

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0086.14
(430 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
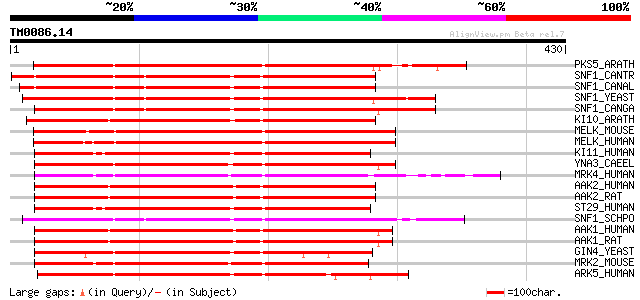
Score E
Sequences producing significant alignments: (bits) Value
PKS5_ARATH (O22932) SOS2-like protein kinase PKS5 (EC 2.7.1.37) 326 6e-89
SNF1_CANTR (O94168) Carbon catabolite derepressing protein kinas... 231 4e-60
SNF1_CANAL (P52497) Carbon catabolite derepressing protein kinas... 226 9e-59
SNF1_YEAST (P06782) Carbon catabolite derepressing protein kinas... 225 1e-58
SNF1_CANGA (Q00372) Carbon catabolite derepressing protein kinas... 225 1e-58
KI10_ARATH (Q38997) SNF1-related protein kinase KIN10 (EC 2.7.1.... 223 6e-58
MELK_MOUSE (Q61846) Maternal embryonic leucine zipper kinase (EC... 217 4e-56
MELK_HUMAN (Q14680) Maternal embryonic leucine zipper kinase (EC... 216 7e-56
KI11_HUMAN (Q8TDC3) Probable serine/threonine-protein kinase KIA... 213 1e-54
YNA3_CAEEL (P45894) Putative serine/threonine-protein kinase PAR... 211 2e-54
MRK4_HUMAN (Q96L34) MAP/microtubule affinity-regulating kinase 4... 208 2e-53
AAK2_HUMAN (P54646) 5'-AMP-activated protein kinase, catalytic a... 206 9e-53
AAK2_RAT (Q09137) 5'-AMP-activated protein kinase, catalytic alp... 206 1e-52
ST29_HUMAN (Q8IWQ3) Serine/threonine-protein kinase 29 (EC 2.7.1... 204 5e-52
SNF1_SCHPO (O74536) SNF1-like protein kinase ssp2 (EC 2.7.1.-) 202 1e-51
AAK1_HUMAN (Q13131) 5'-AMP-activated protein kinase, catalytic a... 201 4e-51
AAK1_RAT (P54645) 5'-AMP-activated protein kinase, catalytic alp... 199 9e-51
GIN4_YEAST (Q12263) Serine/threonine-protein kinase GIN4 (EC 2.7... 197 3e-50
MRK2_MOUSE (Q05512) MAP/microtubule affinity-regulating kinase 2... 196 1e-49
ARK5_HUMAN (O60285) AMPK-related protein kinase 5 (EC 2.7.1.37) 196 1e-49
>PKS5_ARATH (O22932) SOS2-like protein kinase PKS5 (EC 2.7.1.37)
Length = 435
Score = 326 bits (836), Expect = 6e-89
Identities = 179/347 (51%), Positives = 240/347 (68%), Gaps = 22/347 (6%)
Query: 19 ILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTV-DAAMEPRIVREIDAMRR 77
+ GKY++ +LLG G+FAKV+ AR G VAVKI++K K + + A+ I REI MRR
Sbjct: 17 LFGKYELGKLLGCGAFAKVFHARDRRTGQSVAVKILNKKKLLTNPALANNIKREISIMRR 76
Query: 78 LHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRF 137
L H PNI+++HEVMATK+KI +EF GGELF+ IS+ G+L E+ +RRYFQQL+SA+ +
Sbjct: 77 LSH-PNIVKLHEVMATKSKIFFAMEFVKGGELFNKISKHGRLSEDLSRRYFQQLISAVGY 135
Query: 138 CHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQ-NGLLHTACGTPAYTAPEILR 196
CH GV HRDLKP+NLL+D GNLKVSDFGLSAL + ++ +GLLHT CGTPAY APEIL
Sbjct: 136 CHARGVYHRDLKPENLLIDENGNLKVSDFGLSALTDQIRPDGLLHTLCGTPAYVAPEILS 195
Query: 197 LSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVI 256
GY+G+K D WSCG+VLFVL+AGYLPF+D N+ MY+KI + +Y+FP W++ + +
Sbjct: 196 -KKGYEGAKVDVWSCGIVLFVLVAGYLPFNDPNVMNMYKKIYKGEYRFPRWMSPDLKRFV 254
Query: 257 RRLLDPNPETRMSVEDLYGNSWFK----KSLKV---EAESETESSTLNLDSGYGNRVRGL 309
RLLD NPETR++++++ + WF K +K E E + S+L V+ L
Sbjct: 255 SRLLDINPETRITIDEILKDPWFVRGGFKQIKFHDDEIEDQKVESSL-------EAVKSL 307
Query: 310 GVNAFDLISMSSGLDLSGLFQ--DEGKRKEKRFTSGAKLEVVEEKVK 354
NAFDLIS SSGLDLSGLF + +RF S E++ E+V+
Sbjct: 308 --NAFDLISYSSGLDLSGLFAGCSNSSGESERFLSEKSPEMLAEEVE 352
>SNF1_CANTR (O94168) Carbon catabolite derepressing protein kinase
(EC 2.7.1.-)
Length = 619
Score = 231 bits (588), Expect = 4e-60
Identities = 120/283 (42%), Positives = 188/283 (66%), Gaps = 8/283 (2%)
Query: 2 EPPMRQPPPPRSSAATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVD 61
+P P P + A I G+YQ+I+ LG GSF KV A+ + G VA+KII++
Sbjct: 32 QPAQPIPIDPNVNPANRI-GRYQIIKTLGEGSFGKVKLAQHVGTGQKVALKIINRKTLAK 90
Query: 62 AAMEPRIVREIDAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPE 121
+ M+ R+ REI +R L H P+I+++++V+ +K +I +V+EFA G ELF I +RGK+PE
Sbjct: 91 SDMQGRVEREISYLRLLRH-PHIIKLYDVIKSKDEIIMVIEFA-GKELFDYIVQRGKMPE 148
Query: 122 NTARRYFQQLVSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNGLLH 181
+ ARR+FQQ+++A+ +CHR+ + HRDLKP+NLLLD N+K++DFGLS + L
Sbjct: 149 DEARRFFQQIIAAVEYCHRHKIVHRDLKPENLLLDDQLNVKIADFGLSNI--MTDGNFLK 206
Query: 182 TACGTPAYTAPEILRLSGG-YDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRR 240
T+CG+P Y APE+ +SG Y G + D WS G++L+V+L G LPFDD I A+++KIS
Sbjct: 207 TSCGSPNYAAPEV--ISGKLYAGPEVDVWSSGVILYVMLCGRLPFDDEFIPALFKKISNG 264
Query: 241 DYQFPEWITKPARFVIRRLLDPNPETRMSVEDLYGNSWFKKSL 283
Y P +++ A+ ++ R+L NP R+++ ++ + WFK+ +
Sbjct: 265 VYTLPNYLSPGAKHLLTRMLVVNPLNRITIHEIMEDEWFKQDM 307
>SNF1_CANAL (P52497) Carbon catabolite derepressing protein kinase
(EC 2.7.1.-)
Length = 620
Score = 226 bits (576), Expect = 9e-59
Identities = 119/278 (42%), Positives = 187/278 (66%), Gaps = 9/278 (3%)
Query: 8 PPPPRSSAATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPR 67
P P ++ A I G+YQ+++ LG GSF KV A+ L G VA+KII++ + M+ R
Sbjct: 39 PIDPAANPANRI-GRYQILKTLGEGSFGKVKLAQHLGTGQKVALKIINRKTLAKSDMQGR 97
Query: 68 IVREIDAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRY 127
+ REI +R L H P+I+++++V+ +K +I +V+EFA G ELF I +RGK+PE+ ARR+
Sbjct: 98 VEREISYLRLLRH-PHIIKLYDVIKSKDEIIMVIEFA-GKELFDYIVQRGKMPEDEARRF 155
Query: 128 FQQLVSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNGLLHTACGTP 187
FQQ+++A+ +CHR+ + HRDLKP+NLLLD N+K++DFGLS + L T+CG+P
Sbjct: 156 FQQIIAAVEYCHRHKIVHRDLKPENLLLDDQLNVKIADFGLSNI--MTDGNFLKTSCGSP 213
Query: 188 AY-TAPEILRLSGG-YDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFP 245
Y APE+ +SG Y G + D WS G++L+V+L G LPFDD I A+++KIS Y P
Sbjct: 214 NYMPAPEV--ISGKLYAGPEVDVWSAGVILYVMLCGRLPFDDEFIPALFKKISNGVYTLP 271
Query: 246 EWITKPARFVIRRLLDPNPETRMSVEDLYGNSWFKKSL 283
+++ A+ ++ R+L NP R+++ ++ + WFK+ +
Sbjct: 272 NYLSAGAKHLLTRMLVVNPLNRITIHEIMEDDWFKQDM 309
>SNF1_YEAST (P06782) Carbon catabolite derepressing protein kinase
(EC 2.7.1.-)
Length = 633
Score = 225 bits (574), Expect = 1e-58
Identities = 127/331 (38%), Positives = 200/331 (60%), Gaps = 18/331 (5%)
Query: 11 PRSSAATTI-LGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIV 69
P+SS A +G YQ+++ LG GSF KV A G VA+KII+K + M+ RI
Sbjct: 42 PKSSLADGAHIGNYQIVKTLGEGSFGKVKLAYHTTTGQKVALKIINKKVLAKSDMQGRIE 101
Query: 70 REIDAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQ 129
REI +R L H P+I+++++V+ +K +I +V+E+A G ELF I +R K+ E ARR+FQ
Sbjct: 102 REISYLRLLRH-PHIIKLYDVIKSKDEIIMVIEYA-GNELFDYIVQRDKMSEQEARRFFQ 159
Query: 130 QLVSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNGLLHTACGTPAY 189
Q++SA+ +CHR+ + HRDLKP+NLLLD N+K++DFGLS + L T+CG+P Y
Sbjct: 160 QIISAVEYCHRHKIVHRDLKPENLLLDEHLNVKIADFGLSNI--MTDGNFLKTSCGSPNY 217
Query: 190 TAPEILRLSGG-YDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWI 248
APE+ +SG Y G + D WSCG++L+V+L LPFDD +I +++ IS Y P+++
Sbjct: 218 AAPEV--ISGKLYAGPEVDVWSCGVILYVMLCRRLPFDDESIPVLFKNISNGVYTLPKFL 275
Query: 249 TKPARFVIRRLLDPNPETRMSVEDLYGNSWFK---------KSLKVEAESETESSTLNLD 299
+ A +I+R+L NP R+S+ ++ + WFK LK E E E++ D
