

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0082b.2
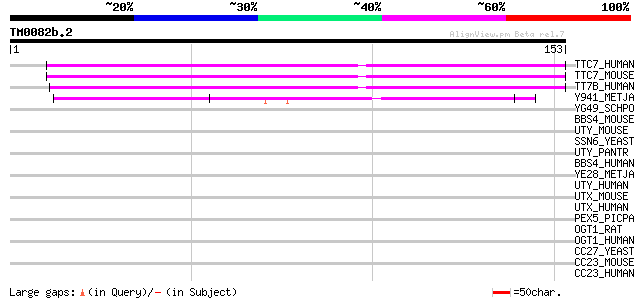
(153 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TTC7_HUMAN (Q9ULT0) Tetratricopeptide repeat protein 7A (TPR rep... 66 3e-11
TTC7_MOUSE (Q8BGB2) Tetratricopeptide repeat protein 7A (TPR rep... 66 3e-11
TT7B_HUMAN (Q86TV6) Tetratricopeptide repeat protein 7B (Tetratr... 64 1e-10
Y941_METJA (Q57711) Hypothetical protein MJ0941 42 4e-04
YG49_SCHPO (O60184) Protein C23E6.09 in chromosome II 40 0.002
BBS4_MOUSE (Q8C1Z7) Bardet-Biedl syndrome 4 protein homolog 38 0.010
UTY_MOUSE (P79457) Ubiquitously transcribed Y chromosome tetratr... 35 0.064
SSN6_YEAST (P14922) Glucose repression mediator protein 35 0.064
UTY_PANTR (Q6B4Z3) Ubiquitously transcribed Y chromosome tetratr... 34 0.14
BBS4_HUMAN (Q96RK4) Bardet-Biedl syndrome 4 protein 34 0.14
YE28_METJA (Q58823) Hypothetical protein MJ1428 33 0.19
UTY_HUMAN (O14607) Ubiquitously transcribed Y chromosome tetratr... 33 0.19
UTX_MOUSE (O70546) Ubiquitously transcribed X chromosome tetratr... 33 0.19
UTX_HUMAN (O15550) Ubiquitously transcribed X chromosome tetratr... 33 0.19
PEX5_PICPA (P33292) Peroxisomal targeting signal receptor (Perox... 33 0.19
OGT1_RAT (P56558) UDP-N-acetylglucosamine--peptide N-acetylgluco... 33 0.19
OGT1_HUMAN (O15294) UDP-N-acetylglucosamine--peptide N-acetylglu... 33 0.19
CC27_YEAST (P38042) Anaphase promoting complex subunit CDC27 (Ce... 33 0.19
CC23_MOUSE (Q8BGZ4) Cell division cycle protein 23 homolog (Anap... 33 0.24
CC23_HUMAN (Q9UJX2) Cell division cycle protein 23 homolog (Anap... 33 0.24
>TTC7_HUMAN (Q9ULT0) Tetratricopeptide repeat protein 7A (TPR repeat
protein 7)
Length = 858
Score = 66.2 bits (160), Expect = 3e-11
Identities = 40/143 (27%), Positives = 73/143 (50%), Gaps = 2/143 (1%)
Query: 11 EEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRD 70
E+IW A ++ +A C+ ++ + S ++ G + E KG +EA + +++
Sbjct: 711 EQIWLQAAELFMEQQHLKEAGFCIQEAAGLFPTSHSVLYMRGRLAEVKGNLEEAKQLYKE 770
Query: 71 ALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGR 130
AL ++P V + S + L R ++S + + L DA+ AW LG + +A+G+
Sbjct: 771 ALTVNPDGVRIMHSLGLMLSRLGHKS--LAQKVLRDAVERQSTCHEAWQGLGEVLQAQGQ 828
Query: 131 VLEAVECFQAANSLEETAPIEPF 153
AV+CF A LE ++P+ PF
Sbjct: 829 NEAAVDCFLTALELEASSPVLPF 851
>TTC7_MOUSE (Q8BGB2) Tetratricopeptide repeat protein 7A (TPR repeat
protein 7)
Length = 858
Score = 65.9 bits (159), Expect = 3e-11
Identities = 39/143 (27%), Positives = 74/143 (51%), Gaps = 2/143 (1%)
Query: 11 EEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRD 70
E+IW A ++ Q +A C+ ++ + S ++ G + E KG ++EA + +++
Sbjct: 711 EQIWLQAAELFMEQRQLKEAGFCIQEAAGLFPTSHSVLYMRGRLAEVKGSFEEAKQLYKE 770
Query: 71 ALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGR 130
AL ++P V + S + L + ++S + + L DA+ AW LG + + +G+
Sbjct: 771 ALTVNPDGVRIMHSLGLMLSQLGHKS--LAQKVLRDAVERQSTFHEAWQGLGEVLQDQGQ 828
Query: 131 VLEAVECFQAANSLEETAPIEPF 153
AV+CF A LE ++P+ PF
