

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0095c.5
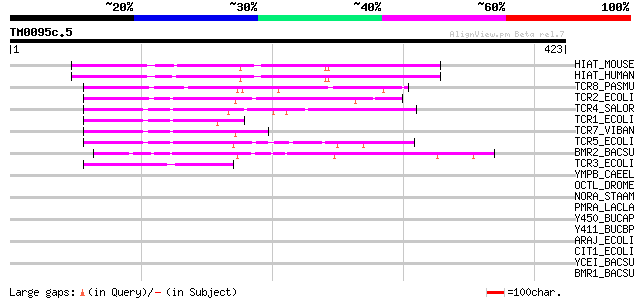
(423 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
HIAT_MOUSE (P70187) Hippocampus abundant transcript 1 protein 84 5e-16
HIAT_HUMAN (Q96MC6) Hippocampus abundant transcript 1 protein (P... 82 2e-15
TCR8_PASMU (P51564) Tetracycline resistance protein, class H (TE... 64 6e-10
TCR2_ECOLI (P02980) Tetracycline resistance protein, class B (TE... 60 1e-08
TCR4_SALOR (P33733) Tetracycline resistance protein, class D (TE... 59 2e-08
TCR1_ECOLI (P02982) Tetracycline resistance protein, class A (TE... 57 7e-08
TCR7_VIBAN (P51563) Tetracycline resistance protein, class G (TE... 55 4e-07
TCR5_ECOLI (Q07282) Tetracycline resistance protein, class E (TE... 53 1e-06
BMR2_BACSU (P39843) Multidrug resistance protein 2 (Multidrug-ef... 50 1e-05
TCR3_ECOLI (P02981) Tetracycline resistance protein, class C (TE... 49 2e-05
YMPB_CAEEL (Q7Z118) Hypothetical protein B0361.11 in chromosome III 42 0.004
OCTL_DROME (Q95R48) Organic cation transporter-like protein 42 0.004
NORA_STAAM (P21191) Quinolone resistance protein norA 40 0.011
PMRA_LACLA (P58120) Multi-drug resistance efflux pump pmrA homolog 39 0.019
Y450_BUCAP (Q8K999) Hypothetical transport protein BUsg450 39 0.025
Y411_BUCBP (Q89AA9) Hypothetical transport protein bbp411 39 0.033
ARAJ_ECOLI (P23910) Protein araJ precursor 38 0.056
CIT1_ECOLI (P07661) Citrate-proton symporter (Citrate transporte... 37 0.096
YCEI_BACSU (O34691) Hypothetical metabolite transport protein yceI 37 0.13
BMR1_BACSU (P33449) Multidrug resistance protein 1 (Multidrug-ef... 36 0.16
>HIAT_MOUSE (P70187) Hippocampus abundant transcript 1 protein
Length = 490
Score = 84.3 bits (207), Expect = 5e-16
Identities = 73/304 (24%), Positives = 132/304 (43%), Gaps = 35/304 (11%)
Query: 48 TCPKVIYI-NGLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQS 106
T PK ++ NGL Q + G+ + PL+G LSD GRK LL+T+ T ++
Sbjct: 65 TFPKHTFLMNGLIQGVKGLLSFLSAPLIGALSDVWGRKSFLLLTVFFTCAPIPLMKISP- 123
Query: 107 EEFVYAYYVLRTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVL 166
+ Y+ + + S + + F + AYVAD+ E +R+ + ++ F+AS V +
Sbjct: 124 ----WWYFAVISVSGVFAV--TFSVVFAYVADITQEHERSMAYGLVSATFAASLVTSPAI 177
Query: 167 ARFLPEEY------IFVVSIALLTFCPVYMQFFLVETVTPAPKKNQVSGFCTKVVDVLHK 220
+L + Y + +IALL C F LV P+K + + + +
Sbjct: 178 GAYLGQMYGDSLVVVLATAIALLDIC-----FILVAVPESLPEKMRPASWGAPISWEQAD 232
Query: 221 RYKSMRNAAEILDNLIIAV-----------QY-----YLKAVFGFNKNQFSELLMMVGIG 264
+ S++ + L+I + QY YLK + F+ + + ++GI
Sbjct: 233 PFASLKKVGQDSIVLLICITVFLSYLPEAGQYSSFFLYLKQIMKFSPESVAAFIAVLGIL 292
