A++AK+ Y+I ++ + F + D
Sbjct: 1 MKKPLGKIVASTALLISVAFSSSIASA----AEEAKEKYLIGFNEQEAVSEFVEQVEAND 56
Query: 60 S-SLQSVSESAEI--LYTYKHVAHGFSTRLTVQEAETLAEQPGVLSVSPEVRYELHTTRT 116
++ S E EI L+ ++ + S L+ ++ + L P + + +
Sbjct: 57 EVAILSEEEEVEIELLHEFETIPV-LSVELSPEDVDALELDPAISYIEEDAEVTTMAQSV 115
Query: 117 PEFLGLLKKTTTLSPGSDKQSQVVIGVLDTGVWPELKSLDDTGLSPVPSTWKGQCEAGNN 176
P + ++ + G S V + VLDTG+
Sbjct: 116 PWGISRVQAPAAHNRGLTG-SGVKVAVLDTGI---------------------------- 146
Query: 177 MNSSSCNRKLIGARFFSKGYEATLGPIDVSTESRSARDDDGHGSHTLTTAAGSAVAGASL 236
S+ + + G F G +T +D +GHG+H AG+ A +
Sbjct: 147 --STHPDLNIRGGASFVPGEPST-------------QDGNGHGTH----VAGTIAALNNS 187
Query: 237 FGLASGTARGMATQARVAAYKVCWLGGCFS-SDIAAGIDKAIEDGVNIISMSIGGSSADY 295
G+ G+A A + A KV G S S IA G++ A +G+++ ++S+G S
Sbjct: 188 IGVL-----GVAPSAELYAVKVLGASGSGSVSSIAQGLEWAGNNGMHVANLSLGSPSPS- 241
Query: 296 FRDIIAIGAFTANSHGILVSTSAGNGGPSPSSLSNTAPWITTVGAGTIDRD 346
+ +A S G+LV ++GN G S+S A + + G D++
Sbjct: 242 --ATLEQAVNSATSRGVLVVAASGNSG--AGSISYPARYANAMAVGATDQN 288
Score = 51.6 bits (122), Expect = 8e-06
Identities = 30/91 (32%), Positives = 48/91 (51%), Gaps = 18/91 (19%)
Query: 511 DLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSP 570
D++APGVN+ + + G+ ++ ++GTSM+ PHV+G AA++K +P WS
Sbjct: 302 DIVAPGVNVQSTYPGS-------------TYASLNGTSMATPHVAGAAALVKQKNPSWSN 348
Query: 571 AAIRSALMTT-----SYTAYKNGQTIQDVAT 596
IR+ L T S Y +G + AT
Sbjct: 349 VQIRNHLKNTATSLGSTNLYGSGLVNAEAAT 379
>ELYA_BACAO (P27693) Alkaline protease precursor (EC 3.4.21.-)
Length = 380
Score = 55.8 bits (133), Expect = 4e-07
Identities = 80/351 (22%), Positives = 144/351 (40%), Gaps = 68/351 (19%)
Query: 1 MKMPIFQLLQS-ALLLLLIFCSSYTIAEKKTAQQAKKTYIIHMDKSTMPATFNDHQHWFD 59
MK P+ +++ S ALL+ + F SS A A++AK+ Y+I ++ + F + D
Sbjct: 1 MKKPLGKIVASTALLISVAFSSSIASA----AEEAKEKYLIGFNEQEAVSEFVEQVEAND 56
Query: 60 S-SLQSVSESAEI--LYTYKHVAHGFSTRLTVQEAETLAEQPGVLSVSPEVRYELHTTRT 116
++ S E EI L+ ++ + S L+ ++ + L P + + +
Sbjct: 57 EVAILSEEEEVEIELLHEFETIPV-LSVELSPEDVDALELDPAISYIEEDAEVTTMAQSV 115
Query: 117 PEFLGLLKKTTTLSPGSDKQSQVVIGVLDTGVWPELKSLDDTGLSPVPSTWKGQCEAGNN 176
P + ++ + G S V + VLDTG+
Sbjct: 116 PWGISRVQAPAAHNRGLTG-SGVKVAVLDTGI---------------------------- 146
Query: 177 MNSSSCNRKLIGARFFSKGYEATLGPIDVSTESRSARDDDGHGSHTLTTAAGSAVAGASL 236
S+ + + G F G +T +D +GHG+H AG+ A +
Sbjct: 147 --STHPDLNIRGGASFVPGEPST-------------QDGNGHGTH----VAGTIAALNNS 187
Query: 237 FGLASGTARGMATQARVAAYKVCWLGGCFS-SDIAAGIDKAIEDGVNIISMSIGGSSADY 295
G+ G+A A + A KV G S S IA G++ A +G+++ ++S+G S
Sbjct: 188 IGVL-----GVAPNAELYAVKVLGASGSGSVSSIAQGLEWAGNNGMHVANLSLGSPSPS- 241