Sbjct: 829 NEAAVDCFLTALELEASSPVLPF 851
Score = 28.5 bits (62), Expect = 6.0
Identities = 26/109 (23%), Positives = 47/109 (42%), Gaps = 14/109 (12%)
Query: 34 LSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWS 93
L+ K QY ++G +H +G Y++AV + A ID V + + L
Sbjct: 111 LNNGKLPPQYMCEAMLILGKLHYVEGSYRDAVSMYARA-GIDDISVENKPLYQMRL---- 165
Query: 94 NQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGRVLEAVECFQAAN 142
+ +F++ L +RL S ++ + + E E V CF+ A+
Sbjct: 166 -----LSEAFVIKGLSLERLPNSVASHIRLTEREE----EVVACFERAS 205
>TT7B_HUMAN (Q86TV6) Tetratricopeptide repeat protein 7B
(Tetratricopeptide repeat protein 7 like-1)
Length = 741
Score = 63.9 bits (154), Expect = 1e-10
Identities = 40/142 (28%), Positives = 67/142 (47%), Gaps = 2/142 (1%)
Query: 12 EIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDA 71
+IW A YI + + +A C ++ + + ++ G + E +G EA + + +A
Sbjct: 595 QIWLHAAEVYIGIGKPAEATACTQEAANLFPMSHNVLYMRGQIAELRGSMDEARRWYEEA 654
Query: 72 LNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGRV 131
L I P HV S+ A+ L + S + L DA++ + W LG + +A+G
Sbjct: 655 LAISPTHVKSMQRLALILHQLGRYS--LAEKILRDAVQVNSTAHEVWNGLGEVLQAQGND 712
Query: 132 LEAVECFQAANSLEETAPIEPF 153
A ECF A LE ++P PF
Sbjct: 713 AAATECFLTALELEASSPAVPF 734
>Y941_METJA (Q57711) Hypothetical protein MJ0941
Length = 338
Score = 42.4 bits (98), Expect = 4e-04
Identities = 27/84 (32%), Positives = 40/84 (47%), Gaps = 2/84 (2%)
Query: 56 EAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNA 115
E G Y E +K + + L P VP + A LR+ D ++ L AL +
Sbjct: 142 EFLGEYDELLKCYNEVLTYTPNFVPMWVKKAEILRKLGRYEDALL--CLNRALELKPHDK 199
Query: 116 SAWYNLGILHKAEGRVLEAVECFQ 139
+A Y G+L K G+ EA+ECF+
Sbjct: 200 NALYLKGVLLKRMGKFREALECFK 223
Score = 41.6 bits (96), Expect = 7e-04
Identities = 30/140 (21%), Positives = 66/140 (46%), Gaps = 15/140 (10%)
Query: 13 IWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFR--- 69
+W A L ++ DA +CL+++ + + + ++ G + + G ++EA++ F+
Sbjct: 167 MWVKKAEILRKLGRYEDALLCLNRALELKPHDKNALYLKGVLLKRMGKFREALECFKKLI 226
Query: 70 DALNID----PRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILH 125
D LN+ RH SL+ L+ ++ L + + + WY G L+
Sbjct: 227 DELNVKWIDAIRHAVSLMLALDDLKD--------AERYINIGLEIRKDDVALWYFKGELY 278
Query: 126 KAEGRVLEAVECFQAANSLE 145
+ G++ EA++C++ L+
Sbjct: 279 ERLGKLDEALKCYEKVIELQ 298
>YG49_SCHPO (O60184) Protein C23E6.09 in chromosome II
Length = 1102
Score = 40.4 bits (93), Expect = 0.002
Identities = 30/117 (25%), Positives = 60/117 (50%), Gaps = 3/117 (2%)
Query: 34 LSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWS 93
L+KS A ++IG + A+ Y +A +A++ A+ D R+ P+ + L
Sbjct: 573 LTKSLEADDTDAQSWYLIGRCYVAQQKYNKAYEAYQQAVYRDGRN-PTFWCSIGVLYYQI 631
Query: 94 NQSDPVVRSFLMDALRYDRLNASAWYNLGILHKA-EGRVLEAVECFQAANSLEETAP 149
NQ + ++ A+R + + WY+LG L+++ ++ +A++ +Q A L+ T P
Sbjct: 632 NQYQDALDAYSR-AIRLNPYISEVWYDLGTLYESCHNQISDALDAYQRAAELDPTNP 687
>BBS4_MOUSE (Q8C1Z7) Bardet-Biedl syndrome 4 protein homolog
Length = 520
