

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
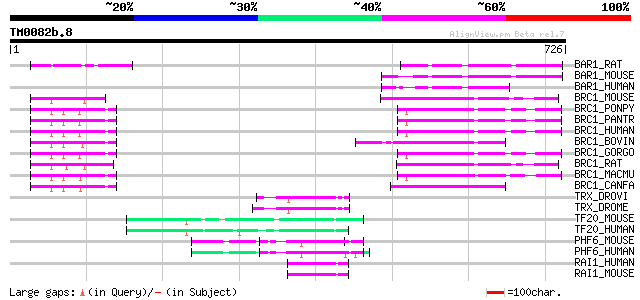
Query= TM0082b.8
(726 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BAR1_RAT (Q9QZH2) BRCA1-associated RING domain protein 1 (BARD-1) 91 1e-17
BAR1_MOUSE (O70445) BRCA1-associated RING domain protein 1 (BARD-1) 87 2e-16
BAR1_HUMAN (Q99728) BRCA1-associated RING domain protein 1 (BARD-1) 86 3e-16
BRC1_MOUSE (P48754) Breast cancer type 1 susceptibility protein ... 78 7e-14
BRC1_PONPY (Q6J6J0) Breast cancer type 1 susceptibility protein ... 78 9e-14
BRC1_PANTR (Q9GKK8) Breast cancer type 1 susceptibility protein ... 78 9e-14
BRC1_HUMAN (P38398) Breast cancer type 1 susceptibility protein 78 9e-14
BRC1_BOVIN (Q864U1) Breast cancer type 1 susceptibility protein ... 77 2e-13
BRC1_GORGO (Q6J6I8) Breast cancer type 1 susceptibility protein ... 76 3e-13
BRC1_RAT (O54952) Breast cancer type 1 susceptibility protein ho... 75 6e-13
BRC1_MACMU (Q6J6I9) Breast cancer type 1 susceptibility protein ... 74 2e-12
BRC1_CANFA (Q95153) Breast cancer type 1 susceptibility protein ... 72 5e-12
TRX_DROVI (Q24742) Trithorax protein 72 7e-12
TRX_DROME (P20659) Trithorax protein 72 7e-12
TF20_MOUSE (Q9EPQ8) Transcription factor 20 (Stromelysin 1 PDGF-... 71 9e-12
TF20_HUMAN (Q9UGU0) Transcription factor 20 (Stromelysin 1 PDGF-... 71 9e-12
PHF6_MOUSE (Q9D4J7) PHD finger protein 6 65 5e-10
PHF6_HUMAN (Q8IWS0) PHD finger protein 6 (PHD-like zinc finger p... 64 1e-09
RAI1_HUMAN (Q7Z5J4) Retinoic acid induced protein 1 62 4e-09
RAI1_MOUSE (Q61818) Retinoic acid induced protein 1 60 2e-08
>BAR1_RAT (Q9QZH2) BRCA1-associated RING domain protein 1 (BARD-1)
Length = 768
Score = 90.5 bits (223), Expect = 1e-17
Identities = 61/213 (28%), Positives = 97/213 (44%), Gaps = 16/213 (7%)
Query: 512 VSGRGEVLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTLKVLMAILNGRWVLKI 571
+S + + LL + + A + + VTHVI + TLK ++ ILNG WVLK
Sbjct: 568 LSSQQQKLLSKLETVLKAKKCAEFDNTVTHVIVPDEE---AQSTLKCMLGILNGCWVLKF 624
Query: 572 DWIKACMKVMSLVEEEPYEISLDNQGCNGGPKAGRLSALANEPKLFSGLKFYFSGDYVSS 631
DW+KAC+ +EE YE+ GGP+ RL+ PKLF G F+ G++
Sbjct: 625 DWVKACLDSQEREQEEKYEVP-------GGPQRSRLNREQLLPKLFDGCYFFLGGNFKHH 677
Query: 632 YKEDLEDLVEVGGGAVLTSKDKLEAKRHEFEVTSNL-LIVYNLDPPQGSKLGDEVSTLWQ 690
KEDL L+ GG +L+ K K ++ +VT + + Y+ P + +
Sbjct: 678 PKEDLLKLIAAAGGRILSRKPKPDS-----DVTQTINTVAYHAKPDSDQRFCTQYIVYED 732
Query: 691 RLNEAEDLAANTGSQVIGHTWILESIAACKLQP 723
N + + TW++ + A +L P
Sbjct: 733 LFNCHPERVRQGKVWMAPSTWLISCVMAFELLP 765
Score = 47.4 bits (111), Expect = 1e-04
Identities = 36/136 (26%), Positives = 62/136 (45%), Gaps = 14/136 (10%)
Query: 28 LKCPLCLSLFEKPVLLP-CDHLFCDSCLVDSSLSGSECAVCKTKYAQTDIRNVPFVENMV 86
L+C C ++ +PV L C+H+FC C+ S GS C VC T D++ +++M+
Sbjct: 42 LRCSRCANILREPVCLGGCEHIFCSGCI--SDCVGSGCPVCHTPAWILDLKINRQLDSMI 99
Query: 87 AIYRSLDASFCASMLQQRSNDDSRVLQQCQTFRDSSYSNIKKADNFSQS--SPNSNGFGV 144
+Y L ++L DS+ T R S + + ++ N + SP S
Sbjct: 100 QLYSKLQ-----NLLHDNKGSDSK----DDTSRASLFGDAERKKNSVKMWFSPRSKKIRC 150
Query: 145 GENRKSMITMHVKPEE 160
N+ S+ T K ++
Sbjct: 151 VVNKVSVQTQPQKAKD 166
>BAR1_MOUSE (O70445) BRCA1-associated RING domain protein 1 (BARD-1)
Length = 765
Score = 86.7 bits (213), Expect = 2e-16
Identities = 64/238 (26%), Positives = 103/238 (42%), Gaps = 34/238 (14%)
Query: 487 VVFCGSALSDEEKGSIENLRVMAVVVSGRGEVLLINFASKVGATVTKFWTSDVTHVIAAT 546
+VF GS LS +++ + L + + A + S VTHVI
Sbjct: 558 LVFIGSGLSSQQQKMLSKLETV------------------LKAKKCMEFDSTVTHVIVPD 599