Query: 265 SIFSQMVLLPILNPKVGEKVILSAALLASIAYAWLYGLAWAPWVPYFSASFGIIYVLVKP 324
SI +Q ++L +L +G K + L I YG PW+ + + + + + P
Sbjct: 293 SIIAQTIVLSLLMRSIGNKNTILLGLGFQILQLAWYGFGSEPWMMWAAGAVAAMSSITFP 352
Query: 325 SGKA 328
+ A
Sbjct: 353 AVSA 356
>HIAT_HUMAN (Q96MC6) Hippocampus abundant transcript 1 protein
(Putative tetracycline transporter-like protein)
Length = 490
Score = 82.4 bits (202), Expect = 2e-15
Identities = 72/304 (23%), Positives = 131/304 (42%), Gaps = 35/304 (11%)
Query: 48 TCPKVIYI-NGLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQS 106
T PK ++ NGL Q + G+ + PL+G LSD GRK LL+T+ T ++
Sbjct: 65 TFPKHTFLMNGLIQGVKGLLSFLSAPLIGALSDVWGRKSFLLLTVFFTCAPIPLMKISP- 123
Query: 107 EEFVYAYYVLRTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVL 166
+ Y+ + + S + + F + AYVAD+ E +R+ + ++ F+AS V +
Sbjct: 124 ----WWYFAVISVSGVFA--VTFSVVFAYVADITQEHERSMAYGLVSATFAASLVTSPAI 177
Query: 167 ARFLPEEY------IFVVSIALLTFCPVYMQFFLVETVTPAPKKNQVSGFCTKVVDVLHK 220
+L Y + +IALL C F LV P+K + + + +
Sbjct: 178 GAYLGRVYGDSLVVVLATAIALLDIC-----FILVAVPESLPEKMRPASWGAPISWEQAD 232
Query: 221 RYKSMRNAAEILDNLIIAV-----------QY-----YLKAVFGFNKNQFSELLMMVGIG 264
+ S++ + L+I + QY YL+ + F+ + + ++GI
Sbjct: 233 PFASLKKVGQDSIVLLICITVFLSYLPEAGQYSSFFLYLRQIMKFSPESVAAFIAVLGIL 292
Query: 265 SIFSQMVLLPILNPKVGEKVILSAALLASIAYAWLYGLAWAPWVPYFSASFGIIYVLVKP 324
SI +Q ++L +L +G K + L I YG PW+ + + + + + P
Sbjct: 293 SIIAQTIVLSLLMRSIGNKNTILLGLGFQILQLAWYGFGSEPWMMWAAGAVAAMSSITFP 352
Query: 325 SGKA 328
+ A
Sbjct: 353 AVSA 356
>TCR8_PASMU (P51564) Tetracycline resistance protein, class H
(TETA(H))
Length = 400
Score = 64.3 bits (155), Expect = 6e-10
Identities = 61/266 (22%), Positives = 117/266 (43%), Gaps = 27/266 (10%)
Query: 57 GLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVL 116
G+ + +++ P+LG+LSD++GRKP+LL ++ + ++A+ + ++ Y+
Sbjct: 44 GVLLALYATMQVIFAPILGRLSDKYGRKPILLFSLLGAALDYLLMAFSTT---LWMLYIG 100
Query: 117 RTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPE---- 172
R + I C S ++DV R F ++ G+F ++G +L L +
Sbjct: 101 RIIAGITGATGAVCASA--MSDVTPAKNRTRYFGFLGGVFGVGLIIGPMLGGLLGDISAH 158
Query: 173 -EYIF-------VVSIALLTFCPVYMQFFLVETVTPAPK--KNQVSGFCTKVVDVLHKRY 222
+IF ++ ++LL F + LV TP + N V+ F K + Y
Sbjct: 159 MPFIFAAISHSILLILSLLFFRETQKREALVANRTPENQTASNTVTVFFKKSLYFWLATY 218
Query: 223 KSMRNAAEILDNLIIAVQYYLKAVFGFNKNQFSELLMMVGIGSIFSQMVLLPILNPKVGE 282
++ +I + + Y F +N L ++G+ IF Q ++ L K GE
Sbjct: 219 FIIQLIGQIPATIWVLFTQYR---FDWNTTSIGMSLAVLGVLHIFFQAIVAGKLAQKWGE 275
Query: 283 K----VILSAALLASIAYAWLYGLAW 304
K + +S ++ + AW+ G W
Sbjct: 276 KTTIMISMSIDMMGCLLLAWI-GHVW 300
>TCR2_ECOLI (P02980) Tetracycline resistance protein, class B