Score = 37.7 bits (86), Expect = 0.010
Identities = 29/143 (20%), Positives = 59/143 (40%), Gaps = 6/143 (4%)
Query: 13 IWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDAL 72
+W+++ + ++ A CL ++ + + +G +H Y A A+
Sbjct: 272 LWNNIGMCFFGKKKYVAAISCLKRANYLAPFDWKILYNLGLVHLTMQQYASAFHFLSAAI 331
Query: 73 NIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGRVL 132
N P+ + AVAL + + R ++A+R D+ N N +L +G
Sbjct: 332 NFQPKMGELYMLLAVALTNLEDIEN--ARRAYVEAVRLDKCNPLVNLNYAVLLYNQGEKK 389
Query: 133 EAVECFQ----AANSLEETAPIE 151
A+ +Q N L++ +P+E
Sbjct: 390 SALAQYQEMEKKVNFLKDNSPLE 412
Score = 32.3 bits (72), Expect = 0.41
Identities = 31/137 (22%), Positives = 61/137 (43%), Gaps = 11/137 (8%)
Query: 15 HDLAYAYISLSQWHDAEVCLSKS-----KA--FRQYTASRCHVIGTMHEAKGLYKEAVKA 67
HDL Y I L + H + L K+ KA F +G ++ G+Y++A +
Sbjct: 167 HDLTY--IMLGKIHLLQGDLDKAIEIYKKAVEFSPENTELLTTLGLLYLQLGVYQKAFEH 224
Query: 68 FRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKA 127
+AL DP + ++++ A ++ + D + + + A + W N+G+
Sbjct: 225 LGNALTYDPANYKAILA-AGSMMQTHGDFDVALTKYRVVACAIPE-SPPLWNNIGMCFFG 282
Query: 128 EGRVLEAVECFQAANSL 144
+ + + A+ C + AN L
Sbjct: 283 KKKYVAAISCLKRANYL 299
>UTY_MOUSE (P79457) Ubiquitously transcribed Y chromosome
tetratricopeptide repeat protein (Ubiquitously
transcribed TPR protein ON the Y chromosome)
(Male-specific histocompatibility antigen H-YDB)
Length = 1212
Score = 35.0 bits (79), Expect = 0.064
Identities = 20/90 (22%), Positives = 50/90 (55%), Gaps = 2/90 (2%)
Query: 49 HVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDAL 108
+ +G + G ++A ++R +++ + S V L + NQ +++++ A+
Sbjct: 283 YFLGRCYSCIGKVQDAFVSYRQSIDKSEASADTWCSIGV-LYQQQNQPMDALQAYIC-AV 340
Query: 109 RYDRLNASAWYNLGILHKAEGRVLEAVECF 138
+ D +A+AW +LGIL+++ + +A++C+
Sbjct: 341 QLDHGHAAAWMDLGILYESCNQPQDAIKCY 370
>SSN6_YEAST (P14922) Glucose repression mediator protein
Length = 966
Score = 35.0 bits (79), Expect = 0.064
Identities = 27/103 (26%), Positives = 48/103 (46%), Gaps = 4/103 (3%)
Query: 51 IGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQS--DPV-VRSFLMDA 107
+G++ E+ G ++ A +A+ L + H ++ L SN DP +L+ +
Sbjct: 230 LGSVLESMGEWQGAKEAYEHVLAQNQHHA-KVLQQLGCLYGMSNVQFYDPQKALDYLLKS 288
Query: 108 LRYDRLNASAWYNLGILHKAEGRVLEAVECFQAANSLEETAPI 150
L D +A+ WY+LG +H A + FQ A + + PI
Sbjct: 289 LEADPSDATTWYHLGRVHMIRTDYTAAYDAFQQAVNRDSRNPI 331
Score = 33.9 bits (76), Expect = 0.14
Identities = 30/124 (24%), Positives = 57/124 (45%), Gaps = 6/124 (4%)
Query: 26 QWHDAEVCLS---KSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSL 82
Q++D + L KS A+ + +G +H + Y A AF+ A+N D R+
Sbjct: 274 QFYDPQKALDYLLKSLEADPSDATTWYHLGRVHMIRTDYTAAYDAFQQAVNRDSRNPIFW 333
Query: 83 ISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHK-AEGRVLEAVECFQAA 141
S V + S D + A+R + + WY+LG L++ ++ +A++ ++ A
Sbjct: 334 CSIGVLYYQISQYRDAL--DAYTRAIRLNPYISEVWYDLGTLYETCNNQLSDALDAYKQA 391
Query: 142 NSLE 145
L+
Sbjct: 392 ARLD 395
Score = 30.0 bits (66), Expect = 2.1
Identities = 15/67 (22%), Positives = 32/67 (47%), Gaps = 1/67 (1%)