Query: 296 FRDIIAIGAFTANSHGILVSTSAGNGGPSPSSLSNTAPWITTVGAGTIDRD 346
+ +A S G+LV ++GN G S+S A + + G D++
Sbjct: 242 --ATLEQAVNSATSRGVLVVAASGNSG--AGSISYPARYANAMAVGATDQN 288
Score = 51.6 bits (122), Expect = 8e-06
Identities = 30/91 (32%), Positives = 48/91 (51%), Gaps = 18/91 (19%)
Query: 511 DLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSP 570
D++APGVN+ + + G+ ++ ++GTSM+ PHV+G AA++K +P WS
Sbjct: 302 DIVAPGVNVQSTYPGS-------------TYASLNGTSMATPHVAGAAALVKQKNPSWSN 348
Query: 571 AAIRSALMTT-----SYTAYKNGQTIQDVAT 596
IR+ L T S Y +G + AT
Sbjct: 349 VQIRNHLKNTATSLGSTNLYGSGLVNAEAAT 379
>ISP6_SCHPO (P40903) Sexual differentiation process putative
subtilase-type proteinase isp6 (EC 3.4.21.-)
Length = 467
Score = 55.1 bits (131), Expect = 7e-07
Identities = 28/68 (41%), Positives = 41/68 (60%), Gaps = 11/68 (16%)
Query: 511 DLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSP 570
D+ APG+NIL+ W G+ + S N ISGTSM+ PHV+GL+A G HP S
Sbjct: 383 DIFAPGLNILSTWIGS-----------NTSTNTISGTSMATPHVAGLSAYYLGLHPAASA 431
Query: 571 AAIRSALM 578
+ ++ A++
Sbjct: 432 SEVKDAII 439
Score = 38.5 bits (88), Expect = 0.068
Identities = 37/150 (24%), Positives = 68/150 (44%), Gaps = 26/150 (17%)
Query: 214 DDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCWLGGCFS-SDIAAG 272
D++GHG+H T A A G+A +A + A KV G + +D+ AG
Sbjct: 249 DNNGHGTHVAGTIASRAY--------------GVAKKAEIVAVKVLRSSGSGTMADVIAG 294
Query: 273 ID------KAIEDGVNIISMSIGGSSADYFRDIIAIGAFTANSHGILVSTSAGNGGPSPS 326
++ K+ ++ +MS+GG ++ ++ + +A ++G++ + +AGN
Sbjct: 295 VEWTVRHHKSSGKKTSVGNMSLGGGNS----FVLDMAVDSAVTNGVIYAVAAGNEYDDAC 350
Query: 327 SLSNTA-PWITTVGAGTIDRDFPAYITLGN 355
S A TVGA TI+ + G+
Sbjct: 351 YSSPAASKKAITVGASTINDQMAYFSNYGS 380
>WPRA_BACSU (P54423) Cell wall-associated protease precursor (EC
3.4.21.-) [Contains: Cell wall-associated polypeptide
CWBP23; Cell wall-associated polypeptide CWBP52]
Length = 894
Score = 54.7 bits (130), Expect = 9e-07
Identities = 50/187 (26%), Positives = 85/187 (44%), Gaps = 21/187 (11%)
Query: 209 SRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCWLGGCFSSD 268
+ +A DD GHG+H A + G S+ GL + +A++ KV G ++
Sbjct: 488 NNNAMDDQGHGTHVAGIIAAQSDNGYSMTGLNA--------KAKIIPVKVLDSAGSGDTE 539
Query: 269 -IAAGIDKAIEDGVNIISMSIGGSSADYFRDIIAIGAFTANSHGILVSTSAGNGGPSPSS 327
IA GI A + G +I++S+GG + ++ A +L++ ++GN G + S
Sbjct: 540 QIALGIKYAADKGAKVINLSLGGG----YSRVLEFALKYAADKNVLIAAASGNDGENALS 595
Query: 328 LSNTAPWITTVGAGTIDRDFPAYITLGNNITHTGASLYRGKPLSDSPLPLVYAGNASNFS 387
++ ++ +VGA T D A + ++ G L P SD P LV GN + S
Sbjct: 596 YPASSKYVMSVGA-TNRMDMTA------DFSNYGKGLDISAPGSDIP-SLVPNGNVTYMS 647