(TETA(B)) (Metal-tetracycline/H(+) antiporter)
Length = 401
Score = 60.1 bits (144), Expect = 1e-08
Identities = 61/257 (23%), Positives = 114/257 (43%), Gaps = 22/257 (8%)
Query: 57 GLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVL 116
G+ + + +++ P LG++SD GR+P+LL+++ + +LA+ + ++ Y+
Sbjct: 44 GVLLALYALMQVIFAPWLGKMSDRFGRRPVLLLSLIGASLDYLLLAFSSA---LWMLYLG 100
Query: 117 RTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFL----PE 172
R S I G+ ++ + +AD + S+R F W+ F + G ++ F P
Sbjct: 101 RLLSGI--TGATGAVAASVIADTTSASQRVKWFGWLGASFGLGLIAGPIIGGFAGEISPH 158
Query: 173 EYIFVVSIA-LLTFCPVYMQFFLVETVTPAPKKNQVSGFCTKVVDVLHKRYKSMRNAAEI 231
F+ ++ ++TF + + F+ ET + G T+ V +K+M I
Sbjct: 159 SPFFIAALLNIVTF--LVVMFWFRETKNTRDNTDTEVGVETQSNSVYITLFKTMPILLII 216
Query: 232 LDNLIIAVQYYLKAVFGFNKNQFSELLMMVG--------IGSIFSQMVLLPILNPKVGEK 283
+ + Q F +N+F MMVG + S+F V I K GEK
Sbjct: 217 YFSAQLIGQIPATVWVLFTENRFGWNSMMVGFSLAGLGLLHSVFQAFVAGRIAT-KWGEK 275
Query: 284 VILSAALLA-SIAYAWL 299
+ +A S A+A+L
Sbjct: 276 TAVLLGFIADSSAFAFL 292
>TCR4_SALOR (P33733) Tetracycline resistance protein, class D
(TETA(D))
Length = 394
Score = 59.3 bits (142), Expect = 2e-08
Identities = 62/265 (23%), Positives = 116/265 (43%), Gaps = 19/265 (7%)
Query: 57 GLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVL 116
G+ + + ++ PLLG+ SD+ GR+P+LL++++ F + +LA ++ Y+
Sbjct: 44 GILLALYAVMQVCFAPLLGRWSDKLGRRPVLLLSLAGAAFDYTLLA---LSNVLWMLYLG 100
Query: 117 RTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNV---LARFLPEE 173
R S I G+ ++ + VAD S+R A F + F A + G LA +
Sbjct: 101 RIISGI--TGATGAVAASVVADSTAVSERTAWFGRLGAAFGAGLIAGPAIGGLAGDISPH 158
Query: 174 YIFVVSIALLTFCPVYMQFFLVETVT-----PAPKKNQVSG--FCTKVVDVLHKRYKSMR 226
FV++ A+L C M FF+ + PA +K + +G F T + + +
Sbjct: 159 LPFVIA-AILNACTFLMVFFIFKPAVQTEEKPADEKQESAGISFITLLKPLALLLFVFF- 216
Query: 227 NAAEILDNLIIAV-QYYLKAVFGFNKNQFSELLMMVGIGSIFSQMVLLPILNPKVGEKVI 285
A+++ + V + ++ F ++ L +G Q V+ L ++ EK I
Sbjct: 217 -TAQLIGQIPATVWVLFTESRFAWDSAAVGFSLAGLGAMHALFQAVVAGALAKRLSEKTI 275
Query: 286 LSAALLASIAYAWLYGLAWAPWVPY 310
+ A +A L + W+ Y
Sbjct: 276 IFAGFIADATAFLLMSAITSGWMVY 300
>TCR1_ECOLI (P02982) Tetracycline resistance protein, class A
(TETA(A))
Length = 399
Score = 57.4 bits (137), Expect = 7e-08
Identities = 33/127 (25%), Positives = 66/127 (50%), Gaps = 9/127 (7%)
Query: 57 GLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVL 116
G+ + + + P+LG LSD GR+P+LL++++ +A++A + F++ Y+
Sbjct: 46 GILLALYALMQFACAPVLGALSDRFGRRPVLLVSLAGAAVDYAIMA---TAPFLWVLYIG 102
Query: 117 RTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFS----ASHVVGNVLARFLPE 172
R + I G+ ++ AY+AD+ + +RA F +++ F A V+G ++ F P