Query: 14 WHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHE-AKGLYKEAVKAFRDAL 72
W + Y +SQ+ DA +++ Y + + +GT++E +A+ A++ A
Sbjct: 333 WCSIGVLYYQISQYRDALDAYTRAIRLNPYISEVWYDLGTLYETCNNQLSDALDAYKQAA 392
Query: 73 NIDPRHV 79
+D +V
Sbjct: 393 RLDVNNV 399
>UTY_PANTR (Q6B4Z3) Ubiquitously transcribed Y chromosome
tetratricopeptide repeat protein (Ubiquitously
transcribed TPR protein on the Y chromosome)
Length = 1079
Score = 33.9 bits (76), Expect = 0.14
Identities = 22/105 (20%), Positives = 54/105 (50%), Gaps = 2/105 (1%)
Query: 34 LSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWS 93
L KS + + +G + + G ++A ++R +++ + S V L +
Sbjct: 270 LQKSLEADPNSGQSWYFLGRCYSSIGKVQDAFVSYRQSIDRSEASADTWCSIGV-LYQQQ 328
Query: 94 NQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGRVLEAVECF 138
NQ +++++ A++ D +A+AW +LG L+++ + +A++C+
Sbjct: 329 NQPIDALQAYIC-AVQLDHGHAAAWMDLGTLYESCNQPQDAIKCY 372
Score = 30.8 bits (68), Expect = 1.2
Identities = 34/147 (23%), Positives = 61/147 (41%), Gaps = 17/147 (11%)
Query: 12 EIWHDLAYAYISLSQWHDA----EVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKE---- 63
EI +A+ Y + ++H A E L Q A+ +G MH L +
Sbjct: 203 EIQFHIAHLYETQRKYHSAKEAYEQLLQTENLPAQVKATVLQQLGWMHHNMDLVGDKATK 262
Query: 64 ---AVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASA--W 118
A+ + +L DP S L R + V +F+ DR ASA W
Sbjct: 263 ESYAIPYLQKSLEADPNSGQSWYF----LGRCYSSIGKVQDAFVSYRQSIDRSEASADTW 318
Query: 119 YNLGILHKAEGRVLEAVECFQAANSLE 145
++G+L++ + + ++A++ + A L+
Sbjct: 319 CSIGVLYQQQNQPIDALQAYICAVQLD 345
>BBS4_HUMAN (Q96RK4) Bardet-Biedl syndrome 4 protein
Length = 519
Score = 33.9 bits (76), Expect = 0.14
Identities = 32/137 (23%), Positives = 61/137 (44%), Gaps = 11/137 (8%)
Query: 15 HDLAYAYISLSQWHDAEVCLSKS-----KA--FRQYTASRCHVIGTMHEAKGLYKEAVKA 67
HDL Y I L + H E L K+ KA F +G ++ G+Y++A +
Sbjct: 167 HDLTY--IMLGKIHLLEGDLDKAIEVYKKAVEFSPENTELLTTLGLLYLQLGIYQKAFEH 224
Query: 68 FRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKA 127
+AL DP + ++++ A ++ + D + + + A + W N+G+
Sbjct: 225 LGNALTYDPTNYKAILA-AGSMMQTHGDFDVALTKYRVVACAVPE-SPPLWNNIGMCFFG 282
Query: 128 EGRVLEAVECFQAANSL 144
+ + + A+ C + AN L
Sbjct: 283 KKKYVAAISCLKRANYL 299
Score = 31.6 bits (70), Expect = 0.71
Identities = 28/118 (23%), Positives = 51/118 (42%), Gaps = 3/118 (2%)
Query: 12 EIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDA 71
EI H+L YI L Q++ A+ L + ++ + ++G +H +G +A++ ++ A
Sbjct: 136 EISHNLGVCYIYLKQFNKAQDQLHNALNLNRHDLTYI-MLGKIHLLEGDLDKAIEVYKKA 194
Query: 72 LNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEG 129
+ P + L T + L L +AL YD N A G + + G
Sbjct: 195 VEFSPENTELL--TTLGLLYLQLGIYQKAFEHLGNALTYDPTNYKAILAAGSMMQTHG 250
Score = 30.8 bits (68), Expect = 1.2
Identities = 23/127 (18%), Positives = 49/127 (38%), Gaps = 2/127 (1%)
Query: 13 IWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDAL 72
+W+++ + ++ A CL ++ + + +G +H Y A A+
Sbjct: 272 LWNNIGMCFFGKKKYVAAISCLKRANYLAPFDWKILYNLGLVHLTMQQYASAFHFLSAAI 331