Query: 388 VGYLCLP 394
+ P
Sbjct: 648 GTSMATP 654
Score = 35.0 bits (79), Expect = 0.75
Identities = 55/213 (25%), Positives = 87/213 (40%), Gaps = 41/213 (19%)
Query: 377 LVYAGNASNFSVGYLCLPDSLVPSKVL-----GKIVICERGGNARVEKGLVV---KRAGG 428
++ A + + +S+ L ++P KVL G G +KG V GG
Sbjct: 504 IIAAQSDNGYSMTGLNAKAKIIPVKVLDSAGSGDTEQIALGIKYAADKGAKVINLSLGGG 563
Query: 429 IGMILANNEEFGEELVADSHLLPAAALGERSSKALKDYVFSSRNPTAKLVFGGTHLQVKP 488
+L EF + AD ++L AAA G AL Y SS+ + G T+
Sbjct: 564 YSRVL----EFALKYAADKNVLIAAASGNDGENALS-YPASSKYVMS---VGATNRM--- 612
Query: 489 SPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTS 548
+ A FS+ G GL D+ APG +I ++ P G + +SGTS
Sbjct: 613 -DMTADFSNYG-KGL-------DISAPGSDI-----PSLVPNG--------NVTYMSGTS 650
Query: 549 MSCPHVSGLAAILKGSHPEWSPAAIRSALMTTS 581
M+ P+ + A +L +P+ + L T+
Sbjct: 651 MATPYAAAAAGLLFAQNPKLKRTEVEDMLKKTA 683
>TKSU_PYRKO (P58502) Tk-subtilisin precursor (EC 3.4.21.-)
Length = 422
Score = 52.0 bits (123), Expect = 6e-06
Identities = 50/162 (30%), Positives = 76/162 (46%), Gaps = 37/162 (22%)
Query: 205 VSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCWLGGC 264
VST+ R D +GHG+H + T A A + G+ G+A ++ + +V G
Sbjct: 164 VSTKLRDCADQNGHGTHVIGTIA----ALNNDIGVV-----GVAPGVQIYSVRVLDARGS 214
Query: 265 FS-SDIAAGIDKAI--------------------EDGVNIISMSIGGSSADYFRDIIAIG 303
S SDIA GI++AI +D +ISMS+GG + D + + I
Sbjct: 215 GSYSDIAIGIEQAILGPDGVADKDGDGIIAGDPDDDAAEVISMSLGGPADDSYLYDMIIQ 274
Query: 304 AFTANSHGILVSTSAGN-GGPSPSSLSNTAPWITTVGAGTID 344
A+ A GI++ ++GN G PSP S A + + G ID
Sbjct: 275 AYNA---GIVIVAASGNEGAPSP---SYPAAYPEVIAVGAID 310
Score = 38.1 bits (87), Expect = 0.089
Identities = 24/74 (32%), Positives = 42/74 (56%), Gaps = 23/74 (31%)
Query: 492 VAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSC 551
+A+FS+R +P++ APGV+IL+ + P D S+ + GTSM+
Sbjct: 315 IASFSNR----------QPEVSAPGVDILSTY---------PDD----SYETLMGTSMAT 351
Query: 552 PHVSGLAAILKGSH 565
PHVSG+ A+++ ++
Sbjct: 352 PHVSGVVALIQAAY 365
>SUBT_BACAM (P00782) Subtilisin BPN' precursor (EC 3.4.21.62)
(Subtilisin Novo) (Subtilisin DFE) (Alkaline protease)
Length = 382
Score = 52.0 bits (123), Expect = 6e-06
Identities = 41/141 (29%), Positives = 66/141 (46%), Gaps = 26/141 (18%)
Query: 441 EELVADSHLLPAAALGERSSKALKDYVFSSRNPTAKLVFGGTHLQVKPSPVVAAFSSRGP 500
++ VA ++ AAA E +S + + + P+ V V S A+FSS GP
Sbjct: 247 DKAVASGVVVVAAAGNEGTSGSSSTVGYPGKYPSVIAVGA-----VDSSNQRASFSSVGP 301
Query: 501 NGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAI 560
+ D++APGV+I + G + +GTSM+ PHV+G AA+
Sbjct: 302 --------ELDVMAPGVSIQSTLPGN-------------KYGAYNGTSMASPHVAGAAAL 340