Query: 547 DANGACSRTLKVLMAILNGRWVLKIDWIKACMKVMSLVEEEPYEISLDNQGCNGGPKAGR 606
+ TLK ++ IL+G W+LK DW+KAC+ +EE YE+ GGP+ R
Sbjct: 600 EE---AQSTLKCMLGILSGCWILKFDWVKACLDSKVREQEEKYEVP-------GGPQRSR 649
Query: 607 LSALANEPKLFSGLKFYFSGDYVSSYKEDLEDLVEVGGGAVLTSKDKLEAKRHEFEVTSN 666
L+ PKLF G F+ G++ ++DL L+ GG VL+ K K ++ +VT
Sbjct: 650 LNREQLLPKLFDGCYFFLGGNFKHHPRDDLLKLIAAAGGKVLSRKPKPDS-----DVTQT 704
Query: 667 L-LIVYNLDPPQGSKLGDEVSTLWQRLNEAEDLAANTGSQVIGHTWILESIAACKLQP 723
+ + Y+ P + + N + + TW++ I A +L P
Sbjct: 705 INTVAYHAKPESDQRFCTQYIVYEDLFNCHPERVRQGKVWMAPSTWLISCIMAFELLP 762
Score = 44.3 bits (103), Expect = 0.001
Identities = 39/132 (29%), Positives = 57/132 (42%), Gaps = 33/132 (25%)
Query: 28 LKCPLCLSLFEKPVLLP-CDHLFCDSCLVDSSLSGSECAVCKTKYAQTDIRNVPFVENMV 86
L+C C ++ ++PV L C+H+FC C+ S GS C VC T D++
Sbjct: 42 LRCSRCANILKEPVCLGGCEHIFCSGCI--SDCVGSGCPVCYTPAWILDLK--------- 90
Query: 87 AIYRSLDASFCASMLQQRSNDDSRVLQQCQTFRDSSYSNIKKADNFSQSSPNSNGFGVGE 146
I R LD SM+Q S ++ + N DN S++S FG E
Sbjct: 91 -INRQLD-----SMIQLSSK-----------LQNLLHDNKDSKDNTSRASL----FGDAE 129
Query: 147 NRKSMITMHVKP 158
+K+ I M P
Sbjct: 130 RKKNSIKMWFSP 141
>BAR1_HUMAN (Q99728) BRCA1-associated RING domain protein 1 (BARD-1)
Length = 777
Score = 86.3 bits (212), Expect = 3e-16
Identities = 59/167 (35%), Positives = 83/167 (49%), Gaps = 28/167 (16%)
Query: 487 VVFCGSALSDEEKGSIENLRVMAVVVSGRGEVLLINFASKVGATVTKFWTSDVTHVIAAT 546
+V GS LS E++ + L AV++ + T+F S VTHV+
Sbjct: 570 LVLIGSGLSSEQQKMLSEL---AVILKAK--------------KYTEF-DSTVTHVVVPG 611
Query: 547 DANGACSRTLKVLMAILNGRWVLKIDWIKACMKVMSLVEEEPYEISLDNQGCNGGPKAGR 606
DA TLK ++ ILNG W+LK +W+KAC++ +EE YEI GP+ R
Sbjct: 612 DA---VQSTLKCMLGILNGCWILKFEWVKACLRRKVCEQEEKYEIP-------EGPRRSR 661
Query: 607 LSALANEPKLFSGLKFYFSGDYVSSYKEDLEDLVEVGGGAVLTSKDK 653
L+ PKLF G FY G + K++L LV GGG +L+ K K
Sbjct: 662 LNREQLLPKLFDGCYFYLWGTFKHHPKDNLIKLVTAGGGQILSRKPK 708
Score = 43.9 bits (102), Expect = 0.002
Identities = 33/132 (25%), Positives = 55/132 (41%), Gaps = 31/132 (23%)
Query: 28 LKCPLCLSLFEKPVLLP-CDHLFCDSCLVDSSLSGSECAVCKTKYAQTDIRNVPFVENMV 86
L+C C ++ +PV L C+H+FC +C+ S G+ C VC T D++
Sbjct: 48 LRCSRCTNILREPVCLGGCEHIFCSNCV--SDCIGTGCPVCYTPAWIQDLK--------- 96
Query: 87 AIYRSLDASFCASMLQQRSNDDSRVLQQCQTFRDSSYSNIKKADNFSQSSPNSNGFGVGE 146
I R LD+ ++Q C R+ + N + + + P + F
Sbjct: 97 -INRQLDS----------------MIQLCSKLRNLLHDN--ELSDLKEDKPRKSLFNDAG 137
Query: 147 NRKSMITMHVKP 158
N+K+ I M P
Sbjct: 138 NKKNSIKMWFSP 149
>BRC1_MOUSE (P48754) Breast cancer type 1 susceptibility protein
homolog
Length = 1812
Score = 78.2 bits (191), Expect = 7e-14
Identities = 62/240 (25%), Positives = 111/240 (45%), Gaps = 28/240 (11%)
Query: 485 KKVVFCGSALSDEEKGSIENL-RVMAVVVSGRG--EVLLIN-FASKVGATVTKFWTSDVT 540
K VV S + E S E R +++VVSG EV+ + FA K T+T T + T
Sbjct: 1569 KAVVGIVSKIKPELTSSEERADRDISMVVSGLTPKEVMTVQKFAEKYRLTLTDAITEETT 1628
Query: 541 HVIAATDANGACSRTLKVLMAILNGRWVLKIDWIKACMKVMSLVEEEPYEISLD--NQGC 598
HVI TDA C RTLK + I G+W++ W+ ++ L+ +E++ D
Sbjct: 1629 HVIIKTDAEFVCERTLKYFLGIAGGKWIVSYSWVVRSIQERRLLNVHEFEVTGDVVTGRN 1688
Query: 599 NGGPKAGRLSALANEPKLFSGLKFYFSGDYVSSYKEDLEDLVEVGGGAVLTSKDKLEAKR 658
+ GP+ R S KLF GL+ Y + + K+DLE ++++ G +V+ L
Sbjct: 1689 HQGPRRSRES----REKLFKGLQVYCCDPFTNMPKDDLERMLQLCGASVVKELPSL---- 1740
Query: 659 HEFEVTSNLLIVYNLDPPQGSKLGDEVSTLWQRLNEAEDLAANTGSQVIGHTWILESIAA 718
+ ++L+++ + W + D+ ++++ W+L+S+++
Sbjct: 1741 -THDTGAHLVVIVQ-------------PSAWTEDSNCPDIGQLCKARLVMWDWVLDSLSS 1786