Query: 73 NIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGRVL 132
N P+ + AVAL + + + +A+ D+ N N +L +G
Sbjct: 332 NFQPKMGELYMLLAVALTNLEDTEN--AKRAYAEAVHLDKCNPLVNLNYAVLLYNQGEKK 389
Query: 133 EAVECFQ 139
A+ +Q
Sbjct: 390 NALAQYQ 396
>YE28_METJA (Q58823) Hypothetical protein MJ1428
Length = 567
Score = 33.5 bits (75), Expect = 0.19
Identities = 16/55 (29%), Positives = 34/55 (61%), Gaps = 1/55 (1%)
Query: 49 HVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSF 103
+V G ++ G YK+A ++F +AL ++P+ + +L S A+ L + S + D + ++
Sbjct: 383 YVKGYIYYKLGNYKDAYESFMNALRVNPKDISTLKSLAIVLEK-SGKIDEAITTY 436
>UTY_HUMAN (O14607) Ubiquitously transcribed Y chromosome
tetratricopeptide repeat protein (Ubiquitously
transcribed TPR protein on the Y chromosome)
Length = 1347
Score = 33.5 bits (75), Expect = 0.19
Identities = 22/105 (20%), Positives = 54/105 (50%), Gaps = 2/105 (1%)
Query: 34 LSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWS 93
L KS + + +G + + G ++A ++R +++ + S V L +
Sbjct: 270 LQKSLEADPNSGQSWYFLGRCYSSIGKVQDAFISYRQSIDKSEASADTWCSIGV-LYQQQ 328
Query: 94 NQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGRVLEAVECF 138
NQ +++++ A++ D +A+AW +LG L+++ + +A++C+
Sbjct: 329 NQPMDALQAYIC-AVQLDHGHAAAWMDLGTLYESCNQPQDAIKCY 372
Score = 30.8 bits (68), Expect = 1.2
Identities = 33/147 (22%), Positives = 62/147 (41%), Gaps = 17/147 (11%)
Query: 12 EIWHDLAYAYISLSQWHDA----EVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKE---- 63
EI +A+ Y + ++H A E L Q A+ +G MH L +
Sbjct: 203 EIQFHIAHLYETQRKYHSAKEAYEQLLQTENLPAQVKATVLQQLGWMHHNMDLVGDKATK 262
Query: 64 ---AVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASA--W 118
A++ + +L DP S L R + V +F+ D+ ASA W
Sbjct: 263 ESYAIQYLQKSLEADPNSGQSWYF----LGRCYSSIGKVQDAFISYRQSIDKSEASADTW 318
Query: 119 YNLGILHKAEGRVLEAVECFQAANSLE 145
++G+L++ + + ++A++ + A L+
Sbjct: 319 CSIGVLYQQQNQPMDALQAYICAVQLD 345
>UTX_MOUSE (O70546) Ubiquitously transcribed X chromosome
tetratricopeptide repeat protein (Ubiquitously
transcribed TPR protein on the X chromosome) (Fragment)
Length = 1333
Score = 33.5 bits (75), Expect = 0.19
Identities = 22/105 (20%), Positives = 54/105 (50%), Gaps = 2/105 (1%)
Query: 34 LSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWS 93
L KS + + +G + + G ++A ++R +++ + S V L +
Sbjct: 207 LQKSLEADPNSGQSWYFLGRCYSSIGKVQDAFISYRQSIDKSEASADTWCSIGV-LYQQQ 265
Query: 94 NQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGRVLEAVECF 138
NQ +++++ A++ D +A+AW +LG L+++ + +A++C+
Sbjct: 266 NQPMDALQAYIC-AVQLDHGHAAAWMDLGTLYESCNQPQDAIKCY 309
Score = 32.0 bits (71), Expect = 0.54
Identities = 33/147 (22%), Positives = 62/147 (41%), Gaps = 17/147 (11%)
Query: 12 EIWHDLAYAYISLSQWHDA----EVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKE---- 63
EI +A+ Y + ++H A E L Q A+ +G MH L +
Sbjct: 140 EIQFHIAHLYETQRKYHSAKEAYEQLLQTENLSAQVKATILQQLGWMHHTVDLLGDKATK 199
Query: 64 ---AVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASA--W 118
