

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
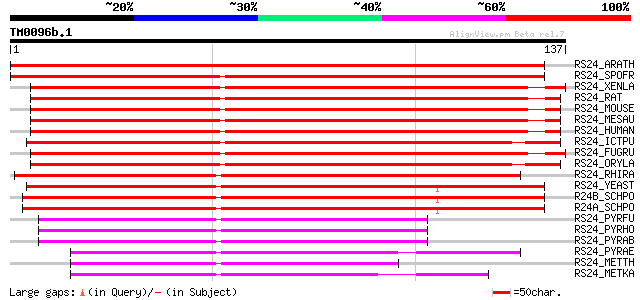
Query= TM0096b.1
(137 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
RS24_ARATH (Q9SS17) 40S ribosomal protein S24 242 2e-64
RS24_SPOFR (Q962Q6) 40S ribosomal protein S24 171 6e-43
RS24_XENLA (P02377) 40S ribosomal protein S24 (S19) 169 2e-42
RS24_RAT (P62850) 40S ribosomal protein S24 168 4e-42
RS24_MOUSE (P62849) 40S ribosomal protein S24 168 4e-42
RS24_MESAU (P62848) 40S ribosomal protein S24 (Ribosomal protein... 168 4e-42
RS24_HUMAN (P62847) 40S ribosomal protein S24 168 4e-42
RS24_ICTPU (Q90YQ0) 40S ribosomal protein S24 166 1e-41
RS24_FUGRU (O42387) 40S ribosomal protein S24 164 4e-41
RS24_ORYLA (Q9W6X9) 40S ribosomal protein S24 159 1e-39
RS24_RHIRA (P14249) 40S ribosomal protein S24 157 5e-39
RS24_YEAST (P26782) 40S ribosomal protein S24 (RP50) 151 5e-37
R24B_SCHPO (O59865) 40S ribosomal protein S24-B 150 8e-37
R24A_SCHPO (O13784) 40S ribosomal protein S24-A 145 3e-35
RS24_PYRFU (Q8U442) 30S ribosomal protein S24e 67 1e-11
RS24_PYRHO (P58746) 30S ribosomal protein S24e 62 5e-10
RS24_PYRAB (Q9UY20) 30S ribosomal protein S24e 60 1e-09
RS24_PYRAE (Q8ZV65) 30S ribosomal protein S24e 57 9e-09
RS24_METTH (O26367) 30S ribosomal protein S24e 47 1e-05
RS24_METKA (Q8TVD8) 30S ribosomal protein S24e 47 1e-05
>RS24_ARATH (Q9SS17) 40S ribosomal protein S24
Length = 133
Score = 242 bits (618), Expect = 2e-64
Identities = 116/132 (87%), Positives = 129/132 (96%)
Query: 1 MADKAVTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVF 60
MA+KAVTIRTRKFMTNRLLSRKQF++DVLHPGRANVSKAELKEKLAR+Y+VKDPN +FVF
Sbjct: 1 MAEKAVTIRTRKFMTNRLLSRKQFVIDVLHPGRANVSKAELKEKLARMYEVKDPNAIFVF 60
Query: 61 KFRTHFGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIR 120
KFRTHFGGGKS+GFGLIYD++E+AKK+EPKYRLIRNGLDTK+EKSRKQ+KERKNRAKKIR
Sbjct: 61 KFRTHFGGGKSSGFGLIYDTVESAKKFEPKYRLIRNGLDTKIEKSRKQIKERKNRAKKIR 120
Query: 121 GVKKTKAADAAK 132
GVKKTKA DA K
Sbjct: 121 GVKKTKAGDAKK 132
>RS24_SPOFR (Q962Q6) 40S ribosomal protein S24
Length = 132
Score = 171 bits (432), Expect = 6e-43
Identities = 85/132 (64%), Positives = 104/132 (78%), Gaps = 1/132 (0%)
Query: 1 MADKAVTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVF 60
M + TIRTRKFMTNRLL+RKQ + DVLHPG+ VSK E++EKLA++Y V P+ VFVF
Sbjct: 1 MTEGTATIRTRKFMTNRLLARKQMVCDVLHPGKPTVSKTEIREKLAKMYKVT-PDVVFVF 59
Query: 61 KFRTHFGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIR 120
F+T+FGGGKSTGF LIYD+L+ AKK+EPK+RL R+GL K +RKQ KERKNR KK+R
Sbjct: 60 GFKTNFGGGKSTGFALIYDTLDLAKKFEPKHRLARHGLYEKKRPTRKQRKERKNRMKKVR 119
Query: 121 GVKKTKAADAAK 132
G KK+K AAK
Sbjct: 120 GTKKSKVGAAAK 131
>RS24_XENLA (P02377) 40S ribosomal protein S24 (S19)
Length = 132
Score = 169 bits (427), Expect = 2e-42
Identities = 85/132 (64%), Positives = 102/132 (76%), Gaps = 5/132 (3%)
Query: 6 VTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVFKFRTH 65