Score = 49.3 bits (116), Expect = 4e-05
Identities = 27/111 (24%), Positives = 50/111 (44%), Gaps = 13/111 (11%)
Query: 28 LKCPLCLSLFEKPVLLPCDHLFCDSC---LVDSSLSGSECAVCKTKYAQTDIRNVPFVEN 84
L+CP+CL L ++PV CDH+FC C L++ S+C +CK + + ++
Sbjct: 22 LECPICLELIKEPVSTKCDHIFCKFCMLKLLNQKKGPSQCPLCKNEITKRSLQGSTRFSQ 81
Query: 85 MVAIYRSLDASF----------CASMLQQRSNDDSRVLQQCQTFRDSSYSN 125
+ + A+F S ++R+N R+ ++ + Y N
Sbjct: 82 LAEELLRIMAAFELDTGMQLTNGFSFSKKRNNSCERLNEEASIIQSVGYRN 132
>BRC1_PONPY (Q6J6J0) Breast cancer type 1 susceptibility protein
homolog
Length = 1863
Score = 77.8 bits (190), Expect = 9e-14
Identities = 58/220 (26%), Positives = 99/220 (44%), Gaps = 26/220 (11%)
Query: 508 MAVVVSGRGE---VLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTLKVLMAILN 564
M++VVSG +L+ FA K T+T T + THV+ TDA C RTLK + I
Sbjct: 1650 MSMVVSGLTPEEFMLVYKFARKHHITLTNLITEETTHVVMKTDAEFVCERTLKYFLGIAG 1709
Query: 565 GRWVLKIDWIKACMKVMSLVEEEPYEISLD--NQGCNGGPKAGRLSALANEPKLFSGLKF 622
G+WV+ W+ +K ++ E +E+ D N + GPK R S + K+F GL+
Sbjct: 1710 GKWVVSYFWVTQSIKERKMLNEHDFEVRGDVVNGRNHQGPKRARES---QDRKIFRGLEI 1766
Query: 623 YFSGDYVSSYKEDLEDLVEVGGGAVLTSKDKLEAKRHEFEVTSNLLIVYNLDPPQGSKLG 682
G + + + LE +V++ G +V+ + F + + + + + P
Sbjct: 1767 CCYGPFTNMPTDQLEWIVQLCGASVV-------KELSSFTLGTGVHPIVVVQP------- 1812
Query: 683 DEVSTLWQRLNEAEDLAANTGSQVIGHTWILESIAACKLQ 722
W N + + V+ W+L+S+A + Q
Sbjct: 1813 ----DAWTEDNGFHAIGQMCEAPVVTREWVLDSVALYQCQ 1848
Score = 49.7 bits (117), Expect = 3e-05
Identities = 32/125 (25%), Positives = 54/125 (42%), Gaps = 15/125 (12%)
Query: 28 LKCPLCLSLFEKPVLLPCDHLFCDSC---LVDSSLSGSECAVCKT----KYAQTDIRNVP 80
L+CP+CL L ++PV CDH+FC C L++ S+C +CK + Q R
Sbjct: 22 LECPICLELIKEPVSTKCDHIFCKFCMLKLLNQKKGPSQCPLCKNDITKRSLQESTRFSQ 81
Query: 81 FVENMVAIY------RSLDASFCASMLQQRSNDDSRVLQQCQTFRDSSYSNIKKADNFSQ 134
VE ++ I L + + ++ +N + + + Y N +A Q
Sbjct: 82 LVEELLKIICAFQLDTGLQYANSYNFAKKENNSPEHLKDEVSIIQSMGYRN--RAKRLLQ 139
Query: 135 SSPNS 139
S P +
Sbjct: 140 SEPEN 144
>BRC1_PANTR (Q9GKK8) Breast cancer type 1 susceptibility protein
homolog
Length = 1863
Score = 77.8 bits (190), Expect = 9e-14
Identities = 58/220 (26%), Positives = 99/220 (44%), Gaps = 26/220 (11%)
Query: 508 MAVVVSGRGE---VLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTLKVLMAILN 564
M++VVSG +L+ FA K T+T T + THV+ TDA C RTLK + I
Sbjct: 1650 MSMVVSGLTPEEFMLVYKFARKHHITLTNLITEETTHVVMKTDAEFVCERTLKYFLGIAG 1709
Query: 565 GRWVLKIDWIKACMKVMSLVEEEPYEISLD--NQGCNGGPKAGRLSALANEPKLFSGLKF 622
G+WV+ W+ +K ++ E +E+ D N + GPK R S + K+F GL+
Sbjct: 1710 GKWVVSYFWVTQSIKERKMLNEHDFEVRGDVVNGRNHQGPKRARES---QDRKIFRGLEI 1766
Query: 623 YFSGDYVSSYKEDLEDLVEVGGGAVLTSKDKLEAKRHEFEVTSNLLIVYNLDPPQGSKLG 682
G + + + LE +V++ G +V+ + F + + + + + P
Sbjct: 1767 CCYGPFTNMPTDQLEWMVQLCGASVV-------KELSSFTLGTGVHPIVVVQP------- 1812
Query: 683 DEVSTLWQRLNEAEDLAANTGSQVIGHTWILESIAACKLQ 722
W N + + V+ W+L+S+A + Q
Sbjct: 1813 ----DAWTEDNGFHAIGQMCEAPVVTREWVLDSVALYQCQ 1848
Score = 50.4 bits (119), Expect = 2e-05
Identities = 32/125 (25%), Positives = 55/125 (43%), Gaps = 15/125 (12%)
Query: 28 LKCPLCLSLFEKPVLLPCDHLFCDSC---LVDSSLSGSECAVCKT----KYAQTDIRNVP 80
L+CP+CL L ++PV CDH+FC C L++ S+C +CK + Q R
Sbjct: 22 LECPICLELIKEPVSTKCDHIFCKFCMLKLLNQKKGPSQCPLCKNDITKRSLQESTRFSQ 81
Query: 81 FVENMVAIY------RSLDASFCASMLQQRSNDDSRVLQQCQTFRDSSYSNIKKADNFSQ 134
VE ++ I L+ + + ++ +N + + + Y N +A Q
Sbjct: 82 LVEELLKIICAFQLDTGLEYANSYNFAKKENNSPEHLKDEVSIIQSMGYRN--RAKRLLQ 139
Query: 135 SSPNS 139
S P +
Sbjct: 140 SEPEN 144