Sbjct: 103 RIVAGI--TGATGAVAGAYIADITDGDERARHFGFMSACFGFGMVAGPVLGGLMGGFSPH 160
Query: 173 EYIFVVS 179
F +
Sbjct: 161 APFFAAA 167
>TCR7_VIBAN (P51563) Tetracycline resistance protein, class G
(TETA(G))
Length = 393
Score = 54.7 bits (130), Expect = 4e-07
Identities = 37/144 (25%), Positives = 68/144 (46%), Gaps = 8/144 (5%)
Query: 57 GLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVL 116
G ++ + ++V P+LGQLSD +GR+P+LL +++ + ++A S ++ Y+
Sbjct: 44 GALLSLYALMQVVFAPMLGQLSDSYGRRPVLLASLAGAAVDYTIMA---SAPVLWVLYIG 100
Query: 117 RTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFL---PEE 173
R S + G+ ++ + +AD E RA F ++ + A + G L L
Sbjct: 101 RLVSGV--TGATGAVAASTIADSTGEGSRARWFGYMGACYGAGMIAGPALGGMLGGISAH 158
Query: 174 YIFVVSIALLTFCPVYMQFFLVET 197
F+ + L F + FL ET
Sbjct: 159 APFIAAALLNGFAFLLACIFLKET 182
>TCR5_ECOLI (Q07282) Tetracycline resistance protein, class E
(TETA(E))
Length = 405
Score = 53.1 bits (126), Expect = 1e-06
Identities = 59/273 (21%), Positives = 119/273 (42%), Gaps = 36/273 (13%)
Query: 57 GLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVL 116
G+ + + +++ PLLG+ SD GR+P+LL+++ +A++A + V+ Y+
Sbjct: 44 GVLLALYAMMQVIFAPLLGRWSDRIGRRPVLLLSLLGATLDYALMA---TASVVWVLYLG 100
Query: 117 RTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARF-----LP 171
R + I G+ ++ + +ADV E R F + F + G V+ F +
Sbjct: 101 RLIAGI--TGATGAVAASTIADVTPEESRTHWFGMMGACFGGGMIAGPVIGGFAGQLSVQ 158
Query: 172 EEYIFVVSIALLTFCPVYMQFFLVETVTPAPKKNQVSGFCTKVVDVLHKRYKSMRNAAEI 231
++F +I L F + F L ET NQVS + +++ S+R
Sbjct: 159 APFMFAAAINGLAF--LVSLFILHET----HNANQVSDELKN--ETINETTSSIREMISP 210
Query: 232 LDNLIIAVQYYLKAVFG---------FNKNQFSELLMMVGIG-SIFS------QMVLLPI 275
L L++ +++ + G F + +F+ +MVG+ ++F Q +
Sbjct: 211 LSGLLVV--FFIIQLIGQIPATLWVLFGEERFAWDGVMVGVSLAVFGLTHALFQGLAAGF 268
Query: 276 LNPKVGEKVILSAALLASIAYAWLYGLAWAPWV 308
+ +GE+ ++ +LA +L + W+
Sbjct: 269 IAKHLGERKAIAVGILADGCGLFLLAVITQSWM 301
>BMR2_BACSU (P39843) Multidrug resistance protein 2
(Multidrug-efflux transporter 2)
Length = 400
Score = 49.7 bits (117), Expect = 1e-05
Identities = 72/333 (21%), Positives = 139/333 (41%), Gaps = 36/333 (10%)
Query: 65 IFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVLRTFSYIIS 124
I +++ P G+ D GRK ++++ + IFS + L + V +Y R + S
Sbjct: 56 ISQLITSPFAGRWVDRFGRKKMIILGL--LIFSLSELIFGLGTH-VSIFYFSRILGGV-S 111
Query: 125 QGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPE-----EYIFVVS 179
I AYVAD+ +R+ +++ S ++G F+ + F +
Sbjct: 112 AAFIMPAVTAYVADITTLKERSKAMGYVSAAISTGFIIGPGAGGFIAGFGIRMPFFFASA 171
Query: 180 IALLTFCPVYMQFFLVETVTPAPKKNQVSGFCTKVVDVLHKRYKSMRNAAEILDNLIIAV 239
IAL+ V F L E+++ +++Q+S TK + + +S+ I ++ +