A++ + +L DP S L R + V +F+ D+ ASA W
Sbjct: 200 ESYAIQYLQKSLEADPNSGQSWYF----LGRCYSSIGKVQDAFISYRQSIDKSEASADTW 255
Query: 119 YNLGILHKAEGRVLEAVECFQAANSLE 145
++G+L++ + + ++A++ + A L+
Sbjct: 256 CSIGVLYQQQNQPMDALQAYICAVQLD 282
>UTX_HUMAN (O15550) Ubiquitously transcribed X chromosome
tetratricopeptide repeat protein (Ubiquitously
transcribed TPR protein on the X chromosome)
Length = 1401
Score = 33.5 bits (75), Expect = 0.19
Identities = 22/105 (20%), Positives = 54/105 (50%), Gaps = 2/105 (1%)
Query: 34 LSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWS 93
L KS + + +G + + G ++A ++R +++ + S V L +
Sbjct: 273 LQKSLEADPNSGQSWYFLGRCYSSIGKVQDAFISYRQSIDKSEASADTWCSIGV-LYQQQ 331
Query: 94 NQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGRVLEAVECF 138
NQ +++++ A++ D +A+AW +LG L+++ + +A++C+
Sbjct: 332 NQPMDALQAYIC-AVQLDHGHAAAWMDLGTLYESCNQPQDAIKCY 375
Score = 32.0 bits (71), Expect = 0.54
Identities = 33/147 (22%), Positives = 62/147 (41%), Gaps = 17/147 (11%)
Query: 12 EIWHDLAYAYISLSQWHDA----EVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKE---- 63
EI +A+ Y + ++H A E L Q A+ +G MH L +
Sbjct: 206 EIQFHIAHLYETQRKYHSAKEAYEQLLQTENLSAQVKATVLQQLGWMHHTVDLLGDKATK 265
Query: 64 ---AVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASA--W 118
A++ + +L DP S L R + V +F+ D+ ASA W
Sbjct: 266 ESYAIQYLQKSLEADPNSGQSWYF----LGRCYSSIGKVQDAFISYRQSIDKSEASADTW 321
Query: 119 YNLGILHKAEGRVLEAVECFQAANSLE 145
++G+L++ + + ++A++ + A L+
Sbjct: 322 CSIGVLYQQQNQPMDALQAYICAVQLD 348
>PEX5_PICPA (P33292) Peroxisomal targeting signal receptor
(Peroxisomal protein PAS8) (Peroxin-5) (PTS1 receptor)
Length = 576
Score = 33.5 bits (75), Expect = 0.19
Identities = 32/143 (22%), Positives = 55/143 (38%), Gaps = 23/143 (16%)
Query: 7 RNLEEEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYK---- 62
+N E++W D+ +Y + +D + K F QY R + +E K ++
Sbjct: 221 QNTFEQVWDDIQVSYADVELTND-QFQAQWEKDFAQYAEGRLNYGEYKYEEKNQFRNDPD 279
Query: 63 ----------------EAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMD 106
EA AF A+ DP+HV + + + +SD + + L
Sbjct: 280 AYEIGMRLMESGAKLSEAGLAFEAAVQQDPKHVDAWLKLGEVQTQNEKESDGI--AALEK 337
Query: 107 ALRYDRLNASAWYNLGILHKAEG 129
L D N +A L I + +G
Sbjct: 338 CLELDPTNLAALMTLAISYINDG 360
Score = 30.0 bits (66), Expect = 2.1
Identities = 24/101 (23%), Positives = 45/101 (43%), Gaps = 2/101 (1%)
Query: 51 IGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRY 110
+G + + + + + F+ A+ ++P + AL + N+ + V ++ AL+
Sbjct: 428 LGVLFYSMEEFDKTIDCFKAAIEVEPDKALNWNRLGAALANY-NKPEEAVEAYSR-ALQL 485
Query: 111 DRLNASAWYNLGILHKAEGRVLEAVECFQAANSLEETAPIE 151
+ A YNLG+ GR EAVE SL E ++
Sbjct: 486 NPNFVRARYNLGVSFINMGRYKEAVEHLLTGISLHEVEGVD 526
>OGT1_RAT (P56558) UDP-N-acetylglucosamine--peptide
N-acetylglucosaminyltransferase 110 kDa subunit (EC
2.4.1.-) (O-GlcNAc transferase p110 subunit)
Length = 1036
Score = 33.5 bits (75), Expect = 0.19
Identities = 25/115 (21%), Positives = 53/115 (45%), Gaps = 9/115 (7%)