>BRC1_HUMAN (P38398) Breast cancer type 1 susceptibility protein
Length = 1863
Score = 77.8 bits (190), Expect = 9e-14
Identities = 58/220 (26%), Positives = 99/220 (44%), Gaps = 26/220 (11%)
Query: 508 MAVVVSGRGE---VLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTLKVLMAILN 564
M++VVSG +L+ FA K T+T T + THV+ TDA C RTLK + I
Sbjct: 1650 MSMVVSGLTPEEFMLVYKFARKHHITLTNLITEETTHVVMKTDAEFVCERTLKYFLGIAG 1709
Query: 565 GRWVLKIDWIKACMKVMSLVEEEPYEISLD--NQGCNGGPKAGRLSALANEPKLFSGLKF 622
G+WV+ W+ +K ++ E +E+ D N + GPK R S + K+F GL+
Sbjct: 1710 GKWVVSYFWVTQSIKERKMLNEHDFEVRGDVVNGRNHQGPKRARES---QDRKIFRGLEI 1766
Query: 623 YFSGDYVSSYKEDLEDLVEVGGGAVLTSKDKLEAKRHEFEVTSNLLIVYNLDPPQGSKLG 682
G + + + LE +V++ G +V+ + F + + + + + P
Sbjct: 1767 CCYGPFTNMPTDQLEWMVQLCGASVV-------KELSSFTLGTGVHPIVVVQP------- 1812
Query: 683 DEVSTLWQRLNEAEDLAANTGSQVIGHTWILESIAACKLQ 722
W N + + V+ W+L+S+A + Q
Sbjct: 1813 ----DAWTEDNGFHAIGQMCEAPVVTREWVLDSVALYQCQ 1848
Score = 50.4 bits (119), Expect = 2e-05
Identities = 32/125 (25%), Positives = 55/125 (43%), Gaps = 15/125 (12%)
Query: 28 LKCPLCLSLFEKPVLLPCDHLFCDSC---LVDSSLSGSECAVCKT----KYAQTDIRNVP 80
L+CP+CL L ++PV CDH+FC C L++ S+C +CK + Q R
Sbjct: 22 LECPICLELIKEPVSTKCDHIFCKFCMLKLLNQKKGPSQCPLCKNDITKRSLQESTRFSQ 81
Query: 81 FVENMVAIY------RSLDASFCASMLQQRSNDDSRVLQQCQTFRDSSYSNIKKADNFSQ 134
VE ++ I L+ + + ++ +N + + + Y N +A Q
Sbjct: 82 LVEELLKIICAFQLDTGLEYANSYNFAKKENNSPEHLKDEVSIIQSMGYRN--RAKRLLQ 139
Query: 135 SSPNS 139
S P +
Sbjct: 140 SEPEN 144
>BRC1_BOVIN (Q864U1) Breast cancer type 1 susceptibility protein
homolog
Length = 1849
Score = 77.0 bits (188), Expect = 2e-13
Identities = 55/198 (27%), Positives = 100/198 (49%), Gaps = 10/198 (5%)
Query: 453 SRPKKQATQEHPASAHLSSQLSNPLGAFHDDGKKVVFCGSALSDEEKGSIENLRVMAVVV 512
S+ + + + ++PA+AH+++ L +K +S E+ S + L ++A +
Sbjct: 1596 SQFRVEESTKNPAAAHIANTTRCNLREESMSKEKP----EVISSTER-SKKRLSMVASGL 1650
Query: 513 SGRGEVLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTLKVLMAILNGRWVLKID 572
+ + +L+ FA K T+T T + THVI TD C RTLK + I G+WV+
Sbjct: 1651 TPKELMLVQKFARKHHVTLTNLITEETTHVIMKTDPEFVCERTLKYFLGIAGGKWVVSYF 1710
Query: 573 WIKACMKVMSLVEEEPYEISLD--NQGCNGGPKAGRLSALANEPKLFSGLKFYFSGDYVS 630
W+ +K +++E +E+ D N + GPK R S + K+F GL+ G + +
Sbjct: 1711 WVTQSIKEGKMLDEHDFEVRGDVVNGRNHQGPKRARES---RDKKIFKGLEICCYGPFTN 1767
Query: 631 SYKEDLEDLVEVGGGAVL 648
+ LE +V++ G +V+
Sbjct: 1768 MPTDQLEWMVQLCGASVV 1785
Score = 51.6 bits (122), Expect = 7e-06
Identities = 35/125 (28%), Positives = 56/125 (44%), Gaps = 15/125 (12%)
Query: 28 LKCPLCLSLFEKPVLLPCDHLFCDSC---LVDSSLSGSECAVCKT----KYAQTDIRNVP 80
L+CP+CL L ++PV CDH+FC C L++ S+C +CK + Q R
Sbjct: 22 LECPICLELIKEPVSTKCDHIFCKFCMLKLLNQKKGPSQCPLCKNDITKRSLQESTRFSQ 81
Query: 81 FVENMVAIYRSLD----ASFCASMLQQRSNDDS--RVLQQCQTFRDSSYSNIKKADNFSQ 134
VE ++ I + + F S R D+S + ++ + Y N +A Q
Sbjct: 82 LVEELLKIIHAFELDTGLQFANSYNFSRKEDNSPEHLKEEVSIIQSMGYRN--RAKRLWQ 139
Query: 135 SSPNS 139
S P +
Sbjct: 140 SEPEN 144
>BRC1_GORGO (Q6J6I8) Breast cancer type 1 susceptibility protein
homolog
Length = 1863
Score = 76.3 bits (186), Expect = 3e-13
Identities = 58/220 (26%), Positives = 98/220 (44%), Gaps = 26/220 (11%)
Query: 508 MAVVVSGRGE---VLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTLKVLMAILN 564
M++VVSG +L+ FA K T+T T + THV+ TDA C RTLK + I
Sbjct: 1650 MSMVVSGLTPEEFMLVYKFARKHHITLTNLITEETTHVVMKTDAEFVCERTLKYFLGIAG 1709
Query: 565 GRWVLKIDWIKACMKVMSLVEEEPYEISLD--NQGCNGGPKAGRLSALANEPKLFSGLKF 622
G+WV+ W+ +K ++ E +E+ D N + GPK R S + K+F GL