VTIRTRKFMTNRLL RKQ ++DVLHPG+A V K E++EKLA++Y P+ +FVF FRTH
Sbjct: 5 VTIRTRKFMTNRLLQRKQMVIDVLHPGKATVPKTEIREKLAKMYKTT-PDVIFVFGFRTH 63
Query: 66 FGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIRGVKKT 125
FGGGK+TGFG+IYDSL+ AKK EPK+RL ++GL K + SRKQ KERKNR KK+RG K
Sbjct: 64 FGGGKTTGFGMIYDSLDYAKKNEPKHRLAKHGLYEKKKTSRKQRKERKNRMKKVRGTAKA 123
Query: 126 KAADAAKAGKKK 137
AGKKK
Sbjct: 124 NVG----AGKKK 131
>RS24_RAT (P62850) 40S ribosomal protein S24
Length = 133
Score = 168 bits (425), Expect = 4e-42
Identities = 85/131 (64%), Positives = 101/131 (76%), Gaps = 5/131 (3%)
Query: 6 VTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVFKFRTH 65
VTIRTRKFMTNRLL RKQ ++DVLHPG+A V K E++EKLA++Y P+ +FVF FRTH
Sbjct: 5 VTIRTRKFMTNRLLQRKQMVIDVLHPGKATVPKTEIREKLAKMYKTT-PDVIFVFGFRTH 63
Query: 66 FGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIRGVKKT 125
FGGGK+TGFG+IYDSL+ AKK EPK+RL R+GL K + SRKQ KERKNR KK+RG K
Sbjct: 64 FGGGKTTGFGMIYDSLDYAKKNEPKHRLARHGLYEKKKTSRKQRKERKNRMKKVRGTAKA 123
Query: 126 KAADAAKAGKK 136
AGKK
Sbjct: 124 NVG----AGKK 130
>RS24_MOUSE (P62849) 40S ribosomal protein S24
Length = 133
Score = 168 bits (425), Expect = 4e-42
Identities = 85/131 (64%), Positives = 101/131 (76%), Gaps = 5/131 (3%)
Query: 6 VTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVFKFRTH 65
VTIRTRKFMTNRLL RKQ ++DVLHPG+A V K E++EKLA++Y P+ +FVF FRTH
Sbjct: 5 VTIRTRKFMTNRLLQRKQMVIDVLHPGKATVPKTEIREKLAKMYKTT-PDVIFVFGFRTH 63
Query: 66 FGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIRGVKKT 125
FGGGK+TGFG+IYDSL+ AKK EPK+RL R+GL K + SRKQ KERKNR KK+RG K
Sbjct: 64 FGGGKTTGFGMIYDSLDYAKKNEPKHRLARHGLYEKKKTSRKQRKERKNRMKKVRGTAKA 123
Query: 126 KAADAAKAGKK 136
AGKK
Sbjct: 124 NVG----AGKK 130
>RS24_MESAU (P62848) 40S ribosomal protein S24 (Ribosomal protein
S19)
Length = 133
Score = 168 bits (425), Expect = 4e-42
Identities = 85/131 (64%), Positives = 101/131 (76%), Gaps = 5/131 (3%)
Query: 6 VTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVFKFRTH 65
VTIRTRKFMTNRLL RKQ ++DVLHPG+A V K E++EKLA++Y P+ +FVF FRTH
Sbjct: 5 VTIRTRKFMTNRLLQRKQMVIDVLHPGKATVPKTEIREKLAKMYKTT-PDVIFVFGFRTH 63
Query: 66 FGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIRGVKKT 125
FGGGK+TGFG+IYDSL+ AKK EPK+RL R+GL K + SRKQ KERKNR KK+RG K
Sbjct: 64 FGGGKTTGFGMIYDSLDYAKKNEPKHRLARHGLYEKKKTSRKQRKERKNRMKKVRGTAKA 123
Query: 126 KAADAAKAGKK 136
AGKK
Sbjct: 124 NVG----AGKK 130
>RS24_HUMAN (P62847) 40S ribosomal protein S24
Length = 133
Score = 168 bits (425), Expect = 4e-42
Identities = 85/131 (64%), Positives = 101/131 (76%), Gaps = 5/131 (3%)
Query: 6 VTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVFKFRTH 65
VTIRTRKFMTNRLL RKQ ++DVLHPG+A V K E++EKLA++Y P+ +FVF FRTH
Sbjct: 5 VTIRTRKFMTNRLLQRKQMVIDVLHPGKATVPKTEIREKLAKMYKTT-PDVIFVFGFRTH 63
Query: 66 FGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIRGVKKT 125
FGGGK+TGFG+IYDSL+ AKK EPK+RL R+GL K + SRKQ KERKNR KK+RG K
