

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
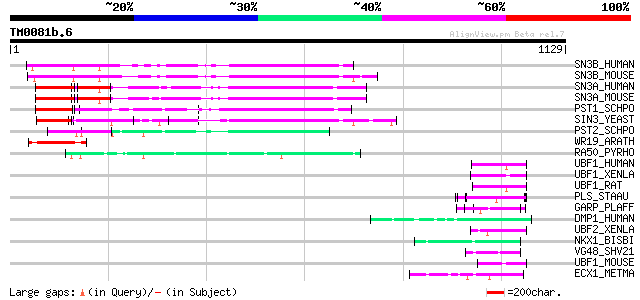
Query= TM0081b.6
(1129 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SN3B_HUMAN (O75182) Paired amphipathic helix protein Sin3b 343 1e-93
SN3B_MOUSE (Q62141) Paired amphipathic helix protein Sin3b 338 4e-92
SN3A_HUMAN (Q96ST3) Paired amphipathic helix protein Sin3a 278 8e-74
SN3A_MOUSE (Q60520) Paired amphipathic helix protein Sin3a 272 3e-72
PST1_SCHPO (Q09750) Paired amphipathic helix protein pst1 (SIN3 ... 192 6e-48
SIN3_YEAST (P22579) Paired amphipathic helix protein SIN3 166 3e-40
PST2_SCHPO (O13919) Paired amphipathic helix protein pst2 (SIN3 ... 143 2e-33
WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY ... 103 3e-21
RA50_PYRHO (O58687) DNA double-strand break repair rad50 ATPase 63 4e-09
UBF1_HUMAN (P17480) Nucleolar transcription factor 1 (Upstream b... 59 1e-07
UBF1_XENLA (P25979) Nucleolar transcription factor 1 (Upstream b... 57 2e-07
UBF1_RAT (P25977) Nucleolar transcription factor 1 (Upstream bin... 56 5e-07
PLS_STAAU (P80544) Surface protein precursor (Plasmin-sensitive ... 55 1e-06
GARP_PLAFF (P13816) Glutamic acid-rich protein precursor 55 1e-06
DMP1_HUMAN (Q13316) Dentin matrix acidic phosphoprotein 1 precur... 53 4e-06
UBF2_XENLA (P25980) Nucleolar transcription factor 2 (Upstream b... 52 9e-06
NKX1_BISBI (O46383) Sodium/potassium/calcium exchanger 1 (Na(+)/... 52 9e-06
VG48_SHV21 (Q01033) Hypothetical gene 48 protein 51 2e-05
UBF1_MOUSE (P25976) Nucleolar transcription factor 1 (Upstream b... 51 2e-05
ECX1_METMA (Q8PTT8) Probable exosome complex exonuclease 1 (EC 3... 50 3e-05
>SN3B_HUMAN (O75182) Paired amphipathic helix protein Sin3b
Length = 1130
Score = 343 bits (881), Expect = 1e-93
Identities = 243/731 (33%), Positives = 346/731 (47%), Gaps = 163/731 (22%)
Query: 35 GGGGGGGGAGG--------------GGEATTSQKLTTNDALSYLKQVKDMFQDQREKYDL 80
GGG GG GAGG G + DAL+YL QVK F Y+
Sbjct: 5 GGGSGGSGAGGPAGRGLSGARWGRSGSAGHEKLPVHVEDALTYLDQVKIRFGSDPATYNG 64
Query: 81 FLEVMKDFKAQRTDTAGVIARVKELFKGHNHLIFGFNTFLPKGYEITL------------ 128
FLE+MK+FK+Q DT GVI RV +LF H LI GFN FLP GY I +