Sbjct: 1710 GKWVVSYFWVTQSIKEGKMLNEHDFEVRGDVVNGRNHQGPKRARES---QDRKIFRGLDI 1766
Query: 623 YFSGDYVSSYKEDLEDLVEVGGGAVLTSKDKLEAKRHEFEVTSNLLIVYNLDPPQGSKLG 682
G + + + LE +V++ G +V+ + F + + + + + P
Sbjct: 1767 CCYGPFTNMPTDQLEWMVQLCGASVV-------KELSSFTLGTGVHPIVVVQP------- 1812
Query: 683 DEVSTLWQRLNEAEDLAANTGSQVIGHTWILESIAACKLQ 722
W N + + V+ W+L+S+A + Q
Sbjct: 1813 ----DAWTEDNGFHAIGQMCEAPVVTREWVLDSVALYQCQ 1848
Score = 49.3 bits (116), Expect = 4e-05
Identities = 31/125 (24%), Positives = 56/125 (44%), Gaps = 15/125 (12%)
Query: 28 LKCPLCLSLFEKPVLLPCDHLFCDSC---LVDSSLSGSECAVCKT----KYAQTDIRNVP 80
L+CP+CL L ++PV CDH+FC C L++ S+C +CK + Q R
Sbjct: 22 LECPICLELIKEPVSTKCDHIFCKFCMLKLLNQKKGPSQCPLCKNDITKRSLQESTRFSQ 81
Query: 81 FVENMVAIY------RSLDASFCASMLQQRSNDDSRVLQQCQTFRDSSYSNIKKADNFSQ 134
VE ++ I L+ + + ++ +N + + + + Y + +A Q
Sbjct: 82 LVEELLKIICAFQLDTGLEYANSYNFAKKENNSPEHLKDEVSIIQSTGYRS--RAKRLLQ 139
Query: 135 SSPNS 139
S P +
Sbjct: 140 SEPEN 144
>BRC1_RAT (O54952) Breast cancer type 1 susceptibility protein homolog
Length = 1817
Score = 75.1 bits (183), Expect = 6e-13
Identities = 54/217 (24%), Positives = 101/217 (45%), Gaps = 27/217 (12%)
Query: 506 RVMAVVVSGRG--EVLLIN-FASKVGATVTKFWTSDVTHVIAATDANGACSRTLKVLMAI 562
R +++VVSG EV+++ FA K +T T + THVI TDA C RTLK + I
Sbjct: 1594 RDISMVVSGLTPKEVMIVQKFAEKYRLALTDVITEETTHVIIKTDAEFVCERTLKYFLGI 1653
Query: 563 LNGRWVLKIDWIKACMKVMSLVEEEPYEISLD--NQGCNGGPKAGRLSALANEPKLFSGL 620
G+W++ W+ ++ L+ +E+ D + GP+ R S + KLF GL
Sbjct: 1654 AGGKWIVSYSWVIKSIQERKLLSVHEFEVKGDVVTGSNHQGPRRSRES----QEKLFEGL 1709
Query: 621 KFYFSGDYVSSYKEDLEDLVEVGGGAVLTSKDKLEAKRHEFEVTSNLLIVYNLDPPQGSK 680
+ Y + + K++LE ++++ G +V+ L + ++ +++
Sbjct: 1710 QIYCCEPFTNMPKDELERMLQLCGASVVKELPLLTR-----DTGAHPIVLVQ-------- 1756
Query: 681 LGDEVSTLWQRLNEAEDLAANTGSQVIGHTWILESIA 717
+ W N+ D+ +++ W+L+SI+
Sbjct: 1757 -----PSAWTEDNDCPDIGQLCKGRLVMWDWVLDSIS 1788
Score = 50.1 bits (118), Expect = 2e-05
Identities = 34/123 (27%), Positives = 55/123 (44%), Gaps = 14/123 (11%)
Query: 28 LKCPLCLSLFEKPVLLPCDHLFCDSC---LVDSSLSGSECAVCKTKYA----QTDIRNVP 80
L+CP+CL L ++PV CDH+FC C L++ S+C +CK + Q R
Sbjct: 22 LECPICLELIKEPVSTQCDHIFCKFCMLKLLNQKKGPSQCPLCKNEITKRSLQGSARFSQ 81
Query: 81 FVENMVAIYRSLDASF---CAS--MLQQRSNDDSRVL-QQCQTFRDSSYSN-IKKADNFS 133
VE ++ I + + CA+ ++ N S +L + + Y N +KK
Sbjct: 82 LVEELLKIIDAFELDTGMQCANGFSFSKKKNSSSELLNEDASIIQSVGYRNRVKKLQQIE 141
Query: 134 QSS 136
S
Sbjct: 142 SGS 144
>BRC1_MACMU (Q6J6I9) Breast cancer type 1 susceptibility protein
homolog
Length = 1863
Score = 73.6 bits (179), Expect = 2e-12
Identities = 56/220 (25%), Positives = 96/220 (43%), Gaps = 26/220 (11%)
Query: 508 MAVVVSGRGE---VLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTLKVLMAILN 564
M++VVSG +L+ FA + +T + + THV+ TDA C RTLK + I
Sbjct: 1650 MSLVVSGLTPEEFMLVYKFARRYHIALTNLISEETTHVVMKTDAEFVCERTLKYFLGIAG 1709
Query: 565 GRWVLKIDWIKACMKVMSLVEEEPYEISLD--NQGCNGGPKAGRLSALANEPKLFSGLKF 622
G+WV+ W+ +K ++ E +E+ D N + GPK R S + K+F GL+
Sbjct: 1710 GKWVVSYFWVTQSIKERKMLNEHDFEVRGDVVNGRNHQGPKRARESP---DRKIFRGLEI 1766
Query: 623 YFSGDYVSSYKEDLEDLVEVGGGAVLTSKDKLEAKRHEFEVTSNLLIVYNLDPPQGSKLG 682
G + + + LE +V++ G +V+ E + ++V D
Sbjct: 1767 CCYGPFTNMPTDQLEWMVQLCGASVVK-----ELSSFTLGTGFHPIVVVQPD-------- 1813
Query: 683 DEVSTLWQRLNEAEDLAANTGSQVIGHTWILESIAACKLQ 722
W N + + V+ W+L+S+A + Q
Sbjct: 1814 -----AWTEDNGFHAIGQMCEAPVVTREWVLDSVALYQCQ 1848
Score = 48.1 bits (113), Expect = 8e-05
Identities = 31/125 (24%), Positives = 55/125 (43%), Gaps = 15/125 (12%)