Sbjct: 64 FGGGKTTGFGMIYDSLDYAKKNEPKHRLARHGLYEKKKTSRKQRKERKNRMKKVRGTAKA 123
Query: 126 KAADAAKAGKK 136
AGKK
Sbjct: 124 NVG----AGKK 130
>RS24_ICTPU (Q90YQ0) 40S ribosomal protein S24
Length = 131
Score = 166 bits (420), Expect = 1e-41
Identities = 84/132 (63%), Positives = 102/132 (76%), Gaps = 4/132 (3%)
Query: 5 AVTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVFKFRT 64
+VT+RTRKFMTNRLL RKQ ++DVLHPG+A V K E++EKLA++Y P+ VFVF F+T
Sbjct: 4 SVTLRTRKFMTNRLLQRKQMVLDVLHPGKATVPKTEIREKLAKMYKTT-PDVVFVFGFKT 62
Query: 65 HFGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIRGVKK 124
FGGGK+TGF ++YDSL+ AKK EPKYRL R+GL K + SRKQ KERKNR KK+RG KK
Sbjct: 63 QFGGGKTTGFAMVYDSLDYAKKNEPKYRLARHGLYEKKKTSRKQRKERKNRMKKVRGTKK 122
Query: 125 TKAADAAKAGKK 136
A AGKK
Sbjct: 123 ---ASVGAAGKK 131
>RS24_FUGRU (O42387) 40S ribosomal protein S24
Length = 132
Score = 164 bits (416), Expect = 4e-41
Identities = 84/132 (63%), Positives = 100/132 (75%), Gaps = 5/132 (3%)
Query: 6 VTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVFKFRTH 65
VT+RTRKFMTNRLL RKQ +VDVLHPG+A V K E++EKLA++Y P+ VFVF FRT
Sbjct: 5 VTVRTRKFMTNRLLQRKQMVVDVLHPGKATVPKTEIREKLAKMYKTT-PDVVFVFGFRTQ 63
Query: 66 FGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIRGVKKT 125
FGGGK+TGF ++YDSL+ AKK EPK+RL R+GL K + SRKQ KERKNR KK+RG KK
Sbjct: 64 FGGGKTTGFAMVYDSLDYAKKNEPKHRLARHGLFEKKKTSRKQRKERKNRMKKVRGTKKA 123
Query: 126 KAADAAKAGKKK 137
A KKK
Sbjct: 124 SVG----ASKKK 131
>RS24_ORYLA (Q9W6X9) 40S ribosomal protein S24
Length = 132
Score = 159 bits (403), Expect = 1e-39
Identities = 82/131 (62%), Positives = 99/131 (74%), Gaps = 4/131 (3%)
Query: 6 VTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVFKFRTH 65
VTIRTRKFMTNRLL RKQ +VDVLHPG+A V K E++EKLA++Y P+ VFVF FRT
Sbjct: 5 VTIRTRKFMTNRLLQRKQMVVDVLHPGKATVPKTEIREKLAKMYKTT-PDVVFVFGFRTQ 63
Query: 66 FGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIRGVKKT 125
FGGGK+TGF ++YDSL+ AKK EPK+RL +GL K + SRKQ ER+NR KK+R +KK
Sbjct: 64 FGGGKTTGFAMVYDSLDYAKKNEPKHRLATHGLYEKKKTSRKQRTERQNRMKKVRSIKK- 122
Query: 126 KAADAAKAGKK 136
A AGKK
Sbjct: 123 --ASVGAAGKK 131
>RS24_RHIRA (P14249) 40S ribosomal protein S24
Length = 148
Score = 157 bits (398), Expect = 5e-39
Identities = 78/125 (62%), Positives = 96/125 (76%), Gaps = 1/125 (0%)
Query: 2 ADKAVTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVFK 61
AD AVTIRTRKF+TNRLL RKQ +VDV+HPG ANVSK EL+ K+ ++Y D V VF
Sbjct: 20 ADAAVTIRTRKFLTNRLLQRKQMVVDVIHPGLANVSKDELRSKIGKMYKA-DKEVVSVFG 78
Query: 62 FRTHFGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIRG 121
F+THFGGGK+TGF LIYD++E K +EPK+RL+R GL + RKQ KE+KNR KK RG
Sbjct: 79 FKTHFGGGKTTGFALIYDNVEALKNFEPKHRLVRIGLAEAPKGGRKQRKEKKNREKKFRG 138
Query: 122 VKKTK 126
V+K+K
Sbjct: 139 VRKSK 143
>RS24_YEAST (P26782) 40S ribosomal protein S24 (RP50)
Length = 134
Score = 151 bits (381), Expect = 5e-37