Sbjct: 172 IALIA--AVTSVFILKESLS-IEERHQLSSH-TKESNFIKDLKRSIHPVYFIAFIIVFVM 227
Query: 240 QYYLKAV-----------FGFNKNQFSELLMMVGIGSIFSQMVLLPILNPKVGEKVILSA 288
+ L A FGF + ++ + I ++ Q++L L K+GEK ++
Sbjct: 228 AFGLSAYETVFSLFSDHKFGFTPKDIAAIITISSIVAVVIQVLLFGKLVNKLGEKRMIQL 287
Query: 289 ALLASIAYAWLYG-LAWAPWVPYFSASFGIIYVLVKP---------SGKAQTFIAGAQSI 338
L+ A++ ++ V + + + L++P +G Q F+AG S
Sbjct: 288 CLITGAILAFVSTVMSGFLTVLLVTCFIFLAFDLLRPALTAHLSNMAGNQQGFVAGMNST 347
Query: 339 SDLLSPIAMSPLTS--WFLSSNAPFDCKGFSII 369
L I L + L+ + PF GF +I
Sbjct: 348 YTSLGNIFGPALGGILFDLNIHYPFLFAGFVMI 380
>TCR3_ECOLI (P02981) Tetracycline resistance protein, class C
(TETA(C))
Length = 396
Score = 48.9 bits (115), Expect = 2e-05
Identities = 29/114 (25%), Positives = 54/114 (46%), Gaps = 5/114 (4%)
Query: 57 GLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVL 116
G+ + + + + P+LG LSD GR+P+LL ++ +A++A +YA ++
Sbjct: 46 GVLLALYALMQFLCAPVLGALSDRFGRRPVLLASLLGATIDYAIMATTPVLWILYAGRIV 105
Query: 117 RTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFL 170
+ G+ ++ AY+AD+ + RA F ++ F V G V L
Sbjct: 106 AGIT-----GATGAVAGAYIADITDGEDRARHFGLMSACFGVGMVAGPVAGGLL 154
>YMPB_CAEEL (Q7Z118) Hypothetical protein B0361.11 in chromosome III
Length = 534
Score = 41.6 bits (96), Expect = 0.004
Identities = 43/157 (27%), Positives = 75/157 (47%), Gaps = 22/157 (14%)
Query: 61 TIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSF-AVLAWDQSEEFVYAYYVLRTF 119
TI I ++ +P + L+D +GRKP++ ++T I +F A +A S F + +LR F
Sbjct: 151 TIFTIGAVIAVPFMSMLADRYGRKPII---VTTAILAFLANMAASFSPNFA-IFLILRAF 206
Query: 120 SYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNV---LARFLPEEY-- 174
S + SVA + ++E RA WIT +++ + +G V L + +++
Sbjct: 207 IGACSDSYLSVASVA-TCEYLSEKARA----WITVVYNVAWSLGMVWTLLVTLMTDDWRW 261
Query: 175 -IFVVSI------ALLTFCPVYMQFFLVETVTPAPKK 204
F+VS+ AL F P + + + T KK
Sbjct: 262 RYFIVSLPGVYGFALWYFLPESPHWLITKNKTEKLKK 298
>OCTL_DROME (Q95R48) Organic cation transporter-like protein
Length = 567
Score = 41.6 bits (96), Expect = 0.004
Identities = 27/98 (27%), Positives = 47/98 (47%), Gaps = 4/98 (4%)
Query: 73 LLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVLRTFSYIISQGSIFCIS 132
+ GQLSD++GRKP+L ++ + F VLA E F Y + L + + +F ++
Sbjct: 147 VFGQLSDKYGRKPILFASLVIQVL-FGVLAGVAPEYFTYTFARLMVGA---TTSGVFLVA 202
Query: 133 VAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFL 170
++V KR ++ FS ++ V A F+
Sbjct: 203 YVVAMEMVGPDKRLYAGIFVMMFFSVGFMLTAVFAYFV 240
>NORA_STAAM (P21191) Quinolone resistance protein norA
Length = 388
Score = 40.0 bits (92), Expect = 0.011
Identities = 37/162 (22%), Positives = 72/162 (43%), Gaps = 22/162 (13%)