Query: 28 LKCPLCLSLFEKPVLLPCDHLFCDSC---LVDSSLSGSECAVCKT----KYAQTDIRNVP 80
L+CP+CL L ++PV CDH+FC C L++ S+C +CK + Q R
Sbjct: 22 LECPICLELIKEPVSTKCDHIFCRFCMLKLLNQKKGPSQCPLCKNDITKRSLQESTRFSQ 81
Query: 81 FVENMVAIYRS------LDASFCASMLQQRSNDDSRVLQQCQTFRDSSYSNIKKADNFSQ 134
VE ++ I + L + + ++ ++ + + + Y N +A Q
Sbjct: 82 LVEELLKIIHAFQLDTGLQFANSYNFAKKENHSPEHLKDEVSIIQSMGYRN--RAKRLLQ 139
Query: 135 SSPNS 139
S P +
Sbjct: 140 SEPEN 144
>BRC1_CANFA (Q95153) Breast cancer type 1 susceptibility protein
homolog
Length = 1878
Score = 72.0 bits (175), Expect = 5e-12
Identities = 44/154 (28%), Positives = 80/154 (51%), Gaps = 4/154 (2%)
Query: 499 KGSIENLRVMAVVVSGRGEVLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTLKV 558
+G + + ++A ++ + +L+ FA K ++T + + THVI TDA C RTLK
Sbjct: 1647 RGVNKRISMVASGLTPKEFMLVHKFARKHHISLTNLISEETTHVIMKTDAEFVCERTLKY 1706
Query: 559 LMAILNGRWVLKIDWIKACMKVMSLVEEEPYEISLD--NQGCNGGPKAGRLS--ALANEP 614
+ I G+WV+ W+ +K +++E +E+ D N + GPK R S + +
Sbjct: 1707 FLGIAGGKWVVSYFWVTQSIKERKILDEHDFEVRGDVVNGRNHQGPKRARESQDRESQDR 1766
Query: 615 KLFSGLKFYFSGDYVSSYKEDLEDLVEVGGGAVL 648
K+F GL+ G + + + LE +V + G +V+
Sbjct: 1767 KIFRGLEICCYGPFTNMPTDQLEWMVHLCGASVV 1800
Score = 50.8 bits (120), Expect = 1e-05
Identities = 32/125 (25%), Positives = 56/125 (44%), Gaps = 15/125 (12%)
Query: 28 LKCPLCLSLFEKPVLLPCDHLFCDSC---LVDSSLSGSECAVCKT----KYAQTDIRNVP 80
L+CP+CL L ++PV CDH+FC C L++ S+C +CK + Q R
Sbjct: 22 LECPICLELIKEPVSTKCDHIFCKFCMLKLLNQRKGPSQCPLCKNDITKRSLQESTRFSQ 81
Query: 81 FVENMVAIYRS------LDASFCASMLQQRSNDDSRVLQQCQTFRDSSYSNIKKADNFSQ 134
VE ++ I + L + + ++ +N + ++ + Y N +A Q
Sbjct: 82 LVEELLKIIHAFELDTGLQFADSYNFSKKENNSPEHLKEEVSIIQSMGYRN--RAKRLRQ 139
Query: 135 SSPNS 139
S P +
Sbjct: 140 SEPEN 144
>TRX_DROVI (Q24742) Trithorax protein
Length = 3828
Score = 71.6 bits (174), Expect = 7e-12
Identities = 42/125 (33%), Positives = 64/125 (50%), Gaps = 20/125 (16%)
Query: 324 TSICSFCQSSETSEATGPMLHYANGKSVIGDAAMQPNVIH---VHRCCVDWAPQVYF-VD 379
T +C FC+ S G+ + G+ A H VH C W+ +V+ +D
Sbjct: 1708 TRVCLFCRKS--------------GEGLSGEEARLLYCGHDCWVHINCAMWSAEVFEEID 1753
Query: 380 ETCKNLKAEVARGAKLKCSTCGLKGAALGCYVKSCKRTYHVPCAMDVSTCRWDQEKYLLL 439
+ +N+ + VARG +KC+ CG +GA +GC VKSC YH PCA + C + +K +
Sbjct: 1754 GSLQNVHSAVARGRMIKCTVCGNRGATVGCNVKSCGEHYHYPCARTID-CAFLTDK-SMY 1811
Query: 440 CPVHS 444
CP H+
Sbjct: 1812 CPAHA 1816
>TRX_DROME (P20659) Trithorax protein
Length = 3726
Score = 71.6 bits (174), Expect = 7e-12
Identities = 42/131 (32%), Positives = 66/131 (50%), Gaps = 20/131 (15%)
Query: 318 IDLNPSTSICSFCQSSETSEATGPMLHYANGKSVIGDAAMQPNVIH---VHRCCVDWAPQ 374
I + T +C FC+ S G+ + G+ A H VH C W+ +
Sbjct: 1728 IKMRLDTRMCLFCRKS--------------GEGLSGEEARLLYCGHDCWVHTNCAMWSAE 1773
Query: 375 VYF-VDETCKNLKAEVARGAKLKCSTCGLKGAALGCYVKSCKRTYHVPCAMDVSTCRWDQ 433
V+ +D + +N+ + VARG +KC+ CG +GA +GC V+SC YH PCA + C +
Sbjct: 1774 VFEEIDGSLQNVHSAVARGRMIKCTVCGNRGATVGCNVRSCGEHYHYPCARSID-CAFLT 1832
Query: 434 EKYLLLCPVHS 444
+K + CP H+
Sbjct: 1833 DK-SMYCPAHA 1842
>TF20_MOUSE (Q9EPQ8) Transcription factor 20 (Stromelysin 1
PDGF-responsive element-binding protein) (SPRE-binding
protein) (Nuclear factor SPBP)
Length = 1983
Score = 71.2 bits (173), Expect = 9e-12
Identities = 78/317 (24%), Positives = 121/317 (37%), Gaps = 24/317 (7%)
Query: 154 MHVKPEELEMSSGGRAGFRNDVKPYPMQRSRVEIGDYVEMDVNQVTQAAVYSPPFCDTKG 213
++ + EE R G R+ P S+V + VT+++V C G
Sbjct: 1678 INAEEEEQTKLVRSRKGQRSLTPPPSSTESKVLPASSFMLQGPVVTESSVMGHLVCCLCG 1737