Identities = 81/129 (62%), Positives = 98/129 (75%), Gaps = 2/129 (1%)
Query: 5 AVTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVFKFRT 64
AVTIRTRK ++N LL+RKQF+VDVLHP RANVSK EL+EKLA +Y + + V VF FRT
Sbjct: 3 AVTIRTRKVISNPLLARKQFVVDVLHPNRANVSKDELREKLAEVYKA-EKDAVSVFGFRT 61
Query: 65 HFGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEK-SRKQLKERKNRAKKIRGVK 123
FGGGKS GFGL+Y+S+ AKK+EP YRL+R GL KVEK SR+Q K++KNR KKI G
Sbjct: 62 QFGGGKSVGFGLVYNSVAEAKKFEPTYRLVRYGLAEKVEKASRQQRKQKKNRDKKIFGTG 121
Query: 124 KTKAADAAK 132
K A A+
Sbjct: 122 KRLAKKVAR 130
>R24B_SCHPO (O59865) 40S ribosomal protein S24-B
Length = 133
Score = 150 bits (379), Expect = 8e-37
Identities = 77/130 (59%), Positives = 97/130 (74%), Gaps = 2/130 (1%)
Query: 4 KAVTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVFKFR 63
+AVTIRTRKFMTNRLL RKQ +VD+LHPG+AN+SK E++EKLA++Y D V F R
Sbjct: 2 EAVTIRTRKFMTNRLLQRKQMVVDILHPGKANLSKNEIREKLAQMYKT-DSECVQAFGLR 60
Query: 64 THFGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEK-SRKQLKERKNRAKKIRGV 122
THFGGG+STGF LIYDS E+ KK+EP YRL+R G ++K +R+Q K+RKNR KK+ G
Sbjct: 61 THFGGGRSTGFALIYDSTESMKKFEPHYRLVRVGQAEPIQKVARQQRKQRKNRGKKVFGT 120
Query: 123 KKTKAADAAK 132
K A +K
Sbjct: 121 GKRLAKRKSK 130
>R24A_SCHPO (O13784) 40S ribosomal protein S24-A
Length = 133
Score = 145 bits (366), Expect = 3e-35
Identities = 73/130 (56%), Positives = 96/130 (73%), Gaps = 2/130 (1%)
Query: 4 KAVTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVFKFR 63
+AVTIRTRKFMTNRLL RKQ +VD+LHPG+AN+SK +L+EKL ++Y D + V F R
Sbjct: 2 EAVTIRTRKFMTNRLLQRKQMVVDILHPGKANISKNDLREKLGQMYKT-DASAVQAFGLR 60
Query: 64 THFGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEK-SRKQLKERKNRAKKIRGV 122
TH+GGG++TGF LIYD +E KK+EP YRL+R G ++K +R+Q K+RKNR KK+ G
Sbjct: 61 THYGGGRTTGFALIYDDVEAMKKFEPHYRLVRVGHAEPIQKVARQQRKQRKNRGKKVFGT 120
Query: 123 KKTKAADAAK 132
K A +K
Sbjct: 121 GKRLAKRKSK 130
>RS24_PYRFU (Q8U442) 30S ribosomal protein S24e
Length = 99
Score = 67.0 bits (162), Expect = 1e-11
Identities = 35/96 (36%), Positives = 54/96 (55%), Gaps = 1/96 (1%)
Query: 8 IRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVFKFRTHFG 67
IR ++ N+L+ RK+ ++ HPG S+ ++K KL + D+ +P T + R++FG
Sbjct: 3 IRIKEIKENKLIGRKEIYFEIYHPGEPTPSRKDVKGKLVAMLDL-NPETTVIQYIRSYFG 61
Query: 68 GGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVE 103
S G+ YDS E EP+Y LIR+GL K E
Sbjct: 62 SYISKGYAKAYDSKERMLYIEPEYILIRDGLIEKKE 97
>RS24_PYRHO (P58746) 30S ribosomal protein S24e
Length = 99
Score = 61.6 bits (148), Expect = 5e-10
Identities = 32/96 (33%), Positives = 52/96 (53%), Gaps = 1/96 (1%)
Query: 8 IRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVFKFRTHFG 67
I+ + N+L+ RK+ ++ HPG S+ ++K KL + D+ +P T + R++FG
Sbjct: 3 IKITEVKENKLIGRKEIYFEIYHPGEPTPSRKDVKGKLVAMLDL-NPETTVIQYIRSYFG 61
Query: 68 GGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVE 103
KS G+ Y E EP+Y LIR+G+ K E
Sbjct: 62 SYKSKGYAKYYYDKERMLYIEPEYILIRDGIIEKKE 97
