

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
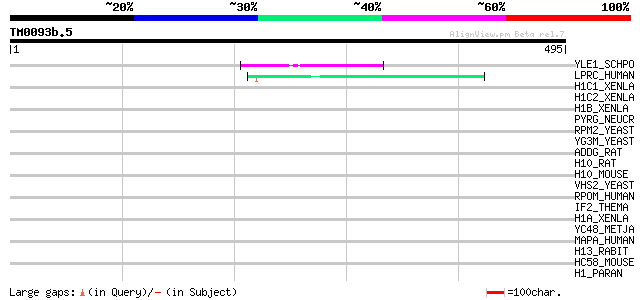
Query= TM0093b.5
(495 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YLE1_SCHPO (Q10451) Hypothetical protein C1093.01 in chromosome I 54 1e-06
LPRC_HUMAN (P42704) 130 kDa leucine-rich protein (LRP 130) (GP13... 45 3e-04
H1C1_XENLA (P15866) Histone H1C (Clone XLHW2) 42 0.003
H1C2_XENLA (P15867) Histone H1C (Clone XLHW8) 41 0.006
H1B_XENLA (P06893) Histone H1B 41 0.006
PYRG_NEUCR (Q7RZV2) CTP synthase (EC 6.3.4.2) (UTP--ammonia liga... 37 0.090
RPM2_YEAST (Q02773) Ribonuclease P protein component, mitochondr... 36 0.20
YG3M_YEAST (P48237) Hypothetical 101.4 kDa protein in RPL24B-RSR... 35 0.45
ADDG_RAT (Q62847) Gamma adducin (Adducin-like protein 70) (Prote... 34 0.99
H10_RAT (P43278) Histone H1.0 (H1(0)) (Histone H1') 33 1.3
H10_MOUSE (P10922) Histone H1' (H1.0) (H1(0)) 33 1.3
VHS2_YEAST (P40463) Protein VHS2 (Viable in a HAL3 SIT4 backgrou... 33 1.7
RPOM_HUMAN (O00411) DNA-directed RNA polymerase, mitochondrial p... 33 1.7
IF2_THEMA (Q9WZN3) Translation initiation factor IF-2 33 1.7
H1A_XENLA (P06892) Histone H1A 33 1.7
YC48_METJA (Q58645) Hypothetical protein MJ1248 32 2.9
MAPA_HUMAN (P78559) Microtubule-associated protein 1A (MAP 1A) (... 32 2.9
H13_RABIT (P02251) Histone H1.3 32 2.9
HC58_MOUSE (Q8BLG0) Hepatocellular carcinoma-associated antigen ... 32 3.8
H1_PARAN (P02256) Histone H1, gonadal 32 3.8
>YLE1_SCHPO (Q10451) Hypothetical protein C1093.01 in chromosome I
Length = 1261
Score = 53.5 bits (127), Expect = 1e-06
Identities = 37/127 (29%), Positives = 57/127 (44%), Gaps = 4/127 (3%)
Query: 207 DKITGALPCIYRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRH 266
+K + I +++ S R K + MK G IP +TF L + R
Sbjct: 846 EKANWFMALILDAMILSSSFARQFKSSNLFCDNMKMLGYIPRA---STFAH-LINNSTRR 901
Query: 267 NPSGLVPETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDW 326
+ LN+ E + H V P+ YN +LS LG+ RR E ++ + MK SG+ P
Sbjct: 902 GDTDDATTALNIFEETKRHNVKPSVFLYNAVLSKLGRARRTTECWKLFQEMKESGLLPTS 961
Query: 327 VSYYLVV 333
V+Y V+
Sbjct: 962 VTYGTVI 968
Score = 37.7 bits (86), Expect = 0.069
Identities = 48/226 (21%), Positives = 95/226 (41%), Gaps = 24/226 (10%)
Query: 123 QDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDE---ALGIFKNLDKYKCSTNE 179
+ + S ++ ++K G + A TF + N + G D+ AL IF+ ++ +
Sbjct: 867 RQFKSSNLFCDNMKMLGYIPRASTFAHLINNSTRRGDTDDATTALNIFEETKRHNVKPSV 926
Query: 180 FTVTAIINALCSKGHAKR-AEGVVLHHKDKITGALPCIYRSLLYGWSVHR-----NVKEA 233
F A+++ L G A+R E L + K +G LP S+ YG ++ + A
Sbjct: 927 FLYNAVLSKL---GRARRTTECWKLFQEMKESGLLP---TSVTYGTVINAACRIGDESLA 980