Query: 38 KAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSD 97
+ F ++ A+ ++ + + +G +EA+ +++A+ I P + + L+ +
Sbjct: 345 EVFPEFAAAHSNLASVLQQ-QGKLQEALMHYKEAIRISPTFADAYSNMGNTLKEMQD--- 400
Query: 98 PVVRSFLMDALRYDRLN---ASAWYNLGILHKAEGRVLEAVECFQAANSLEETAP 149
V+ L R ++N A A NL +HK G + EA+ ++ A L+ P
Sbjct: 401 --VQGALQCYTRAIQINPAFADAHSNLASIHKDSGNIPEAIASYRTALKLKPDFP 453
Score = 32.7 bits (73), Expect = 0.32
Identities = 25/113 (22%), Positives = 50/113 (44%), Gaps = 2/113 (1%)
Query: 29 DAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVA 88
+AE C + + A + + + +G +EAV+ +R AL + P + + A
Sbjct: 301 EAEDCYNTALRLCPTHADSLNNLANIKREQGNIEEAVRLYRKALEVFPEFAAAHSNLASV 360
Query: 89 LRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGRVLEAVECFQAA 141
L++ + ++ +A+R A A+ N+G K V A++C+ A
Sbjct: 361 LQQQGKLQEALMH--YKEAIRISPTFADAYSNMGNTLKEMQDVQGALQCYTRA 411
Score = 31.6 bits (70), Expect = 0.71
Identities = 30/134 (22%), Positives = 59/134 (43%), Gaps = 2/134 (1%)
Query: 16 DLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALNID 75
DL +L + +A+ C K+ + A +G + A+G A+ F A+ +D
Sbjct: 152 DLGNLLKALGRLEEAKACYLKAIETQPNFAVAWSNLGCVFNAQGEIWLAIHHFEKAVTLD 211
Query: 76 PRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGRVLEAV 135
P + + I+ L+ + D V ++L AL +A NL ++ +G + A+
Sbjct: 212 PNFLDAYINLGNVLKE-ARIFDRAVAAYLR-ALSLSPNHAVVHGNLACVYYEQGLIDLAI 269
Query: 136 ECFQAANSLEETAP 149
+ ++ A L+ P
Sbjct: 270 DTYRRAIELQPHFP 283
Score = 28.5 bits (62), Expect = 6.0
Identities = 14/39 (35%), Positives = 26/39 (65%)
Query: 107 ALRYDRLNASAWYNLGILHKAEGRVLEAVECFQAANSLE 145
A++ + L A A+ NLG ++K G++ EA+E ++ A L+
Sbjct: 71 AIKQNPLLAEAYSNLGNVYKERGQLQEAIEHYRHALRLK 109
>OGT1_HUMAN (O15294) UDP-N-acetylglucosamine--peptide
N-acetylglucosaminyltransferase 110 kDa subunit (EC
2.4.1.-) (O-GlcNAc transferase p110 subunit)
Length = 1036
Score = 33.5 bits (75), Expect = 0.19
Identities = 25/115 (21%), Positives = 53/115 (45%), Gaps = 9/115 (7%)
Query: 38 KAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSD 97
+ F ++ A+ ++ + + +G +EA+ +++A+ I P + + L+ +
Sbjct: 345 EVFPEFAAAHSNLASVLQQ-QGKLQEALMHYKEAIRISPTFADAYSNMGNTLKEMQD--- 400
Query: 98 PVVRSFLMDALRYDRLN---ASAWYNLGILHKAEGRVLEAVECFQAANSLEETAP 149
V+ L R ++N A A NL +HK G + EA+ ++ A L+ P
Sbjct: 401 --VQGALQCYTRAIQINPAFADAHSNLASIHKDSGNIPEAIASYRTALKLKPDFP 453
Score = 32.7 bits (73), Expect = 0.32
Identities = 25/113 (22%), Positives = 50/113 (44%), Gaps = 2/113 (1%)
Query: 29 DAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVA 88
+AE C + + A + + + +G +EAV+ +R AL + P + + A
Sbjct: 301 EAEDCYNTALRLCPTHADSLNNLANIKREQGNIEEAVRLYRKALEVFPEFAAAHSNLASV 360
Query: 89 LRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGRVLEAVECFQAA 141
L++ + ++ +A+R A A+ N+G K V A++C+ A
Sbjct: 361 LQQQGKLQEALMH--YKEAIRISPTFADAYSNMGNTLKEMQDVQGALQCYTRA 411
Score = 31.6 bits (70), Expect = 0.71
Identities = 30/134 (22%), Positives = 59/134 (43%), Gaps = 2/134 (1%)
Query: 16 DLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALNID 75