>RS24_PYRAB (Q9UY20) 30S ribosomal protein S24e
Length = 99
Score = 60.5 bits (145), Expect = 1e-09
Identities = 31/96 (32%), Positives = 52/96 (53%), Gaps = 1/96 (1%)
Query: 8 IRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVFKFRTHFG 67
I+ + N+L+ RK+ ++ HPG S+ ++K KL + D+ +P T + R++FG
Sbjct: 3 IKITEVKENKLIGRKEIYFEIYHPGEPTPSRKDVKGKLVAMLDL-NPETTVIQYIRSYFG 61
Query: 68 GGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVE 103
KS G+ Y + EP+Y LIR+G+ K E
Sbjct: 62 SYKSKGYAKYYYDKDRMLYIEPEYILIRDGIIEKKE 97
>RS24_PYRAE (Q8ZV65) 30S ribosomal protein S24e
Length = 121
Score = 57.4 bits (137), Expect = 9e-09
Identities = 35/111 (31%), Positives = 62/111 (55%), Gaps = 5/111 (4%)
Query: 16 NRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVFKFRTHFGGGKSTGFG 75
N+LL+R++ +V+ +H + ++ ++E +A+ + D + VFV + +T FG G+S
Sbjct: 14 NKLLARRELLVEAVHQNASTPTRQSVREWVAKQLGI-DISNVFVRRIKTEFGRGRSLAEV 72
Query: 76 LIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIRGVKKTK 126
+Y+ + A+ EP Y L RN E+ +K L+E K R + R KK K
Sbjct: 73 HVYNDSKIARVIEPLYILARN----LGEEGKKLLEEAKKRRNERREKKKRK 119
>RS24_METTH (O26367) 30S ribosomal protein S24e
Length = 100
Score = 47.0 bits (110), Expect = 1e-05
Identities = 26/81 (32%), Positives = 44/81 (54%), Gaps = 1/81 (1%)
Query: 16 NRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVFKFRTHFGGGKSTGFG 75
N LL+RK+ + L+ G + E+K KL + D D + + V K FG ++TG+
Sbjct: 11 NPLLNRKEIRFECLYEGESTPKVLEVKNKLVAMLDA-DKDLLVVDKIDQGFGEPRATGYA 69
Query: 76 LIYDSLENAKKYEPKYRLIRN 96
IY+S E + EP++ + +N
Sbjct: 70 KIYESAEKLTEIEPEHVIKKN 90
>RS24_METKA (Q8TVD8) 30S ribosomal protein S24e
Length = 116
Score = 47.0 bits (110), Expect = 1e-05
Identities = 30/103 (29%), Positives = 53/103 (51%), Gaps = 10/103 (9%)
Query: 16 NRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVFKFRTHFGGGKSTGFG 75
N LL RK+ V H K+E+ KLA + DV D V + + + FG ++ G+
Sbjct: 11 NPLLYRKEVKFVVRHEDSGTPQKSEVLRKLAAILDV-DKEVVLIDRMESEFGKRETKGYA 69
Query: 76 LIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKK 118
IY S+E+ + EP++ VE+ +K L+E ++ +++
Sbjct: 70 KIYKSMEHLEDIEPEH---------MVERHKKVLEELESESEE 103
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.134 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,530,933
Number of Sequences: 164201
Number of extensions: 584642
Number of successful extensions: 2779
Number of sequences better than 10.0: 85
Number of HSP's better than 10.0 without gapping: 50
Number of HSP's successfully gapped in prelim test: 35
Number of HSP's that attempted gapping in prelim test: 2690
Number of HSP's gapped (non-prelim): 96
length of query: 137
length of database: 59,974,054
effective HSP length: 98
effective length of query: 39
effective length of database: 43,882,356
effective search space: 1711411884
effective search space used: 1711411884
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0096b.1