Query: 234 RRIIKEMKSN-GVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTSI 292
++ EM++ P + YNT ++ + + L + + + P+S
Sbjct: 981 EKLFAEMENQPNYQPRVAPYNTMIQFEVQTMFNRE------KALFYYNRLCATDIEPSSH 1034
Query: 293 SYNILLSCLGKTRRVKESC--QILEAMKTSGVSPDWVSYYLVVRVL 336
+Y +L+ G + V +LE M+ + V + Y + +L
Sbjct: 1035 TYKLLMDAYGTLKPVNVGSVKAVLELMERTDVPILSMHYAAYIHIL 1080
>LPRC_HUMAN (P42704) 130 kDa leucine-rich protein (LRP 130) (GP130)
(Leucine-rich PPR-motif containing protein)
Length = 1273
Score = 45.4 bits (106), Expect = 3e-04
Identities = 40/215 (18%), Positives = 83/215 (38%), Gaps = 11/215 (5%)
Query: 213 LPCIYR----SLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNP 268
+PC Y SLL + + A RI ++ G + D+ YN L+ + + +P
Sbjct: 1 MPCFYLRSCGSLLPELKLEERTEFAHRIWDTLQKLGAVYDVSHYNALLKVYLQNEYKFSP 60
Query: 269 SGLVPETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVS 328
+ + + +M + P ++Y L++ ++ + +IL MKT +
Sbjct: 61 T-------DFLAKMEEANIQPNRVTYQRLIASYCNVGDIEGASKILGFMKTKDLPVTEAV 113
Query: 329 YYLVVRVLFLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMK 388
+ +V +G + I+ M G+ P Y +L+ ++H + EK++
Sbjct: 114 FSALVTGHARAGDMENAENILTVMRDAGIEPGPDTYLALLNAYAEKGDIDHVKQTLEKVE 173
Query: 389 SSSLGGYGPVYDVLIPKLCRGGDFEKGRELWDEAT 423
L +I + G ++ W + T
Sbjct: 174 KFELHLMDRDLLQIIFSFSKAGYLSMSQKFWKKFT 208
Score = 33.5 bits (75), Expect = 1.3
Identities = 31/132 (23%), Positives = 53/132 (39%), Gaps = 2/132 (1%)
Query: 297 LLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFG-KGKEIVDQMIGK 355
LL L R + + +I + ++ G D VS+Y + ++L + + + +M
Sbjct: 12 LLPELKLEERTEFAHRIWDTLQKLGAVYD-VSHYNALLKVYLQNEYKFSPTDFLAKMEEA 70
Query: 356 GLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGDFEKG 415
+ PN Y LI C V + A ++ MK+ L V+ L+ R GD E
Sbjct: 71 NIQPNRVTYQRLIASYCNVGDIEGASKILGFMKTKDLPVTEAVFSALVTGHARAGDMENA 130
Query: 416 RELWDEATSMGI 427
+ GI
Sbjct: 131 ENILTVMRDAGI 142
>H1C1_XENLA (P15866) Histone H1C (Clone XLHW2)
Length = 216
Score = 42.4 bits (98), Expect = 0.003
Identities = 23/47 (48%), Positives = 27/47 (56%)
Query: 440 ITEVFKPVRPEKISLADSPIAKSPKKAKKLLGKVKMRKKPTAVKKKK 486
+T KP P+K + AKSPKKAKK + K KKP AVK KK
Sbjct: 136 VTAKKKPKSPKKPKKVSAAAAKSPKKAKKPVKAAKSPKKPKAVKSKK 182
>H1C2_XENLA (P15867) Histone H1C (Clone XLHW8)
Length = 220
Score = 41.2 bits (95), Expect = 0.006
Identities = 22/42 (52%), Positives = 25/42 (59%)
Query: 445 KPVRPEKISLADSPIAKSPKKAKKLLGKVKMRKKPTAVKKKK 486
KP P+K + AKSPKKAKK + K KKP AVK KK
Sbjct: 140 KPKSPKKPKKVSAAAAKSPKKAKKPVKAAKSPKKPKAVKPKK 181
Score = 30.8 bits (68), Expect = 8.4
Identities = 16/46 (34%), Positives = 25/46 (53%)