Query: 57 GLQQTIVGIFKMVVLPLLGQLSDEHGRKPLL---LITMSTTIFSFAVLAWDQSEEFVYAY 113
GL + +M++ P G L+D+ G+K ++ LI S + F FAV ++
Sbjct: 41 GLLVAAFALSQMIISPFGGTLADKLGKKLIICIGLILFSVSEFMFAV-------GHNFSV 93
Query: 114 YVLRTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPE- 172
+L +S G + +AD+ ++A F +++ + ++ ++G + F+ E
Sbjct: 94 LMLSRVIGGMSAGMVMPGVTGLIADISPSHQKAKNFGYMSAIINSGFILGPGIGGFMAEV 153
Query: 173 ----EYIFVVSIALLTFCPVYMQFFLVETVTPAPKKNQVSGF 210
+ F ++ +L F M L+ PKK+ SGF
Sbjct: 154 SHRMPFYFAGALGILAF---IMSIVLIHD----PKKSTTSGF 188
>PMRA_LACLA (P58120) Multi-drug resistance efflux pump pmrA homolog
Length = 405
Score = 39.3 bits (90), Expect = 0.019
Identities = 37/159 (23%), Positives = 72/159 (45%), Gaps = 13/159 (8%)
Query: 56 NGLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYV 115
+GL ++ + +V P+ G+L+DEHGRK +++ + +A+ + ++ +
Sbjct: 52 SGLAFSLPALASGLVAPIWGRLADEHGRKVMMVRASIVMTLTMGGIAFAPNVWWLLGLRL 111
Query: 116 LRTF--SYIISQGSIFCISVAYVADVVNESKRA-AVFSWITGLFSASHV---VGNVLARF 169
L F YI + S A +A + K A+ + T + S + + +G +LA +
Sbjct: 112 LMGFFSGYIPN-------STAMIASQAPKDKSGYALGTLATAMVSGTLIGPSLGGLLAEW 164
Query: 170 LPEEYIFVVSIALLTFCPVYMQFFLVETVTPAPKKNQVS 208
+F++ ALL + FF+ E P K +S
Sbjct: 165 FGMANVFLIVGALLALATLLTIFFVHENFEPIAKGEMLS 203
Score = 34.7 bits (78), Expect = 0.48
Identities = 23/91 (25%), Positives = 46/91 (50%)
Query: 16 LLFPLSIHWIAEEMTVSVLVDVTTSALCPGGSTCPKVIYINGLQQTIVGIFKMVVLPLLG 75
+LF L + ++T + T + ++ +++I+GL + VG+ M+ LG
Sbjct: 216 ILFGLLVTTFIIQITSQSIEPFVTLYIKTLTTSTNNLMFISGLIVSAVGLSAMLSSSFLG 275
Query: 76 QLSDEHGRKPLLLITMSTTIFSFAVLAWDQS 106
+L D++G L+LI + T + +A+ QS
Sbjct: 276 RLGDKYGSHRLILIGLVFTFIIYLPMAFVQS 306
>Y450_BUCAP (Q8K999) Hypothetical transport protein BUsg450
Length = 391
Score = 38.9 bits (89), Expect = 0.025
Identities = 38/172 (22%), Positives = 74/172 (42%), Gaps = 11/172 (6%)
Query: 54 YINGLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAY 113
++ GL I G ++V G LSD GRK +++ + ++ S ++
Sbjct: 48 FLIGLAVGIYGATQIVFQIPFGILSDRFGRKQIIIFGLFIFFIGSLIVV---STNSIFGL 104
Query: 114 YVLRTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPEE 173
+ R I G+I +S+A ++D++ + R S I F+ S ++ V A + E
Sbjct: 105 IIGRA---IQGSGAISGVSMALLSDLIRKENRIKSISIIGVSFAVSFLISVVSAPIIAEN 161
Query: 174 Y----IFVVSIALLTFCPVYMQFFLVETVTPAPKKNQVSGFCTKVVDVLHKR 221
+ IF +S A+ + + + FFL+ + KN K + + +
Sbjct: 162 FGFFSIFWIS-AVFSIFSILIVFFLIPSSQNEILKNYKKNIYQKKIKFIFNK 212
>Y411_BUCBP (Q89AA9) Hypothetical transport protein bbp411
Length = 394
Score = 38.5 bits (88), Expect = 0.033
Identities = 44/210 (20%), Positives = 92/210 (42%), Gaps = 23/210 (10%)