Query: 561 LKGSHPEWSPAAIRSALMTTS 581
+ HP W+ +RS+L T+
Sbjct: 341 ILSKHPNWTNTQVRSSLENTT 361
Score = 47.4 bits (111), Expect = 1e-04
Identities = 42/143 (29%), Positives = 70/143 (48%), Gaps = 15/143 (10%)
Query: 205 VSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCWLGGC 264
V +E+ +D++ HG+H AG+ A + G+ G+A A + A KV G
Sbjct: 158 VPSETNPFQDNNSHGTHV----AGTVAALNNSIGVL-----GVAPSASLYAVKVLGADGS 208
Query: 265 FS-SDIAAGIDKAIEDGVNIISMSIGGSSADYFRDIIAIGAFTANSHGILVSTSAGNGGP 323
S I GI+ AI + +++I+MS+GG S + A + G++V +AGN G
Sbjct: 209 GQYSWIINGIEWAIANNMDVINMSLGGPSGS---AALKAAVDKAVASGVVVVAAAGNEGT 265
Query: 324 SPSSLSNTAP--WITTVGAGTID 344
S SS + P + + + G +D
Sbjct: 266 SGSSSTVGYPGKYPSVIAVGAVD 288
>PRTM_BACSK (Q99405) M-protease (EC 3.4.21.-)
Length = 269
Score = 52.0 bits (123), Expect = 6e-06
Identities = 25/71 (35%), Positives = 43/71 (60%), Gaps = 13/71 (18%)
Query: 511 DLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSP 570
D++APGVN+ + + G+ ++ ++GTSM+ PHV+G+AA++K +P WS
Sbjct: 191 DIVAPGVNVQSTYPGS-------------TYASLNGTSMATPHVAGVAALVKQKNPSWSN 237
Query: 571 AAIRSALMTTS 581
IR+ L T+
Sbjct: 238 VQIRNHLKNTA 248
Score = 48.9 bits (115), Expect = 5e-05
Identities = 40/137 (29%), Positives = 67/137 (48%), Gaps = 15/137 (10%)
Query: 211 SARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCWLGGCFS-SDI 269
S +D +GHG+H AG+ A + G+ G+A A + A KV G S S I
Sbjct: 55 STQDGNGHGTH----VAGTIAALNNSIGVL-----GVAPSAELYAVKVLGASGSGSVSSI 105
Query: 270 AAGIDKAIEDGVNIISMSIGGSSADYFRDIIAIGAFTANSHGILVSTSAGNGGPSPSSLS 329
A G++ A +G+++ ++S+G S + +A S G+LV ++GN G S+S
Sbjct: 106 AQGLEWAGNNGMHVANLSLGSPSPS---ATLEQAVNSATSRGVLVVAASGNSG--AGSIS 160
Query: 330 NTAPWITTVGAGTIDRD 346
A + + G D++
Sbjct: 161 YPARYANAMAVGATDQN 177
>SUBT_BACSU (P04189) Subtilisin E precursor (EC 3.4.21.62)
Length = 381
Score = 51.6 bits (122), Expect = 8e-06
Identities = 40/141 (28%), Positives = 67/141 (47%), Gaps = 26/141 (18%)
Query: 441 EELVADSHLLPAAALGERSSKALKDYVFSSRNPTAKLVFGGTHLQVKPSPVVAAFSSRGP 500
++ V+ ++ AAA E SS + + ++ P+ V V S A+FSS G
Sbjct: 246 DKAVSSGIVVAAAAGNEGSSGSTSTVGYPAKYPSTIAVGA-----VNSSNQRASFSSAGS 300
Query: 501 NGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAI 560
+ D++APGV+I + G ++ +GTSM+ PHV+G AA+
Sbjct: 301 --------ELDVMAPGVSIQSTLPGG-------------TYGAYNGTSMATPHVAGAAAL 339
Query: 561 LKGSHPEWSPAAIRSALMTTS 581
+ HP W+ A +R L +T+
Sbjct: 340 ILSKHPTWTNAQVRDRLESTA 360
Score = 44.7 bits (104), Expect = 0.001
Identities = 41/143 (28%), Positives = 70/143 (48%), Gaps = 15/143 (10%)
Query: 205 VSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCWLGGC 264