Sbjct: 276 SPGAAGLIKRMLIVNPLNRISIHEIMQDDWFKVDLPEYLLPPDLKPHPEEENENNDSKKD 335
Query: 300 SGYGNRVRGLGVNAFDLISMSSGLDLSGLFQ 330
+ + N +++S + G + +++
Sbjct: 336 GSSPDNDE-IDDNLVNILSSTMGYEKDEIYE 365
>SNF1_CANGA (Q00372) Carbon catabolite derepressing protein kinase
(EC 2.7.1.-)
Length = 612
Score = 225 bits (574), Expect = 1e-58
Identities = 122/314 (38%), Positives = 192/314 (60%), Gaps = 9/314 (2%)
Query: 20 LGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRLH 79
+G YQ+++ LG GSF KV A + G VA+KII+K + M+ RI REI +R L
Sbjct: 36 VGNYQIVKTLGEGSFGKVKLAYHVTTGQKVALKIINKKVLAKSDMQGRIEREISYLRLLR 95
Query: 80 HHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFCH 139
H P+I+++++V+ +K +I +V+E+A G ELF I +R K+ E ARR+FQQ++SA+ +CH
Sbjct: 96 H-PHIIKLYDVIKSKDEIIMVIEYA-GNELFDYIVQRNKMSEQEARRFFQQIISAVEYCH 153
Query: 140 RNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNGLLHTACGTPAYTAPEILRLSG 199
R+ + HRDLKP+NLLLD N+K++DFGLS + L T+CG+P Y APE+ +SG
Sbjct: 154 RHKIVHRDLKPENLLLDEHLNVKIADFGLSNI--MTDGNFLKTSCGSPNYAAPEV--ISG 209
Query: 200 G-YDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVIRR 258
Y G + D WSCG++L+V+L LPFDD +I +++ IS Y P++++ A +I+R
Sbjct: 210 KLYAGPEVDVWSCGVILYVMLCRRLPFDDESIPVLFKNISNGVYTLPKFLSPGASDLIKR 269
Query: 259 LLDPNPETRMSVEDLYGNSWFKKSLK--VEAESETESSTLNLDSGYGNRVRGLGVNAFDL 316
+L NP R+S+ ++ + WFK L + + + N SG V +
Sbjct: 270 MLIVNPLNRISIHEIMQDEWFKVDLAEYLVPQDLKQQEQFNKKSGNEENVEEIDDEMVVT 329
Query: 317 ISMSSGLDLSGLFQ 330
+S + G D +++
Sbjct: 330 LSKTMGYDKDEIYE 343
>KI10_ARATH (Q38997) SNF1-related protein kinase KIN10 (EC 2.7.1.-)
(AKIN10)
Length = 535
Score = 223 bits (569), Expect = 6e-58
Identities = 118/272 (43%), Positives = 179/272 (65%), Gaps = 8/272 (2%)
Query: 14 SAATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREID 73
S +IL Y++ R LG GSF +V A + G VA+KI+++ K + ME ++ REI
Sbjct: 33 SGVESILPNYKLGRTLGIGSFGRVKIAEHALTGHKVAIKILNRRKIKNMEMEEKVRREIK 92
Query: 74 AMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVS 133
+R L HP+I+R++EV+ T T I+LV+E+ GELF I +G+L E+ AR +FQQ++S
Sbjct: 93 ILR-LFMHPHIIRLYEVIETPTDIYLVMEYVNSGELFDYIVEKGRLQEDEARNFFQQIIS 151
Query: 134 ALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNG-LLHTACGTPAYTAP 192
+ +CHRN V HRDLKP+NLLLD+ N+K++DFGLS + +++G L T+CG+P Y AP
Sbjct: 152 GVEYCHRNMVVHRDLKPENLLLDSKCNVKIADFGLSNI---MRDGHFLKTSCGSPNYAAP 208
Query: 193 EILRLSGG-YDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKP 251
E+ +SG Y G + D WSCG++L+ LL G LPFDD NI +++KI Y P ++
Sbjct: 209 EV--ISGKLYAGPEVDVWSCGVILYALLCGTLPFDDENIPNLFKKIKGGIYTLPSHLSPG 266
Query: 252 ARFVIRRLLDPNPETRMSVEDLYGNSWFKKSL 283
AR +I R+L +P R+++ ++ + WF+ L
Sbjct: 267 ARDLIPRMLVVDPMKRVTIPEIRQHPWFQAHL 298
>MELK_MOUSE (Q61846) Maternal embryonic leucine zipper kinase (EC
2.7.1.37) (Protein kinase PK38) (mPK38)
Length = 643
Score = 217 bits (553), Expect = 4e-56
Identities = 110/281 (39%), Positives = 179/281 (63%), Gaps = 4/281 (1%)
Query: 19 ILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRL 78
+L Y++ +G G FAKV A ++ G +VA+KI+DK+ + PR+ EIDA++ L
Sbjct: 7 LLKYYELYETIGTGGFAKVKLACHVLTGEMVAIKIMDKNAL--GSDLPRVKTEIDALKSL 64
Query: 79 HHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFC 138
H +I +++ V+ TK KI +V+E+ GGELF I + +L E R F+Q++SA+ +
Sbjct: 65 RHQ-HICQLYHVLETKNKIFMVLEYCPGGELFDYIISQDRLSEEETRVVFRQILSAVAYV 123
Query: 139 HRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNGLLHTACGTPAYTAPEILRLS 198
H G AHRDLKP+NLL D LK+ DFGL A P+ ++ L T CG+ AY APE+++