DL +L + +A+ C K+ + A +G + A+G A+ F A+ +D
Sbjct: 152 DLGNLLKALGRLEEAKACYLKAIETQPNFAVAWSNLGCVFNAQGEIWLAIHHFEKAVTLD 211
Query: 76 PRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGRVLEAV 135
P + + I+ L+ + D V ++L AL +A NL ++ +G + A+
Sbjct: 212 PNFLDAYINLGNVLKE-ARIFDRAVAAYLR-ALSLSPNHAVVHGNLACVYYEQGLIDLAI 269
Query: 136 ECFQAANSLEETAP 149
+ ++ A L+ P
Sbjct: 270 DTYRRAIELQPHFP 283
Score = 28.5 bits (62), Expect = 6.0
Identities = 14/39 (35%), Positives = 26/39 (65%)
Query: 107 ALRYDRLNASAWYNLGILHKAEGRVLEAVECFQAANSLE 145
A++ + L A A+ NLG ++K G++ EA+E ++ A L+
Sbjct: 71 AIKQNPLLAEAYSNLGNVYKERGQLQEAIEHYRHALRLK 109
>CC27_YEAST (P38042) Anaphase promoting complex subunit CDC27 (Cell
division control protein 27) (Anaphase promoting complex
subunit 3)
Length = 758
Score = 33.5 bits (75), Expect = 0.19
Identities = 26/94 (27%), Positives = 41/94 (42%), Gaps = 2/94 (2%)
Query: 51 IGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRY 110
IG + + + A+KAF A +DP + T SN S ++ AL
Sbjct: 546 IGNLLSLQKDHDAAIKAFEKATQLDPNF--AYAYTLQGHEHSSNDSSDSAKTCYRKALAC 603
Query: 111 DRLNASAWYNLGILHKAEGRVLEAVECFQAANSL 144
D + +A+Y LG G+ EA+ F+ A S+
Sbjct: 604 DPQHYNAYYGLGTSAMKLGQYEEALLYFEKARSI 637
>CC23_MOUSE (Q8BGZ4) Cell division cycle protein 23 homolog
(Anaphase promoting complex subunit 8) (APC8) (Cyclosome
subunit 8)
Length = 597
Score = 33.1 bits (74), Expect = 0.24
Identities = 19/81 (23%), Positives = 36/81 (43%), Gaps = 2/81 (2%)
Query: 42 QYTASRCHVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVR 101
+Y C VIG + + +++A F+ AL ++PR++ + N S +
Sbjct: 328 KYRVETCCVIGNYYSLRSQHEKAALYFQRALKLNPRYLGAWTLMGHEYMEMKNTSAAI-- 385
Query: 102 SFLMDALRYDRLNASAWYNLG 122
A+ ++ + AWY LG
Sbjct: 386 QAYRHAIEVNKRDYRAWYGLG 406
>CC23_HUMAN (Q9UJX2) Cell division cycle protein 23 homolog
(Anaphase promoting complex subunit 8) (APC8) (Cyclosome
subunit 8)
Length = 591
Score = 33.1 bits (74), Expect = 0.24
Identities = 19/81 (23%), Positives = 36/81 (43%), Gaps = 2/81 (2%)
Query: 42 QYTASRCHVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVR 101
+Y C VIG + + +++A F+ AL ++PR++ + N S +
Sbjct: 322 KYRVETCCVIGNYYSLRSQHEKAALYFQRALKLNPRYLGAWTLMGHEYMEMKNTSAAI-- 379
Query: 102 SFLMDALRYDRLNASAWYNLG 122
A+ ++ + AWY LG
Sbjct: 380 QAYRHAIEVNKRDYRAWYGLG 400
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.133 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,791,715
Number of Sequences: 164201
Number of extensions: 583013
Number of successful extensions: 1669
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 36
Number of HSP's that attempted gapping in prelim test: 1564
Number of HSP's gapped (non-prelim): 126
length of query: 153
length of database: 59,974,054
effective HSP length: 101
effective length of query: 52
effective length of database: 43,389,753
effective search space: 2256267156
effective search space used: 2256267156
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0082b.2