V +E+ +D HG+H AG+ A + G+ G++ A + A KV G
Sbjct: 157 VPSETNPYQDGSSHGTHV----AGTIAALNNSIGVL-----GVSPSASLYAVKVLDSTGS 207
Query: 265 FS-SDIAAGIDKAIEDGVNIISMSIGGSSADYFRDIIAIGAFTANSHGILVSTSAGNGGP 323
S I GI+ AI + +++I+MS+GG + + A S GI+V+ +AGN G
Sbjct: 208 GQYSWIINGIEWAISNNMDVINMSLGGPTGSTALKTVVDKAV---SSGIVVAAAAGNEGS 264
Query: 324 --SPSSLSNTAPWITTVGAGTID 344
S S++ A + +T+ G ++
Sbjct: 265 SGSTSTVGYPAKYPSTIAVGAVN 287
>SUBS_BACLE (P29600) Subtilisin Savinase (EC 3.4.21.62) (Alkaline
protease)
Length = 269
Score = 51.6 bits (122), Expect = 8e-06
Identities = 30/91 (32%), Positives = 48/91 (51%), Gaps = 18/91 (19%)
Query: 511 DLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSP 570
D++APGVN+ + + G+ ++ ++GTSM+ PHV+G AA++K +P WS
Sbjct: 191 DIVAPGVNVQSTYPGS-------------TYASLNGTSMATPHVAGAAALVKQKNPSWSN 237
Query: 571 AAIRSALMTT-----SYTAYKNGQTIQDVAT 596
IR+ L T S Y +G + AT
Sbjct: 238 VQIRNHLKNTATSLGSTNLYGSGLVNAEAAT 268
Score = 48.9 bits (115), Expect = 5e-05
Identities = 40/137 (29%), Positives = 67/137 (48%), Gaps = 15/137 (10%)
Query: 211 SARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCWLGGCFS-SDI 269
S +D +GHG+H AG+ A + G+ G+A A + A KV G S S I
Sbjct: 55 STQDGNGHGTH----VAGTIAALNNSIGVL-----GVAPSAELYAVKVLGASGSGSVSSI 105
Query: 270 AAGIDKAIEDGVNIISMSIGGSSADYFRDIIAIGAFTANSHGILVSTSAGNGGPSPSSLS 329
A G++ A +G+++ ++S+G S + +A S G+LV ++GN G S+S
Sbjct: 106 AQGLEWAGNNGMHVANLSLGSPSPS---ATLEQAVNSATSRGVLVVAASGNSG--AGSIS 160
Query: 330 NTAPWITTVGAGTIDRD 346
A + + G D++
Sbjct: 161 YPARYANAMAVGATDQN 177
>SUBB_BACLE (P29599) Subtilisin BL (EC 3.4.21.62) (Alkaline
protease)
Length = 269
Score = 51.6 bits (122), Expect = 8e-06
Identities = 30/91 (32%), Positives = 48/91 (51%), Gaps = 18/91 (19%)
Query: 511 DLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSP 570
D++APGVN+ + + G+ ++ ++GTSM+ PHV+G AA++K +P WS
Sbjct: 191 DIVAPGVNVQSTYPGS-------------TYASLNGTSMATPHVAGAAALVKQKNPSWSN 237
Query: 571 AAIRSALMTT-----SYTAYKNGQTIQDVAT 596
IR+ L T S Y +G + AT
Sbjct: 238 VQIRNHLKNTATSLGSTNLYGSGLVNAEAAT 268
Score = 48.1 bits (113), Expect = 9e-05
Identities = 40/137 (29%), Positives = 68/137 (49%), Gaps = 15/137 (10%)
Query: 211 SARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCWLGGCFS-SDI 269
S +D +GHG+H AG+ A + G+ G+A A + A KV G + S I
Sbjct: 55 STQDGNGHGTHV----AGTIAALNNSIGVL-----GVAPSAELYAVKVLGADGRGAISSI 105
Query: 270 AAGIDKAIEDGVNIISMSIGGSSADYFRDIIAIGAFTANSHGILVSTSAGNGGPSPSSLS 329
A G++ A +G+++ ++S+G S + +A S G+LV ++GN G SS+S
Sbjct: 106 AQGLEWAGNNGMHVANLSLGSPSPSA---TLEQAVNSATSRGVLVVAASGNSG--ASSIS 160