Sbjct: 124 HSQGYAHRDLKPENLLFDENHKLKLIDFGLCAKPKGNKDYHLQTCCGSLAYAAPELIQ-G 182
Query: 199 GGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVIRR 258
Y GS+AD WS G++L+VL+ G+LPFDD N+ A+Y+KI R Y+ P+W++ + ++++
Sbjct: 183 KSYLGSEADVWSMGILLYVLMCGFLPFDDDNVMALYKKIMRGKYEVPKWLSPSSILLLQQ 242
Query: 259 LLDPNPETRMSVEDLYGNSWFKKSLKVEAESETESSTLNLD 299
+L +P+ R+S+ +L + W + E ++++ +LD
Sbjct: 243 MLQVDPKKRISMRNLLNHPWVMQDYSCPVEWQSKTPLTHLD 283
>MELK_HUMAN (Q14680) Maternal embryonic leucine zipper kinase (EC
2.7.1.37) (hMELK) (Protein kinase PK38) (hPK38)
Length = 651
Score = 216 bits (551), Expect = 7e-56
Identities = 111/281 (39%), Positives = 182/281 (64%), Gaps = 4/281 (1%)
Query: 19 ILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRL 78
+L Y++ +G G FAKV A ++ G +VA+KI+DK+ T+ + + PRI EI+A++ L
Sbjct: 7 LLKYYELHETIGTGGFAKVKLACHILTGEMVAIKIMDKN-TLGSDL-PRIKTEIEALKNL 64
Query: 79 HHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFC 138
H +I +++ V+ T KI +V+E+ GGELF I + +L E R F+Q+VSA+ +
Sbjct: 65 RHQ-HICQLYHVLETANKIFMVLEYCPGGELFDYIISQDRLSEEETRVVFRQIVSAVAYV 123
Query: 139 HRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNGLLHTACGTPAYTAPEILRLS 198
H G AHRDLKP+NLL D LK+ DFGL A P+ ++ L T CG+ AY APE+++
Sbjct: 124 HSQGYAHRDLKPENLLFDEYHKLKLIDFGLCAKPKGNKDYHLQTCCGSLAYAAPELIQ-G 182
Query: 199 GGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVIRR 258
Y GS+AD WS G++L+VL+ G+LPFDD N+ A+Y+KI R Y P+W++ + ++++
Sbjct: 183 KSYLGSEADVWSMGILLYVLMCGFLPFDDDNVMALYKKIMRGKYDVPKWLSPSSILLLQQ 242
Query: 259 LLDPNPETRMSVEDLYGNSWFKKSLKVEAESETESSTLNLD 299
+L +P+ R+S+++L + W + E ++++ ++LD
Sbjct: 243 MLQVDPKKRISMKNLLNHPWIMQDYNYPVEWQSKNPFIHLD 283
>KI11_HUMAN (Q8TDC3) Probable serine/threonine-protein kinase
KIAA1811 (EC 2.7.1.37)
Length = 794
Score = 213 bits (541), Expect = 1e-54
Identities = 101/260 (38%), Positives = 169/260 (64%), Gaps = 5/260 (1%)
Query: 20 LGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRLH 79
+G Y++ + LG+G V I G VA+KI+++ K ++ + ++ REI A+ +L
Sbjct: 47 VGPYRLEKTLGKGQTGLVKLGVHCITGQKVAIKIVNREKLSESVLM-KVEREI-AILKLI 104
Query: 80 HHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFCH 139
HP++L++H+V K ++LV+E +GGELF + ++G+L AR++F+Q+VSAL FCH
Sbjct: 105 EHPHVLKLHDVYENKKYLYLVLEHVSGGELFDYLVKKGRLTPKEARKFFRQIVSALDFCH 164
Query: 140 RNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNGLLHTACGTPAYTAPEILRLSG 199
+ HRDLKP+NLLLD N++++DFG+++L + + LL T+CG+P Y PE+++
Sbjct: 165 SYSICHRDLKPENLLLDEKNNIRIADFGMASL--QVGDSLLETSCGSPHYACPEVIK-GE 221
Query: 200 GYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVIRRL 259
YDG +AD WSCG++LF LL G LPFDD N+ + K+ R + P +I + ++R +
Sbjct: 222 KYDGRRADMWSCGVILFALLVGALPFDDDNLRQLLEKVKRGVFHMPHFIPPDCQSLLRGM 281
Query: 260 LDPNPETRMSVEDLYGNSWF 279
++ PE R+S+E + + W+
Sbjct: 282 IEVEPEKRLSLEQIQKHPWY 301
>YNA3_CAEEL (P45894) Putative serine/threonine-protein kinase PAR2.3
(EC 2.7.1.37)
Length = 622
Score = 211 bits (538), Expect = 2e-54
Identities = 112/283 (39%), Positives = 176/283 (61%), Gaps = 8/283 (2%)
Query: 20 LGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRLH 79
+G + + +G+G+F V + + G VA+KI+++ + + EID +++L
Sbjct: 21 IGNFVIKETIGKGAFGAVKRGTHIQTGYDVAIKILNRGRMKGLGTVNKTRNEIDNLQKLT 80
Query: 80 HHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFCH 139
H P+I R+ V++T + I LV+E +GGELFS I+R+G LP +RRYFQQ++S + +CH
Sbjct: 81 H-PHITRLFRVISTPSDIFLVMELVSGGELFSYITRKGALPIRESRRYFQQIISGVSYCH 139