Query: 445 KPVRPEKISLADSPIAKSPKKAKKLLGKVKMRKKPTAVKKKKKRQK 490
K V+P+K++ + + A PK AK + K K+ K A KK +K
Sbjct: 175 KAVKPKKVTKSPAKKATKPKAAKAKIAKPKIAKAKAAKGKKAAAKK 220
>H1B_XENLA (P06893) Histone H1B
Length = 219
Score = 41.2 bits (95), Expect = 0.006
Identities = 23/41 (56%), Positives = 26/41 (63%), Gaps = 4/41 (9%)
Query: 446 PVRPEKISLADSPIAKSPKKAKKLLGKVKMRKKPTAVKKKK 486
P +P+K+S A AKSPKK KKL K KKP AVK KK
Sbjct: 145 PKKPKKVSAA----AKSPKKVKKLAKAAKSPKKPKAVKAKK 181
>PYRG_NEUCR (Q7RZV2) CTP synthase (EC 6.3.4.2) (UTP--ammonia ligase)
(CTP synthetase)
Length = 568
Score = 37.4 bits (85), Expect = 0.090
Identities = 35/149 (23%), Positives = 63/149 (41%), Gaps = 19/149 (12%)
Query: 228 RNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNV-MMEMRSHK 286
+ K + +K ++S G+IPDL+ C CE+ L V + V + ++ + +
Sbjct: 179 QKTKPTQHAVKSVRSAGLIPDLIA------CRCEKPLEQGTINKVASSCQVEVNQVLAVR 232
Query: 287 VLPTSISYNILLSCLGKTRRVKESCQI----LEAMKTSGVSPDWVSYYLVVRVLFLSGRF 342
+PT +LL G R +KE+ ++ L + S W + +V + +
Sbjct: 233 DMPTIYQVPLLLEEQGLLRELKETLKLDDVKLSPARVSQGQEVWAKWQKIVPLGY----- 287
Query: 343 GKGKEIVDQMIGKGLVPNHKFYFSLIGIL 371
E VD ++ V H Y S+I L
Sbjct: 288 ---AETVDIVLVGKYVELHDAYLSVIKAL 313
>RPM2_YEAST (Q02773) Ribonuclease P protein component, mitochondrial
precursor (EC 3.1.26.5) (RNase P)
Length = 1202
Score = 36.2 bits (82), Expect = 0.20
Identities = 21/77 (27%), Positives = 38/77 (49%), Gaps = 1/77 (1%)
Query: 228 RNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNP-SGLVPETLNVMMEMRSHK 286
+N + + + +K N ++P L + L LC+R L +N + E L +M +++
Sbjct: 201 KNYNKINSLYQSLKRNDIVPPLEIFTKVLDSLCKRPLDNNDLDNKMYELLTCYQDMINNR 260
Query: 287 VLPTSISYNILLSCLGK 303
+ P YNI+L L K
Sbjct: 261 LKPPDEIYNIVLLSLFK 277
>YG3M_YEAST (P48237) Hypothetical 101.4 kDa protein in RPL24B-RSR1
intergenic region
Length = 864
Score = 35.0 bits (79), Expect = 0.45
Identities = 24/77 (31%), Positives = 37/77 (47%), Gaps = 11/77 (14%)
Query: 231 KEARRIIKEMK--SNGVIPDLVCYNTFLR-CLCERNLRHNPSGLVPETLNVMMEMRSHKV 287
K+A MK S PD+ YNT LR C ERN P+ L++ E++ H +
Sbjct: 337 KQAWNTFDTMKFLSTKHFPDICTYNTMLRICEKERNF--------PKALDLFQEIQDHNI 388
Query: 288 LPTSISYNILLSCLGKT 304
PT+ +Y ++ L +
Sbjct: 389 KPTTNTYIMMARVLASS 405
>ADDG_RAT (Q62847) Gamma adducin (Adducin-like protein 70) (Protein
kinase C binding protein 35H)
Length = 705
Score = 33.9 bits (76), Expect = 0.99
Identities = 34/116 (29%), Positives = 51/116 (43%), Gaps = 19/116 (16%)
Query: 375 ERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGDFEKGRELWDEATSMGITLQCSKD 434
ER+ ELF K +S V +P + G D + ++ DE L S
Sbjct: 606 ERLEENHELFSKSFTS----------VDVPVIVNGKD--EMHDVEDELAQRVSRLTTSTT 653