Query: 330 NTAPWITTVGAGTIDRD 346
A + + G D++
Sbjct: 161 YPARYANAMAVGATDQN 177
>SUBT_BACST (P29142) Subtilisin J precursor (EC 3.4.21.62)
Length = 381
Score = 51.2 bits (121), Expect = 1e-05
Identities = 40/141 (28%), Positives = 67/141 (47%), Gaps = 26/141 (18%)
Query: 441 EELVADSHLLPAAALGERSSKALKDYVFSSRNPTAKLVFGGTHLQVKPSPVVAAFSSRGP 500
++ V+ ++ AAA E SS + + ++ P+ V V S A+FSS G
Sbjct: 246 DKAVSSGIVVAAAAGNEGSSGSSSTVGYPAKYPSTIAVGA-----VNSSNQRASFSSAGS 300
Query: 501 NGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAI 560
+ D++APGV+I + G ++ +GTSM+ PHV+G AA+
Sbjct: 301 --------ELDVMAPGVSIQSTLPGG-------------TYGAYNGTSMATPHVAGAAAL 339
Query: 561 LKGSHPEWSPAAIRSALMTTS 581
+ HP W+ A +R L +T+
Sbjct: 340 ILSKHPTWTNAQVRDRLESTA 360
Score = 46.6 bits (109), Expect = 2e-04
Identities = 43/143 (30%), Positives = 70/143 (48%), Gaps = 15/143 (10%)
Query: 205 VSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCWLGGC 264
V +E+ +D HG+H AG+ A + G+ G++ A + A KV G
Sbjct: 157 VPSETNPYQDGSSHGTHV----AGTIAALNNSIGVL-----GVSPSASLYAVKVLDSTGS 207
Query: 265 FS-SDIAAGIDKAIEDGVNIISMSIGGSSADYFRDIIAIGAFTANSHGILVSTSAGNGGP 323
S I GI+ AI + +++I+MS+GG S + A S GI+V+ +AGN G
Sbjct: 208 GQYSWIINGIEWAISNNMDVINMSLGGPSGSTALKTVVDKAV---SSGIVVAAAAGNEGS 264
Query: 324 SPSS--LSNTAPWITTVGAGTID 344
S SS + A + +T+ G ++
Sbjct: 265 SGSSSTVGYPAKYPSTIAVGAVN 287
>SUBT_BACSA (P00783) Subtilisin amylosacchariticus precursor (EC
3.4.21.62)
Length = 381
Score = 51.2 bits (121), Expect = 1e-05
Identities = 40/141 (28%), Positives = 67/141 (47%), Gaps = 26/141 (18%)
Query: 441 EELVADSHLLPAAALGERSSKALKDYVFSSRNPTAKLVFGGTHLQVKPSPVVAAFSSRGP 500
++ V+ ++ AAA E SS + + ++ P+ V V S A+FSS G
Sbjct: 246 DKAVSSGIVVAAAAGNEGSSGSSSTVGYPAKYPSTIAVGA-----VNSSNQRASFSSAGS 300
Query: 501 NGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAI 560
+ D++APGV+I + G ++ +GTSM+ PHV+G AA+
Sbjct: 301 --------ELDVMAPGVSIQSTLPGG-------------TYGAYNGTSMATPHVAGAAAL 339
Query: 561 LKGSHPEWSPAAIRSALMTTS 581
+ HP W+ A +R L +T+
Sbjct: 340 ILSKHPTWTNAQVRDRLESTA 360
Score = 46.6 bits (109), Expect = 2e-04
Identities = 43/143 (30%), Positives = 70/143 (48%), Gaps = 15/143 (10%)
Query: 205 VSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCWLGGC 264
V +E+ +D HG+H AG+ A + G+ G++ A + A KV G
Sbjct: 157 VPSETNPYQDGSSHGTHV----AGTIAALNNSIGVL-----GVSPSASLYAVKVLDSTGS 207
Query: 265 FS-SDIAAGIDKAIEDGVNIISMSIGGSSADYFRDIIAIGAFTANSHGILVSTSAGNGGP 323
S I GI+ AI + +++I+MS+GG S + A S GI+V+ +AGN G
Sbjct: 208 GQYSWIINGIEWAISNNMDVINMSLGGPSGSTALKTVVDKAV---SSGIVVAAAAGNEGS 264
Query: 324 SPSS--LSNTAPWITTVGAGTID 344
S SS + A + +T+ G ++