Query: 140 RNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNG-LLHTACGTPAYTAPEILRLS 198
+ + HRDLKP+NLLLDA N+K++DFGLS ++ +G LL TACG+P Y APE++ +
Sbjct: 140 NHMIVHRDLKPENLLLDANKNIKIADFGLS---NYMTDGDLLSTACGSPNYAAPELIS-N 195
Query: 199 GGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVIRR 258
Y G + D WSCG++L+ +L G LPFDD N+ ++ KI Y P + K A +I
Sbjct: 196 KLYVGPEVDLWSCGVILYAMLCGTLPFDDQNVPTLFAKIKSGRYTVPYSMEKQAADLIST 255
Query: 259 LLDPNPETRMSVEDLYGNSWFKKSLK--VEAESETESSTLNLD 299
+L +P R V+ + +SWF+ L + E E ESS +++D
Sbjct: 256 MLQVDPVKRADVKRIVNHSWFRIDLPYYLFPECENESSIVDID 298
>MRK4_HUMAN (Q96L34) MAP/microtubule affinity-regulating kinase 4
(EC 2.7.1.37) (MAP/microtubule affinity-regulating
kinase like 1)
Length = 752
Score = 208 bits (529), Expect = 2e-53
Identities = 124/361 (34%), Positives = 205/361 (56%), Gaps = 29/361 (8%)
Query: 20 LGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRLH 79
+G Y+++R +G+G+FAKV AR ++ G VA+KIIDK++ ++++ ++ RE+ M+ L+
Sbjct: 56 VGNYRLLRTIGKGNFAKVKLARHILTGREVAIKIIDKTQLNPSSLQ-KLFREVRIMKGLN 114
Query: 80 HHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFCH 139
H PNI+++ EV+ T+ ++LV+E+A+ GE+F + G++ E AR F+Q+VSA+ +CH
Sbjct: 115 H-PNIVKLFEVIETEKTLYLVMEYASAGEVFDYLVSHGRMKEKEARAKFRQIVSAVHYCH 173
Query: 140 RNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNGLLHTACGTPAYTAPEILRLSG 199
+ + HRDLK +NLLLDA N+K++DFG S E L T CG+P Y APE+ +
Sbjct: 174 QKNIVHRDLKAENLLLDAEANIKIADFGFS--NEFTLGSKLDTFCGSPPYAAPELFQ-GK 230
Query: 200 GYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVIRRL 259
YDG + D WS G++L+ L++G LPFD N+ + ++ R Y+ P +++ ++RR
Sbjct: 231 KYDGPEVDIWSLGVILYTLVSGSLPFDGHNLKELRERVLRGKYRVPFYMSTDCESILRRF 290
Query: 260 LDPNPETRMSVEDLYGNSWFKKSLKVEAESETESSTLNLDSGYGNRVRGLGVNAFDLISM 319
L NP R ++E + + W + + E E + +G+ R I +
Sbjct: 291 LVLNPAKRCTLEQIMKDKW----INIGYEGEELKPYTEPEEDFGDTKR---------IEV 337
Query: 320 SSGLDLSGLFQDEGKRKEKRFTSGAKLEVVEEKVKEIGGVLGFKVEVGKDSTAIGLVKGK 379
G+ G ++E K + TS EV + +LG K E G D A GL +
Sbjct: 338 MVGM---GYTREEIK---ESLTSQKYNEVTATYL-----LLGRKTEEGGDRGAPGLALAR 386
Query: 380 V 380
V
Sbjct: 387 V 387
>AAK2_HUMAN (P54646) 5'-AMP-activated protein kinase, catalytic
alpha-2 chain (EC 2.7.1.-) (AMPK alpha-2 chain)
Length = 552
Score = 206 bits (524), Expect = 9e-53
Identities = 105/265 (39%), Positives = 168/265 (62%), Gaps = 6/265 (2%)
Query: 20 LGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRLH 79
+G Y + LG G+F KV + G VAVKI+++ K + +I REI ++ L
Sbjct: 13 IGHYVLGDTLGVGTFGKVKIGEHQLTGHKVAVKILNRQKIRSLDVVGKIKREIQNLK-LF 71
Query: 80 HHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFCH 139
HP+I+++++V++T T +V+E+ +GGELF I + G++ E ARR FQQ++SA+ +CH
Sbjct: 72 RHPHIIKLYQVISTPTDFFMVMEYVSGGELFDYICKHGRVEEMEARRLFQQILSAVDYCH 131
Query: 140 RNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNGLLHTACGTPAYTAPEILRLSG 199
R+ V HRDLKP+N+LLDA N K++DFGLS + L T+CG+P Y APE+ +SG
Sbjct: 132 RHMVVHRDLKPENVLLDAHMNAKIADFGLSNMMS--DGEFLRTSCGSPNYAAPEV--ISG 187
Query: 200 G-YDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVIRR 258
Y G + D WSCG++L+ LL G LPFDD ++ +++KI + PE++ + ++
Sbjct: 188 RLYAGPEVDIWSCGVILYALLCGTLPFDDEHVPTLFKKIRGGVFYIPEYLNRSVATLLMH 247
Query: 259 LLDPNPETRMSVEDLYGNSWFKKSL 283
+L +P R +++D+ + WFK+ L
Sbjct: 248 MLQVDPLKRATIKDIREHEWFKQDL 272
>AAK2_RAT (Q09137) 5'-AMP-activated protein kinase, catalytic
alpha-2 chain (EC 2.7.1.-) (AMPK alpha-2 chain)
Length = 552
Score = 206 bits (523), Expect = 1e-52