Query: 435 ILDPSITEVFKPVRPEKISLADSPIAKSPKKAKKLLGKVKMRKKPTAVKKKKKRQK 490
I + IT + P R E++ D +KSP K KK + P+ +KK KK++K
Sbjct: 654 IENIEIT-IKSPDRTEEVLSPDGSPSKSPSKKKKKF------RTPSFLKKNKKKEK 702
>H10_RAT (P43278) Histone H1.0 (H1(0)) (Histone H1')
Length = 193
Score = 33.5 bits (75), Expect = 1.3
Identities = 18/49 (36%), Positives = 24/49 (48%)
Query: 442 EVFKPVRPEKISLADSPIAKSPKKAKKLLGKVKMRKKPTAVKKKKKRQK 490
EV K P+K + +K+P K K K +KKP A KK K+ K
Sbjct: 112 EVKKVATPKKAAKPKKAASKAPSKKPKATPVKKAKKKPAATPKKAKKPK 160
>H10_MOUSE (P10922) Histone H1' (H1.0) (H1(0))
Length = 193
Score = 33.5 bits (75), Expect = 1.3
Identities = 18/49 (36%), Positives = 24/49 (48%)
Query: 442 EVFKPVRPEKISLADSPIAKSPKKAKKLLGKVKMRKKPTAVKKKKKRQK 490
EV K P+K + +K+P K K K +KKP A KK K+ K
Sbjct: 112 EVKKVATPKKAAKPKKAASKAPSKKPKATPVKKAKKKPAATPKKAKKPK 160
>VHS2_YEAST (P40463) Protein VHS2 (Viable in a HAL3 SIT4 background
protein 2)
Length = 436
Score = 33.1 bits (74), Expect = 1.7
Identities = 22/75 (29%), Positives = 37/75 (49%), Gaps = 15/75 (20%)
Query: 40 PPHLQELCGIVTSNVGGLDDLELSLNKFKGSL--------TSSLVAQAIESSKHEAQTR- 90
PP L C IVT + GLDD+++ ++ ++ T SL +Q+ ++ A R
Sbjct: 130 PPALDAGCSIVTDDTTGLDDVDMVYSRRPSTIGLDRALGRTRSLSSQSFDNETSPAHPRS 189
Query: 91 ------RLLRFFLWS 99
RLLRF+ ++
Sbjct: 190 PNDHGSRLLRFYSYA 204
>RPOM_HUMAN (O00411) DNA-directed RNA polymerase, mitochondrial
precursor (EC 2.7.7.6) (MtRPOL)
Length = 1230
Score = 33.1 bits (74), Expect = 1.7
Identities = 17/61 (27%), Positives = 32/61 (51%), Gaps = 2/61 (3%)
Query: 201 VVLHHKDKITGALPCI--YRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRC 258
+V+HH + L + Y +++ GW+ KE ++ +K G+ PDL+ Y L+C
Sbjct: 246 LVVHHGQRQKRKLLTLDMYNAVMLGWARQGAFKELVYVLFMVKDAGLTPDLLSYAAALQC 305
Query: 259 L 259
+
Sbjct: 306 M 306
>IF2_THEMA (Q9WZN3) Translation initiation factor IF-2
Length = 690
Score = 33.1 bits (74), Expect = 1.7
Identities = 20/72 (27%), Positives = 34/72 (46%), Gaps = 5/72 (6%)
Query: 416 RELWDEATSMGITLQCSKDILDPSITEVFKPVRPEKISLADSPIAKSPKKAKKLLGKVKM 475
+EL E +G+ ++ +D + + + E D+ AK P K KK G+ ++
Sbjct: 18 KELLQELEELGVNVKSHMSYVDEEMANIIIDLLEE-----DNRKAKQPSKPKKEKGEEEV 72
Query: 476 RKKPTAVKKKKK 487
K+ KKKKK
Sbjct: 73 EKEVVEKKKKKK 84
>H1A_XENLA (P06892) Histone H1A
Length = 209
Score = 33.1 bits (74), Expect = 1.7
Identities = 20/40 (50%), Positives = 23/40 (57%), Gaps = 4/40 (10%)
Query: 446 PVRPEKISLADSPIAKSPKKAKKLLGKVKMRKKPTAVKKK 485
P +P+K+S A AKSPKK KK K K TAVK K
Sbjct: 147 PKKPKKVSAA----AKSPKKLKKPAKAAKSPAKKTAVKPK 182
>YC48_METJA (Q58645) Hypothetical protein MJ1248
Length = 154
Score = 32.3 bits (72), Expect = 2.9