Sbjct: 65 FLEIMKEFKSQSIDTPGVIRRVSQLFHEHPDLIVGFNAFLPLGYRIDIPKNGKLNIQSPL 124
Query: 129 ------------------------DEDEAPAKK-TVEFEEAISFVNKIKKRFQSDEHVYK 163
D+ + P + +VEF AIS+VNKIK RF +Y+
Sbjct: 125 TSQENSHNHGDGAEDFKQQVPYKEDKPQVPLESDSVEFNNAISYVNKIKTRFLDHPEIYR 184
Query: 164 SFLDILNMYRKEHKDIG----------EVYSEVATLFKDHRDLLEEFTRFLPDTSAAPST 213
SFL+IL+ Y+KE + EV++EVA LF+ DLL EF +FLP+ + T
Sbjct: 185 SFLEILHTYQKEQLNTRGRPFRGMSEEEVFTEVANLFRGQEDLLSEFGQFLPEAKRSLFT 244
Query: 214 QHAPFGRNSLQRFNERNSMTPMMRQMQVDKQRYRRDRLPSHDRDRDLSVEHPEMDDDKTM 273
+ P +S+Q+ NE D+ P H R R ++
Sbjct: 245 GNGPCEMHSVQK-NEH-------------------DKTPEHSRKRSRP----------SL 274
Query: 274 INLHKEQRKRDRRIRDQDERDPDLDNSRDLTSQRFRDKKKTVKKAEGMYG--EAFSFCEK 331
+ K+ ++R ++DL+ A G YG + FSF +K
Sbjct: 275 LRPVSAPAKKKMKLR----------GTKDLSIA-----------AVGKYGTLQEFSFFDK 313
Query: 332 VKEKLSSSDDYQTFLKCLNIFNNGIIKKNDLQNLVTDLLGKHSDLMDEFKDFLERCENIE 391
V+ L S + Y+ FL+C+ +FN ++ ++L LV+ LGK +L +FK FL
Sbjct: 314 VRRVLKSQEVYENFLRCIALFNQELVSGSELLQLVSPFLGKFPELFAQFKSFL------- 366
Query: 392 GFLAGVMSKKSLSTDAHLSRSSKLEDKDKDQKREMDGAKEKDRYKEKYMGKSIQELDLSD 451
GV LS + + D+ D G S +E+D +
Sbjct: 367 ----GVKE---------LSFAPPMSDRSGD-------------------GIS-REIDYAS 393
Query: 452 CKRCTPSYRLLPSDYPIPTASQRSELGAQVLNDHWVSVTSGSEDYSFKHMRKNQYEESLF 511
CKR SYR LP Y P S R+ + +VLND WVS S SED +F +K YEE L
Sbjct: 394 CKRIGSSYRALPKTYQQPKCSGRTAICKEVLNDTWVSFPSWSEDSTFVSSKKTPYEEQLH 453
Query: 512 RCEDDRFELDLLLESVSSASKRAEELYNNINENKISVEALSRIEDHF----TVLNLRCIE 567
RCED+RFELD++LE+ + + E + ++ + R++D V+ R I
Sbjct: 454 RCEDERFELDVVLETNLATIRVLESVQKKLSRMAPEDQEKFRLDDSLGGTSEVIQRRAIY 513
Query: 568 RLYGDHGLDVIDILRKNPTHALPVILTRLKQKQEEWNRCRSDFNKVWAEIYAKNHYKSLD 627
R+YGD ++I+ L+KNP A+PV+L RLK K+EEW + FNK+W E Y K + KSLD
Sbjct: 514 RIYGDKAPEIIESLKKNPVTAVPVVLKRLKAKEEEWREAQQGFNKIWREQYEKAYLKSLD 573
Query: 628 HRSFYFKQQDSKNLSTKSLVAEIKEIKEKLQKEDHIIQSIAAENRQPLIPHLEFEYSDGG 687
H++ FKQ D+K L +KSL+ EI+ + ++ Q++ +S + PHL F Y D
Sbjct: 574 HQAVNFKQNDTKALRSKSLLNEIESVYDEHQEQHSEGRSAPSSE-----PHLIFVYEDRQ 628
Query: 688 IHEDLYKLVQY 698
I ED L+ Y
Sbjct: 629 ILEDAAALISY 639
>SN3B_MOUSE (Q62141) Paired amphipathic helix protein Sin3b