Identities = 105/265 (39%), Positives = 168/265 (62%), Gaps = 6/265 (2%)
Query: 20 LGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRLH 79
+G Y + LG G+F KV + G VAVKI+++ K + +I REI ++ L
Sbjct: 13 IGHYVLGDTLGVGTFGKVKIGEHQLTGHKVAVKILNRQKIRSLDVVGKIKREIQNLK-LF 71
Query: 80 HHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFCH 139
HP+I+++++V++T T +V+E+ +GGELF I + G++ E ARR FQQ++SA+ +CH
Sbjct: 72 RHPHIIKLYQVISTPTDFFMVMEYVSGGELFDYICKHGRVEEVEARRLFQQILSAVDYCH 131
Query: 140 RNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNGLLHTACGTPAYTAPEILRLSG 199
R+ V HRDLKP+N+LLDA N K++DFGLS + L T+CG+P Y APE+ +SG
Sbjct: 132 RHMVVHRDLKPENVLLDAQMNAKIADFGLSNMMS--DGEFLRTSCGSPNYAAPEV--ISG 187
Query: 200 G-YDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVIRR 258
Y G + D WSCG++L+ LL G LPFDD ++ +++KI + PE++ + ++
Sbjct: 188 RLYAGPEVDIWSCGVILYALLCGTLPFDDEHVPTLFKKIRGGVFYIPEYLNRSIATLLMH 247
Query: 259 LLDPNPETRMSVEDLYGNSWFKKSL 283
+L +P R +++D+ + WFK+ L
Sbjct: 248 MLQVDPLKRATIKDIREHEWFKQDL 272
>ST29_HUMAN (Q8IWQ3) Serine/threonine-protein kinase 29 (EC
2.7.1.37) (BRSK2) (HUSSY-12)
Length = 736
Score = 204 bits (518), Expect = 5e-52
Identities = 97/260 (37%), Positives = 167/260 (63%), Gaps = 5/260 (1%)
Query: 20 LGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRLH 79
+G Y++ + LG+G V + VA+KI+++ K ++ + ++ REI A+ +L
Sbjct: 16 VGPYRLEKTLGKGQTGLVKLGVHCVTCQKVAIKIVNREKLSESVLM-KVEREI-AILKLI 73
Query: 80 HHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFCH 139
HP++L++H+V K ++LV+E +GGELF + ++G+L AR++F+Q++SAL FCH
Sbjct: 74 EHPHVLKLHDVYENKKYLYLVLEHVSGGELFDYLVKKGRLTPKEARKFFRQIISALDFCH 133
Query: 140 RNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNGLLHTACGTPAYTAPEILRLSG 199
+ + HRDLKP+NLLLD N++++DFG+++L + + LL T+CG+P Y PE++R
Sbjct: 134 SHSICHRDLKPENLLLDEKNNIRIADFGMASL--QVGDSLLETSCGSPHYACPEVIR-GE 190
Query: 200 GYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVIRRL 259
YDG KAD WSCG++LF LL G LPFDD N+ + K+ R + P +I + ++R +
Sbjct: 191 KYDGRKADVWSCGVILFALLVGALPFDDDNLRQLLEKVKRGVFHMPHFIPPDCQSLLRGM 250
Query: 260 LDPNPETRMSVEDLYGNSWF 279
+ + R+++E + + W+
Sbjct: 251 SEVDAARRLTLEHIQKHIWY 270
>SNF1_SCHPO (O74536) SNF1-like protein kinase ssp2 (EC 2.7.1.-)
Length = 576
Score = 202 bits (515), Expect = 1e-51
Identities = 117/342 (34%), Positives = 193/342 (56%), Gaps = 12/342 (3%)
Query: 11 PRSSAATTILGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVR 70
P + + +G Y + LG GSF KV A VA+K I + + M R+ R
Sbjct: 22 PPEAISKRHIGPYIIRETLGEGSFGKVKLATHYKTQQKVALKFISRQLLKKSDMHMRVER 81
Query: 71 EIDAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQ 130
EI ++ L H P+I+++++V+ T T I +V+E+A GGELF I + ++ E+ RR+FQQ
Sbjct: 82 EISYLKLLRH-PHIIKLYDVITTPTDIVMVIEYA-GGELFDYIVEKKRMTEDEGRRFFQQ 139
Query: 131 LVSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNGLLHTACGTPAYT 190
++ A+ +CHR+ + HRDLKP+NLLLD N+K++DFGLS + L T+CG+P Y
Sbjct: 140 IICAIEYCHRHKIVHRDLKPENLLLDDNLNVKIADFGLSNI--MTDGNFLKTSCGSPNYA 197
Query: 191 APEILRLSGGYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITK 250
APE++ Y G + D WSCG+VL+V+L G LPFDD I +++K++ Y P++++
Sbjct: 198 APEVIN-GKLYAGPEVDVWSCGIVLYVMLVGRLPFDDEFIPNLFKKVNSCVYVMPDFLSP 256
Query: 251 PARFVIRRLLDPNPETRMSVEDLYGNSWFKKSLKVEAESETESSTLNLDSGYGNRVRGLG 310
A+ +IRR++ +P R++++++ + WF +L E DS V LG
Sbjct: 257 GAQSLIRRMIVADPMQRITIQEIRRDPWFNVNLPDYLRPMEEVQGSYADS---RIVSKLG 313
Query: 311 VNAFDLISMSSGLDLSGLFQDEGKRKEKRFTSGAKLEVVEEK 352
+ + S + L DE ++ + + +V++EK