Sbjct: 265 SGSSSTVGYPAKYPSTIAVGAVN 287
>SUBN_BACNA (P35835) Subtilisin NAT precursor (EC 3.4.21.62)
Length = 381
Score = 50.8 bits (120), Expect = 1e-05
Identities = 40/141 (28%), Positives = 67/141 (47%), Gaps = 26/141 (18%)
Query: 441 EELVADSHLLPAAALGERSSKALKDYVFSSRNPTAKLVFGGTHLQVKPSPVVAAFSSRGP 500
++ V+ ++ AAA E SS + + ++ P+ V V S A+FSS G
Sbjct: 246 DKAVSSGIVVAAAAGNEGSSGSTSTVGYPAKYPSTIAVGA-----VNSSNQRASFSSVGS 300
Query: 501 NGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAI 560
+ D++APGV+I + G ++ +GTSM+ PHV+G AA+
Sbjct: 301 --------ELDVMAPGVSIQSTLPGG-------------TYGAYNGTSMATPHVAGAAAL 339
Query: 561 LKGSHPEWSPAAIRSALMTTS 581
+ HP W+ A +R L +T+
Sbjct: 340 ILSKHPTWTNAQVRDRLESTA 360
Score = 45.8 bits (107), Expect = 4e-04
Identities = 42/143 (29%), Positives = 70/143 (48%), Gaps = 15/143 (10%)
Query: 205 VSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCWLGGC 264
V +E+ +D HG+H AG+ A + G+ G+A A + A KV G
Sbjct: 157 VPSETNPYQDGSSHGTHV----AGTIAALNNSIGVL-----GVAPSASLYAVKVLDSTGS 207
Query: 265 FS-SDIAAGIDKAIEDGVNIISMSIGGSSADYFRDIIAIGAFTANSHGILVSTSAGNGGP 323
S I GI+ AI + +++I+MS+GG + + A S GI+V+ +AGN G
Sbjct: 208 GQYSWIINGIEWAISNNMDVINMSLGGPTGSTALKTVVDKAV---SSGIVVAAAAGNEGS 264
Query: 324 --SPSSLSNTAPWITTVGAGTID 344
S S++ A + +T+ G ++
Sbjct: 265 SGSTSTVGYPAKYPSTIAVGAVN 287
>PLS_PYRFU (P72186) Pyrolysin precursor (EC 3.4.21.-)
Length = 1398
Score = 50.8 bits (120), Expect = 1e-05
Identities = 60/223 (26%), Positives = 95/223 (41%), Gaps = 32/223 (14%)
Query: 406 IVICERGGNARVEKGL---------VVKRAGGIGMILANNEEFGEELVADSHLLPAAALG 456
++ GGNA G + ++ G + +I A NE G +V + A
Sbjct: 452 VISMSLGGNAPYLDGTDPESVAVDELTEKYGVVFVIAAGNEGPGINIVGSPGVATKAITV 511
Query: 457 ERSSKALKDYVFSSRNPTAKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPG 516
++ + V+ S+ +G + + +A FSSRGP +KP+++APG
Sbjct: 512 GAAAVPINVGVYVSQALGYPDYYGFYYFPAYTNVRIAFFSSRGPR--IDGEIKPNVVAPG 569
Query: 517 VNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAIL-KGSHPE---WSPAA 572
I + IG + +SGTSM+ PHVSG+ A+L G+ E ++P
Sbjct: 570 YGIYSSLPMWIGGA-----------DFMSGTSMATPHVSGVVALLISGAKAEGIYYNPDI 618
Query: 573 IRSALMTTSYTAYKNGQTIQDVATGKPATPLDFGAGHVDPVAS 615
I+ L S + G D TG+ T LD G G V+ S
Sbjct: 619 IKKVL--ESGATWLEG----DPYTGQKYTELDQGHGLVNVTKS 655
Score = 48.1 bits (113), Expect = 9e-05
Identities = 45/167 (26%), Positives = 73/167 (42%), Gaps = 30/167 (17%)
Query: 214 DDDGHGSHTLTTAAG---------------------SAVAGASLFGLASGTARGMATQAR 252
D GHG+H T AG S + G + + T +G+A A+
Sbjct: 361 DGHGHGTHVAGTVAGYDSNNDAWDWLSMYSGEWEVFSRLYGWDYTNVTTDTVQGVAPGAQ 420