Query: 53 IYINGLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFA-VLAWDQSEEFVY 111
I++ G+ I GIF+++ G LSD++G+K L+I + F ++AW S ++
Sbjct: 47 IFLVGVAIGIYGIFQIIFQIPYGWLSDKYGQK--LIINIGLLCFLLGNIIAW--SSNSIW 102
Query: 112 AYYVLRTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLP 171
+ R G+I + + ++++V R + + F S + +L+ +
Sbjct: 103 GIILGRGLQ---GSGAISSVCMTLLSELVLPHNRIKIMGLLGVSFGISFFLAVILSPIIV 159
Query: 172 EEY----IFVVSIALLTFCPVYMQFFLVETVTPAPKKNQVSGFCTKVVD---VLHKRYKS 224
+ +F+++ L FC + F++ ++ KN V F +++ + +L R
Sbjct: 160 NMFGFYCLFLINSLLSIFCLFFGMFYIPASLL---NKNVVCNFRSEISNFFKILSNRVLC 216
Query: 225 MRNAAEILDNL-----IIAVQYYLKAVFGF 249
N + L + I + LK +F F
Sbjct: 217 QINLSVFLIHFFLMCNFIIIPVELKKIFEF 246
>ARAJ_ECOLI (P23910) Protein araJ precursor
Length = 394
Score = 37.7 bits (86), Expect = 0.056
Identities = 40/216 (18%), Positives = 89/216 (40%), Gaps = 25/216 (11%)
Query: 68 MVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVLRTFSYIIS--- 124
+V P++ S + K +LL ++ + A+ S Y++ ++S
Sbjct: 52 VVGAPIIALFSSRYSLKHILLFLVALCVIGNAMFTLSSS-------YLMLAIGRLVSGFP 104
Query: 125 QGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPEE----YIFVVSI 180
G+ F + ++ ++ K A + + + ++++G L +L +E Y F++ I
Sbjct: 105 HGAFFGVGAIVLSKIIKPGKVTAAVAGMVSGMTVANLLGIPLGTYLSQEFSWRYTFLL-I 163
Query: 181 ALLTFCPVYMQFFLVETVTPAPKKNQVSGFCTKVVDVLHKRYKSMRNAAEILDNLIIA-- 238
A+ + +F V + K N F L + AA + N +
Sbjct: 164 AVFNIAVMASVYFWVPDIRDEAKGNLREQF-----HFLRSPAPWLIFAATMFGNAGVFAW 218
Query: 239 ---VQYYLKAVFGFNKNQFSELLMMVGIGSIFSQMV 271
V+ Y+ + GF++ + ++M+VG+G + M+
Sbjct: 219 FSYVKPYMMFISGFSETAMTFIMMLVGLGMVLGNML 254
>CIT1_ECOLI (P07661) Citrate-proton symporter (Citrate transporter)
(Citrate carrier protein) (Citrate utilization
determinant) (Citrate utilization protein A)
Length = 431
Score = 37.0 bits (84), Expect = 0.096
Identities = 30/99 (30%), Positives = 48/99 (48%), Gaps = 4/99 (4%)
Query: 62 IVGIFKMVVLPLLGQLSDEHGRKPLLL-ITMSTTIFSFAVLAW-DQSEEFVYAYYVLRTF 119
+VGI + LP+ G +SD GR+P+L+ IT+ + + V+ W + +F VL F
Sbjct: 279 LVGISNFIWLPIGGAISDRIGRRPVLMGITLLALVTTLPVMNWLTAAPDFTRMTLVLLWF 338
Query: 120 SYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSA 158
S+ G VA + +V+ R FS L +A
Sbjct: 339 SFFF--GMYNGAMVAALTEVMPVYVRTVGFSLAFSLATA 375
>YCEI_BACSU (O34691) Hypothetical metabolite transport protein yceI
Length = 400
Score = 36.6 bits (83), Expect = 0.13
Identities = 70/350 (20%), Positives = 140/350 (40%), Gaps = 47/350 (13%)
Query: 73 LLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVLRTFSYIISQGSIFCIS 132
L G L+D GRK + +IT+ + A+ S + A+ +LR F + G ++
Sbjct: 65 LFGLLADRIGRKKVFIITLLCFSIGSGISAFVTS---LSAFLILR-FVIGMGLGGELPVA 120