Query: 214 SDNDCSEL-DSDHPFNPE-----ILENSSLKRASTGKGNLKERKSQFRSESSASETDKPT 267
+ D PF P+ + +N KR+S + +K R +S S+ S+TD
Sbjct: 1738 KWASYRNMGDLFGPFYPQDYAATLPKNPPPKRSSEMQSKVKVRH---KSASNGSKTDTEE 1794
Query: 268 RDLKRKKYLTKGDDHIQHVSTHHSKLVDSHCGLDLKSG-KEPGELLPANIPIDLNPSTSI 326
+ ++++ Q H + H D G + LP S
Sbjct: 1795 EEEQQQQ-------KEQRSLAAHPRFKRRHRSEDCGGGPRSLSRGLPCKKAATEGSSEKT 1847
Query: 327 CSFCQSS-ETSEATGPMLHYANGKSVIGDAAMQPNVIHVHRCCVDWAPQVYFVDETCKNL 385
S + S T+ GP L I + + N VH C+ WA +Y V L
Sbjct: 1848 VSDTKPSVPTTSEGGPELELQ-----IPELPLDSNEFWVHEGCILWANGIYLVCGRLYGL 1902
Query: 386 KAEVARGAKLKCSTCGLKGAALGCYVKSCKRTYHVPCAMDVSTCRWDQEKYLLLCPVHSN 445
+ + ++KCS C GA LGCY K C YH PCA+D + C +E + + CP H
Sbjct: 1903 QEALEIAREMKCSHCQEAGATLGCYNKGCSFRYHYPCAID-ADCLLHEENFSVRCPKHKC 1961
Query: 446 AKFPHEKSRPKKQATQE 462
P + K + E
Sbjct: 1962 PLPPLQNKTAKGSLSTE 1978
>TF20_HUMAN (Q9UGU0) Transcription factor 20 (Stromelysin 1
PDGF-responsive element-binding protein) (SPRE-binding
protein) (Nuclear factor SPBP) (AR1)
Length = 1960
Score = 71.2 bits (173), Expect = 9e-12
Identities = 75/303 (24%), Positives = 115/303 (37%), Gaps = 33/303 (10%)
Query: 154 MHVKPEELEMSSGGRAGFRNDVKPYPMQRSRVEIGDYVEMDVNQVTQAAVYSPPFCDTKG 213
++ + EE GR G R+ P S+ + VT+++V C G
Sbjct: 1650 INAEEEEQTKLVRGRKGQRSLTPPPSSTESKALPASSFMLQGPVVTESSVMGHLVCCLCG 1709
Query: 214 SDNDCSEL-DSDHPFNPE-----ILENSSLKRASTGKGNLKERKSQFRSESSASETDKPT 267
+ D PF P+ + +N KRA+ + +K R S S ++T++
Sbjct: 1710 KWASYRNMGDLFGPFYPQDYAATLPKNPPPKRATEMQSKVKVRHKS-ASNGSKTDTEEEE 1768
Query: 268 RDLKRKKYLTKGDDHIQHVSTHHSKLVDSHCG-------LDLKSGKEPGELLPANIPIDL 320
+++K H + H S+ CG L K E +D
Sbjct: 1769 EQQQQQKEQRSLAAHPRFKRRHRSE----DCGGGPRSLSRGLPCKKAATEGSSEKTVLDS 1824
Query: 321 NPSTSICSFCQSSETSEATGPMLHYANGKSVIGDAAMQPNVIHVHRCCVDWAPQVYFVDE 380
PS T+ GP L I + + N VH C+ WA +Y V
Sbjct: 1825 KPSVP---------TTSEGGPELELQ-----IPELPLDSNEFWVHEGCILWANGIYLVCG 1870
Query: 381 TCKNLKAEVARGAKLKCSTCGLKGAALGCYVKSCKRTYHVPCAMDVSTCRWDQEKYLLLC 440
L+ + ++KCS C GA LGCY K C YH PCA+D + C +E + + C
Sbjct: 1871 RLYGLQEALEIAREMKCSHCQEAGATLGCYNKGCSFRYHYPCAID-ADCLLHEENFSVRC 1929
Query: 441 PVH 443
P H
Sbjct: 1930 PKH 1932
>PHF6_MOUSE (Q9D4J7) PHD finger protein 6
Length = 364
Score = 65.5 bits (158), Expect = 5e-10
Identities = 50/200 (25%), Positives = 81/200 (40%), Gaps = 18/200 (9%)
Query: 238 KRASTGKGNLKERKSQFRSESSASETDKPTRDLKRKKYLTKGDDHIQHVSTHHSKLVDSH 297
K A + +L+E ++ E S+ +T K +R + +K KG S+ SH
Sbjct: 133 KTAHNSEADLEESFNEHELEPSSPKTKKKSRKGRPRKTNLKG-------LPEDSRSTSSH 185
Query: 298 CGLDLKSGKEPGELLPANIPIDLNPSTSICSFCQSSETSEATGPMLHYANGKSVIGDAAM 357
+++S + P D P C FC E LH N K A
Sbjct: 186 GTDEMESSSYRDRSPHRSSPNDTRPK---CGFCHVGEEENEARGKLHIFNAKKAA--AHY 240
Query: 358 QPNVIHVHRCCVDWAPQVYFVDETCKNLKAEVARGAKLKCSTCGLKGAALGCYVKSCKRT 417
+ + + + F D K + E+ RG ++KC+ C GA +GC +K+C +T
Sbjct: 241 KCMLFSSGTVQLTTTSRAEFGDFDIKTVLQEIKRGKRMKCTLCSQPGATIGCEIKACVKT 300
Query: 418 YHVPCAMDVSTCRWDQEKYL 437
YH C + D+ KY+
Sbjct: 301 YHYHCGVQ------DKAKYI 314
Score = 58.2 bits (139), Expect = 8e-08
Identities = 44/147 (29%), Positives = 66/147 (43%), Gaps = 24/147 (16%)
Query: 327 CSFCQSSETSEATGPMLHYANGKSVIGDAAMQPNVIHVHRCCVDWAPQVYFVDE------ 380
C FC+S+ E G +L N K V H+C + + V +
Sbjct: 17 CGFCKSNRDKEC-GQLLISENQK-----------VAAHHKCMLFSSALVSSHSDNESLGG 64
Query: 381 -TCKNLKAEVARGAKLKCSTCGLKGAALGCYVKSCKRTYHVPCAM-DVSTCRW--DQEKY 436
+ ++++ E+ RG KL CS C GA +GC VK+C RTYH CA+ D + R Q Y
Sbjct: 65 FSIEDVQKEIKRGTKLMCSLCHCPGATIGCDVKTCHRTYHYHCALHDKAQIREKPSQGIY 124