Query: 253 VAAYKVCWLGGCFSS-DIAAGIDKAIEDGVNIISMSIGGSSADYF----RDIIAIGAFTA 307
+ A +V G S DI G+ A G ++ISMS+GG +A Y + +A+ T
Sbjct: 421 IMAIRVLRSDGRGSMWDIIEGMTYAATHGADVISMSLGG-NAPYLDGTDPESVAVDELT- 478
Query: 308 NSHGILVSTSAGNGGPSPSSLSN--TAPWITTVGAGTIDRDFPAYIT 352
+G++ +AGN GP + + + A TVGA + + Y++
Sbjct: 479 EKYGVVFVIAAGNEGPGINIVGSPGVATKAITVGAAAVPINVGVYVS 525
>ELYA_BACHD (P41363) Thermostable alkaline protease precursor (EC
3.4.21.-)
Length = 361
Score = 50.8 bits (120), Expect = 1e-05
Identities = 61/230 (26%), Positives = 99/230 (42%), Gaps = 65/230 (28%)
Query: 383 ASNFSVGYLCLPDSLVPSKVLGKIVICERGGNARVEKGLVVKRAGGIGMILANNEEFGEE 442
A N S+G L + PS L + + +R G+ G + A GI + NN
Sbjct: 164 ALNNSIGVL----GVAPSADLYAVKVLDRNGS-----GSLASVAQGIEWAINNN------ 208
Query: 443 LVADSHLLPAAALGERSSKALKDYVFSSRNPTAKLVFG--------GTHLQVKPSPVVA- 493
H++ + LG S + + + N L+ G G + + S V+A
Sbjct: 209 ----MHIINMS-LGSTSGSSTLELAVNRANNAGILLVGAAGNTGRQGVNYPARYSGVMAV 263
Query: 494 ----------AFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNI 543
+FS+ GP + ++ APGVN+ + +TG R+VS
Sbjct: 264 AAVDQNGQRASFSTYGP--------EIEISAPGVNVNSTYTG----------NRYVS--- 302
Query: 544 ISGTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTT-----SYTAYKNG 588
+SGTSM+ PHV+G+AA++K +P ++ IR + T S + Y NG
Sbjct: 303 LSGTSMATPHVAGVAALVKSRYPSYTNNQIRQRINQTATYLGSPSLYGNG 352
Score = 48.5 bits (114), Expect = 7e-05
Identities = 38/113 (33%), Positives = 60/113 (52%), Gaps = 13/113 (11%)
Query: 211 SARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCWLGGCFS-SDI 269
S D++GHG+H AG+ A + G+ G+A A + A KV G S + +
Sbjct: 147 SYHDNNGHGTHV----AGTIAALNNSIGVL-----GVAPSADLYAVKVLDRNGSGSLASV 197
Query: 270 AAGIDKAIEDGVNIISMSIGGSSADYFRDIIAIGAFTANSHGILVSTSAGNGG 322
A GI+ AI + ++II+MS+G +S + + AN+ GIL+ +AGN G
Sbjct: 198 AQGIEWAINNNMHIINMSLGSTSGS---STLELAVNRANNAGILLVGAAGNTG 247
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.132 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 92,388,396
Number of Sequences: 164201
Number of extensions: 4146634
Number of successful extensions: 10912
Number of sequences better than 10.0: 94
Number of HSP's better than 10.0 without gapping: 34
Number of HSP's successfully gapped in prelim test: 60
Number of HSP's that attempted gapping in prelim test: 10729
Number of HSP's gapped (non-prelim): 168
length of query: 772
length of database: 59,974,054
effective HSP length: 118
effective length of query: 654
effective length of database: 40,598,336
effective search space: 26551311744
effective search space used: 26551311744
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0096b.8