Query: 133 VAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPEEYIFVVSIALLTFCPVYMQF 192
V++ V KR V + ++ + +++ F+ + + ++ LLT +
Sbjct: 121 STLVSEAVVPEKRGRVIVLLESFWAVGWLAAALISYFVIPSFGWQAAL-LLTALTAFYAL 179
Query: 193 FLVETVTPAPKKNQVS----GFCTKVVDVLHKRYKSMRNAAEILDNLIIAVQY----YLK 244
+L ++ +PK +S V V ++Y I+ ++ Y +L
Sbjct: 180 YLRTSLPDSPKYESLSAKKRSMWENVKSVWARQYIRPTVMLSIVWFCVVFSYYGMFLWLP 239
Query: 245 AVF---GFNKNQFSELLMMVGIGSI---FSQMVLLPILNPKVGEKVILSAALLASIAYAW 298
+V GF+ Q E ++++ + + FS L+ K G K IL L+ + A+
Sbjct: 240 SVMLLKGFSMIQSFEYVLLMTLAQLPGYFSAAWLI----EKAGRKWILVVYLIGTAGSAY 295
Query: 299 LYG--------LAWAPWVPYFS-ASFGIIY--------VLVKPSGKAQTFIAGAQSISDL 341
+G L + +F+ ++G++Y ++ +G T G I +
Sbjct: 296 FFGTADSLSLLLTAGVLLSFFNLGAWGVLYAYTPEQYPTAIRATGSGTTAAFG--RIGGI 353
Query: 342 LSPIAMSPLTSWFLSSNAPFDCKGFSIICASVSMVAETENEKLKENLQDE 391
P+ + L + +S + FSI C ++ + KE Q E
Sbjct: 354 FGPLLVGTLAARHISFSVI-----FSIFCIAILLAVACILIMGKETKQTE 398
>BMR1_BACSU (P33449) Multidrug resistance protein 1
(Multidrug-efflux transporter 1)
Length = 389
Score = 36.2 bits (82), Expect = 0.16
Identities = 49/253 (19%), Positives = 102/253 (39%), Gaps = 18/253 (7%)
Query: 57 GLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVL 116
G I +++V P+ G+ D GRK +++I + S + ++ E ++ +L
Sbjct: 44 GYMVACFAITQLIVSPIAGRWVDRFGRKIMIVIGLLFFSVSEFLFGIGKTVEMLFISRML 103
Query: 117 RTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPE---- 172
S + A++AD+ R +++ S ++G + FL E
Sbjct: 104 GGISAPFIMPGV----TAFIADITTIKTRPKALGYMSAAISTGFIIGPGIGGFLAEVHSR 159
Query: 173 -EYIFVVSIALLTFCPVYMQFFLVETVTPAPKKNQVSGFCTKVVDVLHKRYKSMRNAAEI 231
+ F + ALL + + P+ ++ G T + Y + +
Sbjct: 160 LPFFFAAAFALLA---AILSILTLREPERNPENQEIKGQKTGFKRIFAPMY-FIAFLIIL 215
Query: 232 LDNLIIAVQYYLKAVFGFNKNQF--SELLMMVGIGSI---FSQMVLLPILNPKVGEKVIL 286
+ + +A L A+F +K F S++ +M+ G+I +Q+VL GE ++
Sbjct: 216 ISSFGLASFESLFALFVDHKFGFTASDIAIMITGGAIVGAITQVVLFDRFTRWFGEIHLI 275
Query: 287 SAALLASIAYAWL 299
+L+ S + +L
Sbjct: 276 RYSLILSTSLVFL 288
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.324 0.137 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 45,963,265
Number of Sequences: 164201
Number of extensions: 1787107
Number of successful extensions: 7424
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 42
Number of HSP's that attempted gapping in prelim test: 7387
Number of HSP's gapped (non-prelim): 66
length of query: 423
length of database: 59,974,054
effective HSP length: 113
effective length of query: 310
effective length of database: 41,419,341
effective search space: 12839995710
effective search space used: 12839995710
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0095c.5