Sbjct: 314 ----EAMGFSEDYIVEALRSDENNEVKEAYNLLHENQVIQEK 351
>AAK1_HUMAN (Q13131) 5'-AMP-activated protein kinase, catalytic
alpha-1 chain (EC 2.7.1.-) (AMPK alpha-1 chain)
Length = 550
Score = 201 bits (510), Expect = 4e-51
Identities = 107/280 (38%), Positives = 175/280 (62%), Gaps = 8/280 (2%)
Query: 20 LGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRLH 79
+G Y + LG G+F KV + + G VAVKI+++ K + +I REI ++ L
Sbjct: 15 IGHYILGDTLGVGTFGKVKVGKHELTGHKVAVKILNRQKIRSLDVVGKIRREIQNLK-LF 73
Query: 80 HHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFCH 139
HP+I+++++V++T + I +V+E+ +GGELF I + G+L E +RR FQQ++S + +CH
Sbjct: 74 RHPHIIKLYQVISTPSDIFMVMEYVSGGELFDYICKNGRLDEKESRRLFQQILSGVDYCH 133
Query: 140 RNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNGLLHTACGTPAYTAPEILRLSG 199
R+ V HRDLKP+N+LLDA N K++DFGLS + L T+CG+P Y APE+ +SG
Sbjct: 134 RHMVVHRDLKPENVLLDAHMNAKIADFGLSNMMS--DGEFLRTSCGSPNYAAPEV--ISG 189
Query: 200 G-YDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVIRR 258
Y G + D WS G++L+ LL G LPFDD ++ +++KI + P+++ +++
Sbjct: 190 RLYAGPEVDIWSSGVILYALLCGTLPFDDDHVPTLFKKICDGIFYTPQYLNPSVISLLKH 249
Query: 259 LLDPNPETRMSVEDLYGNSWFKKSLK--VEAESETESSTL 296
+L +P R S++D+ + WFK+ L + E + SST+
Sbjct: 250 MLQVDPMKRASIKDIREHEWFKQDLPKYLFPEDPSYSSTM 289
>AAK1_RAT (P54645) 5'-AMP-activated protein kinase, catalytic
alpha-1 chain (EC 2.7.1.-) (AMPK alpha-1 chain)
Length = 548
Score = 199 bits (507), Expect = 9e-51
Identities = 106/280 (37%), Positives = 175/280 (61%), Gaps = 8/280 (2%)
Query: 20 LGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRLH 79
+G Y + LG G+F KV + + G VAVKI+++ K + +I REI ++ L
Sbjct: 13 IGHYILGDTLGVGTFGKVKVGKHELTGHKVAVKILNRQKIRSLDVVGKIRREIQNLK-LF 71
Query: 80 HHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFCH 139
HP+I+++++V++T + I +V+E+ +GGELF I + G+L E +RR FQQ++S + +CH
Sbjct: 72 RHPHIIKLYQVISTPSDIFMVMEYVSGGELFDYICKNGRLDEKESRRLFQQILSGVDYCH 131
Query: 140 RNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNGLLHTACGTPAYTAPEILRLSG 199
R+ V HRDLKP+N+LLDA N K++DFGLS + L T+CG+P Y APE+ +SG
Sbjct: 132 RHMVVHRDLKPENVLLDAHMNAKIADFGLSNMMS--DGEFLRTSCGSPNYAAPEV--ISG 187
Query: 200 G-YDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVIRR 258
Y G + D WS G++L+ LL G LPFDD ++ +++KI + P+++ +++
Sbjct: 188 RLYAGPEVDIWSSGVILYALLCGTLPFDDDHVPTLFKKICDGIFYTPQYLNPSVISLLKH 247
Query: 259 LLDPNPETRMSVEDLYGNSWFKKSLK--VEAESETESSTL 296
+L +P R +++D+ + WFK+ L + E + SST+
Sbjct: 248 MLQVDPMKRATIKDIREHEWFKQDLPKYLFPEDPSYSSTM 287
>GIN4_YEAST (Q12263) Serine/threonine-protein kinase GIN4 (EC
2.7.1.37)
Length = 1142
Score = 197 bits (502), Expect = 3e-50
Identities = 113/281 (40%), Positives = 172/281 (60%), Gaps = 24/281 (8%)
Query: 20 LGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKS--------------KTVDAAME 65
+G +++ LG GS KV AR+ G AVK+I K+ T A+
Sbjct: 16 IGPWKLGETLGLGSTGKVQLARNGSTGQEAAVKVISKAVFNTGNVSGTSIVGSTTPDALP 75
Query: 66 PRIVREIDAMRRLHHHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTAR 125
I REI M+ L+H PN+LR+++V T T ++LV+E+A GELF+ + RG LPE+ A
Sbjct: 76 YGIEREIIIMKLLNH-PNVLRLYDVWETNTDLYLVLEYAEKGELFNLLVERGPLPEHEAI 134
Query: 126 RYFQQLVSALRFCHRNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNGLLHTACG 185
R+F+Q++ + +CH G+ HRDLKP+NLLLD N+K++DFG++AL + LL T+CG
Sbjct: 135 RFFRQIIIGVSYCHALGIVHRDLKPENLLLDHKYNIKIADFGMAAL--ETEGKLLETSCG 192
Query: 186 TPAYTAPEILRLSG-GYDGSKADAWSCGLVLFVLLAGYLPFD--DSNIAAMYRKISRRDY 242