Identities = 41/146 (28%), Positives = 61/146 (41%), Gaps = 20/146 (13%)
Query: 43 LQELCGIVTSNVGG-LDDLELSLNKFKGSLTSSLVAQAIESSKHEAQTRRLLRFFLWSSK 101
L EL GI ++ L LE SL K GS TS I+S L +
Sbjct: 9 LSELSGIDKKSLRRILIILEFSLRKKDGSPTSFAEKFNIKSFGD------LYNYIRDVKS 62
Query: 102 NLRHDLEDNDYNYALRVF---AEKQDYTSMDILIGDLKKEG----RVMDAQTFGLVAENL 154
NL+ D E +N ++ A + Y MD + ++ V +TFG++ +NL
Sbjct: 63 NLKRDHEIEGFNGLTEMWKSVAPRAQYWIMDTFGEENPRDALFSASVFTMRTFGIMLDNL 122
Query: 155 IKLGKEDEALGIFKNLDKYKCSTNEF 180
+ L K I K LD+Y+ E+
Sbjct: 123 LLLKK------IIKTLDEYQKEVTEY 142
>MAPA_HUMAN (P78559) Microtubule-associated protein 1A (MAP 1A)
(Proliferation-related protein p80) [Contains: MAP1
light chain LC2]
Length = 2805
Score = 32.3 bits (72), Expect = 2.9
Identities = 15/49 (30%), Positives = 30/49 (60%), Gaps = 2/49 (4%)
Query: 445 KPVRPEKISLADSPIAKSPKKA--KKLLGKVKMRKKPTAVKKKKKRQKK 491
KP +PE++ S K+ K+ K +GK +++K + +++KK ++KK
Sbjct: 373 KPAKPERVKTESSEALKAEKRKLIKDKVGKKHLKEKISKLEEKKDKEKK 421
>H13_RABIT (P02251) Histone H1.3
Length = 213
Score = 32.3 bits (72), Expect = 2.9
Identities = 23/52 (44%), Positives = 26/52 (49%), Gaps = 9/52 (17%)
Query: 442 EVFKPVRPEKISLADSPIAKSPKKAKKLLGKVKMRK--KPTAVKKKKKRQKK 491
+V KP P K+ AKSPKKAK + K K KP A K KK KK
Sbjct: 167 KVAKPKSPAKV-------AKSPKKAKAVKPKAAKPKAPKPKAAKAKKTAAKK 211
>HC58_MOUSE (Q8BLG0) Hepatocellular carcinoma-associated antigen 58
homolog
Length = 1010
Score = 32.0 bits (71), Expect = 3.8
Identities = 17/36 (47%), Positives = 21/36 (58%)
Query: 455 ADSPIAKSPKKAKKLLGKVKMRKKPTAVKKKKKRQK 490
A SP AK +K K+ K +R KP KKKKK+ K
Sbjct: 520 ASSPTAKEKEKTKEKKFKELVRVKPKKKKKKKKKTK 555
>H1_PARAN (P02256) Histone H1, gonadal
Length = 248
Score = 32.0 bits (71), Expect = 3.8
Identities = 22/64 (34%), Positives = 32/64 (49%), Gaps = 5/64 (7%)
Query: 430 QCSKDILDPSITEVFKPVRPEKISLADSPIAKSPKKAKKLLGKVKMRKKPTAVKKKKKRQ 489
+ +K P+ K +P K S +SPKKAKK GK +KP A KK ++
Sbjct: 180 KAAKKAKKPAKKSPKKAKKPAKKSPKKKKAKRSPKKAKKAAGK----RKP-AAKKARRSP 234
Query: 490 KKSG 493
+K+G
Sbjct: 235 RKAG 238
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 58,481,153
Number of Sequences: 164201
Number of extensions: 2512295
Number of successful extensions: 7325
Number of sequences better than 10.0: 33
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 7224
Number of HSP's gapped (non-prelim): 104
length of query: 495
length of database: 59,974,054
effective HSP length: 114
effective length of query: 381
effective length of database: 41,255,140
effective search space: 15718208340
effective search space used: 15718208340
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0093b.5