Query: 437 LLLCPVHSNAKFPHEKSRPKKQATQEH 463
++ C H K H +++ EH
Sbjct: 125 MVYCRKHK--KTAHNSEADLEESFNEH 149
>PHF6_HUMAN (Q8IWS0) PHD finger protein 6 (PHD-like zinc finger
protein)
Length = 365
Score = 63.9 bits (154), Expect = 1e-09
Identities = 59/249 (23%), Positives = 96/249 (37%), Gaps = 34/249 (13%)
Query: 238 KRASTGKGNLKERKSQFRSESSASETDKPTRDLKRKKYLTKGDDHIQHVSTHHSKLVDSH 297
K A + +L+E ++ E S+ ++ K +R + +K KG + ++ SH
Sbjct: 133 KTAHNSEADLEESFNEHELEPSSPKSKKKSRKGRPRKTNFKG-------LSEDTRSTSSH 185
Query: 298 CGLDLKSGKEPGELLPANIPIDLNPSTSICSFCQSSETSEATGPMLHYANGKSVIGDAAM 357
+++S + P D P C FC E LH N K A
Sbjct: 186 GTDEMESSSYRDRSPHRSSPSDTRPK---CGFCHVGEEENEARGKLHIFNAKKAA--AHY 240
Query: 358 QPNVIHVHRCCVDWAPQVYFVDETCKNLKAEVARGAKLKCSTCGLKGAALGCYVKSCKRT 417
+ + + + F D K + E+ RG ++KC+ C GA +GC +K+C +T
Sbjct: 241 KCMLFSSGTVQLTTTSRAEFGDFDIKTVLQEIKRGKRMKCTLCSQPGATIGCEIKACVKT 300
Query: 418 YHVPCAMDVSTCRWDQEKYL---------LLCPVHSNAKFPHE-------KSRPKKQATQ 461
YH C + D+ KY+ L C HS E KSR K + Q
Sbjct: 301 YHYHCGVQ------DKAKYIENMSRGIYKLYCKNHSGNDERDEEDEERESKSRGKVEIDQ 354
Query: 462 EHPASAHLS 470
+ L+
Sbjct: 355 QQLTQQQLN 363
Score = 58.2 bits (139), Expect = 8e-08
Identities = 44/147 (29%), Positives = 66/147 (43%), Gaps = 24/147 (16%)
Query: 327 CSFCQSSETSEATGPMLHYANGKSVIGDAAMQPNVIHVHRCCVDWAPQVYFVDE------ 380
C FC+S+ E G +L N K V H+C + + V +
Sbjct: 17 CGFCKSNRDKEC-GQLLISENQK-----------VAAHHKCMLFSSALVSSHSDNESLGG 64
Query: 381 -TCKNLKAEVARGAKLKCSTCGLKGAALGCYVKSCKRTYHVPCAM-DVSTCRW--DQEKY 436
+ ++++ E+ RG KL CS C GA +GC VK+C RTYH CA+ D + R Q Y
Sbjct: 65 FSIEDVQKEIKRGTKLMCSLCHCPGATIGCDVKTCHRTYHYHCALHDKAQIREKPSQGIY 124
Query: 437 LLLCPVHSNAKFPHEKSRPKKQATQEH 463
++ C H K H +++ EH
Sbjct: 125 MVYCRKHK--KTAHNSEADLEESFNEH 149
>RAI1_HUMAN (Q7Z5J4) Retinoic acid induced protein 1
Length = 1906
Score = 62.4 bits (150), Expect = 4e-09
Identities = 30/80 (37%), Positives = 38/80 (47%), Gaps = 1/80 (1%)
Query: 364 VHRCCVDWAPQVYFVDETCKNLKAEVARGAKLKCSTCGLKGAALGCYVKSCKRTYHVPCA 423
VH C W VY V L+ + + CS+C GA +GC K C TYH PCA
Sbjct: 1824 VHEACAVWTGGVYLVAGKLFGLQEAMKVAVDMMCSSCQEAGATIGCCHKGCLHTYHYPCA 1883
Query: 424 MDVSTCRWDQEKYLLLCPVH 443
D C + +E + L CP H
Sbjct: 1884 SDAG-CIFIEENFSLKCPKH 1902
>RAI1_MOUSE (Q61818) Retinoic acid induced protein 1
Length = 1889
Score = 60.5 bits (145), Expect = 2e-08
Identities = 28/80 (35%), Positives = 37/80 (46%), Gaps = 1/80 (1%)
Query: 364 VHRCCVDWAPQVYFVDETCKNLKAEVARGAKLKCSTCGLKGAALGCYVKSCKRTYHVPCA 423
VH C W VY V L+ + + C++C GA + C K C TYH PCA
Sbjct: 1806 VHEACAVWTSGVYLVAGKLFGLQEAMKVAVDMPCTSCHEPGATISCSYKGCIHTYHYPCA 1865
Query: 424 MDVSTCRWDQEKYLLLCPVH 443
D C + +E + L CP H
Sbjct: 1866 NDTG-CTFIEENFTLKCPKH 1884
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.132 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 87,089,006
Number of Sequences: 164201
Number of extensions: 3726083
Number of successful extensions: 9607
Number of sequences better than 10.0: 209
Number of HSP's better than 10.0 without gapping: 153
Number of HSP's successfully gapped in prelim test: 56
Number of HSP's that attempted gapping in prelim test: 9341
Number of HSP's gapped (non-prelim): 289
length of query: 726
length of database: 59,974,054
effective HSP length: 118
effective length of query: 608
effective length of database: 40,598,336
effective search space: 24683788288
effective search space used: 24683788288
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0082b.8