+P Y APEI +SG Y G +D WSCG++LF LL G LPFD D NI + K+ + ++
Sbjct: 193 SPHYAAPEI--VSGIPYQGFASDVWSCGVILFALLTGRLPFDEEDGNIRTLLLKVQKGEF 250
Query: 243 QFP--EWITKPARFVIRRLLDPNPETRMSVEDLYGNSWFKK 281
+ P + I++ A+ +IR++L +PE R+ D+ + +K
Sbjct: 251 EMPSDDEISREAQDLIRKILTVDPERRIKTRDILKHPLLQK 291
>MRK2_MOUSE (Q05512) MAP/microtubule affinity-regulating kinase 2
(EC 2.7.1.37) (Serine/threonine-protein kinase Emk)
Length = 774
Score = 196 bits (498), Expect = 1e-49
Identities = 97/259 (37%), Positives = 168/259 (64%), Gaps = 5/259 (1%)
Query: 20 LGKYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRLH 79
+G Y++++ +G+G+FAKV AR ++ G VAVKIIDK++ ++++ ++ RE+ M+ L+
Sbjct: 50 IGNYRLLKTIGKGNFAKVKLARHILTGKEVAVKIIDKTQLNSSSLQ-KLFREVRIMKVLN 108
Query: 80 HHPNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFCH 139
H PNI+++ EV+ T+ ++LV+E+A+GGE+F + G++ E AR F+Q+V +++CH
Sbjct: 109 H-PNIVKLFEVIETEKTLYLVMEYASGGEVFDYLVAHGRMKEKEARAKFRQIVLHVQYCH 167
Query: 140 RNGVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNGLLHTACGTPAYTAPEILRLSG 199
+ + HRDLK +NLLLDA N+K++DFG S E L T CG+P Y APE+ +
Sbjct: 168 QKFIVHRDLKAENLLLDADMNIKIADFGFS--NEFTFGNKLDTFCGSPPYAAPELFQ-GK 224
Query: 200 GYDGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKPARFVIRRL 259
DG + D WS G++L+ L++G LPFD N+ + ++ R Y+ P +++ ++++
Sbjct: 225 KIDGPEVDVWSLGVILYTLVSGSLPFDGQNLKELRERVLRGKYRIPFYMSTDCENLLKKF 284
Query: 260 LDPNPETRMSVEDLYGNSW 278
L NP R ++E + + W
Sbjct: 285 LILNPSKRGTLEQIMKDRW 303
>ARK5_HUMAN (O60285) AMPK-related protein kinase 5 (EC 2.7.1.37)
Length = 661
Score = 196 bits (497), Expect = 1e-49
Identities = 115/295 (38%), Positives = 181/295 (60%), Gaps = 14/295 (4%)
Query: 22 KYQVIRLLGRGSFAKVYQARSLIDGTIVAVKIIDKSKTVDAAMEPRIVREIDAMRRLHHH 81
+Y++ LG+G++ KV +A G +VA+K I K K D I REI+ M L+H
Sbjct: 54 RYELQETLGKGTYGKVKRATERFSGRVVAIKSIRKDKIKDEQDMVHIRREIEIMSSLNH- 112
Query: 82 PNILRIHEVMATKTKIHLVVEFAAGGELFSAISRRGKLPENTARRYFQQLVSALRFCHRN 141
P+I+ I+EV K KI +++E+A+ GEL+ IS R +L E R +F+Q+VSA+ +CH+N
Sbjct: 113 PHIISIYEVFENKDKIVIIMEYASKGELYDYISERRRLSERETRHFFRQIVSAVHYCHKN 172
Query: 142 GVAHRDLKPQNLLLDAAGNLKVSDFGLSALPEHLQNGLLHTACGTPAYTAPEILRLSGGY 201
GV HRDLK +N+LLD N+K++DFGLS L + ++ L T CG+P Y +PEI+ Y
Sbjct: 173 GVVHRDLKLENILLDDNCNIKIADFGLSNL--YQKDKFLQTFCGSPLYASPEIVN-GRPY 229
Query: 202 DGSKADAWSCGLVLFVLLAGYLPFDDSNIAAMYRKISRRDYQFPEWITKP--ARFVIRRL 259
G + D+W+ G++L+ L+ G +PFD + + R+IS +Y+ P T+P AR +IR +
Sbjct: 230 RGPEVDSWALGVLLYTLVYGTMPFDGFDHKNLIRQISSGEYREP---TQPSDARGLIRWM 286
Query: 260 LDPNPETRMSVEDLYGNSW----FKKSL-KVEAESETESSTLNLDSGYGNRVRGL 309
L NP+ R ++ED+ + W +K S+ +A ++ES L + +R GL
Sbjct: 287 LMVNPDRRATIEDIANHWWVNWGYKSSVCDCDALHDSESPLLARIIDWHHRSTGL 341
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.138 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 50,895,767
Number of Sequences: 164201
Number of extensions: 2159893
Number of successful extensions: 10381
Number of sequences better than 10.0: 1684
Number of HSP's better than 10.0 without gapping: 1479
Number of HSP's successfully gapped in prelim test: 205
Number of HSP's that attempted gapping in prelim test: 5926
Number of HSP's gapped (non-prelim): 1955
length of query: 430
length of database: 59,974,054
effective HSP length: 113
effective length of query: 317
effective length of database: 41,419,341
effective search space: 13129931097
effective search space used: 13129931097
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0086.14