Length = 954
Score = 338 bits (868), Expect = 4e-92
Identities = 252/780 (32%), Positives = 362/780 (46%), Gaps = 167/780 (21%)
Query: 36 GGGGGGGAGGGGE------ATTSQKLTTN--DALSYLKQVKDMFQDQREKYDLFLEVMKD 87
G GG G G GG + +KL + DAL+YL QVK F Y+ FLE+MK+
Sbjct: 5 GSGGSAGRGFGGSRWGRSGSGGHEKLPVHVEDALTYLDQVKIRFGSDPATYNGFLEIMKE 64
Query: 88 FKAQRTDTAGVIARVKELFKGHNHLIFGFNTFLPKGYEITL------------------- 128
FK+Q DT GVI RV +LF H LI GFN FLP GY I +
Sbjct: 65 FKSQSIDTPGVIRRVSQLFHEHPDLIVGFNAFLPLGYRIDIPKNGKLNIQSPLSSQDNSH 124
Query: 129 ----------------DEDEAPAKK-TVEFEEAISFVNKIKKRFQSDEHVYKSFLDILNM 171
D + P + +VEF AIS+VNKIK RF +Y+SFL+IL+
Sbjct: 125 SHGDCGEDFKQMSYKEDRGQVPLESDSVEFNNAISYVNKIKTRFLDHPEIYRSFLEILHT 184
Query: 172 YRKEHKDIG----------EVYSEVATLFKDHRDLLEEFTRFLPDTSAAPSTQHAPFGRN 221
Y+KE EV++EVA LF+ DLL EF +FLP+ + T + N
Sbjct: 185 YQKEQLHTKGRPFRGMSEEEVFTEVANLFRGQEDLLSEFGQFLPEAKRSLFTGNGSCEMN 244
Query: 222 SLQRFNERNSMTPMMRQMQVDKQRYRRDRLPSHDRDRDLSVEHPEMDDDKTMINLHKEQR 281
S Q+ E+ S+EH + +++
Sbjct: 245 SGQKNEEK-------------------------------SLEHNKKRSRPSLLRPVSAPA 273
Query: 282 KRDRRIRDQDERDPDLDNSRDLTSQRFRDKKKTVKKAEGMYG--EAFSFCEKVKEKLSSS 339
K+ ++R ++DL+ A G YG + FSF +KV+ L S
Sbjct: 274 KKKMKLR----------GTKDLSIA-----------AVGKYGTLQEFSFFDKVRRVLKSQ 312
Query: 340 DDYQTFLKCLNIFNNGIIKKNDLQNLVTDLLGKHSDLMDEFKDFLERCENIEGFLAGVMS 399
+ Y+ FL+C+ +FN ++ ++L LV+ LGK +L +FK FL GV
Sbjct: 313 EVYENFLRCIALFNQELVSGSELLQLVSPFLGKFPELFAQFKSFL-----------GVKE 361
Query: 400 KKSLSTDAHLSRSSKLEDKDKDQKREMDGAKEKDRYKEKYMGKSIQELDLSDCKRCTPSY 459
LS + + D+ D G S +E+D + CKR SY
Sbjct: 362 ---------LSFAPPMSDRSGD-------------------GIS-REIDYASCKRIGSSY 392
Query: 460 RLLPSDYPIPTASQRSELGAQVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFE 519
R LP Y P S R+ + +VLND WVS S SED +F +K YEE L RCED+RFE
Sbjct: 393 RALPKTYQQPKCSGRTAICKEVLNDTWVSFPSWSEDSTFVSSKKTPYEEQLHRCEDERFE 452
Query: 520 LDLLLESVSSASKRAEELYNNINENKISVEALSRIEDHF----TVLNLRCIERLYGDHGL 575
LD++LE+ + + E + ++ + R++D V+ R I R+YGD
Sbjct: 453 LDVVLETNLATIRVLESVQKKLSRMAPEDQEKLRLDDCLGGTSEVIQRRAIHRIYGDKAP 512
Query: 576 DVIDILRKNPTHALPVILTRLKQKQEEWNRCRSDFNKVWAEIYAKNHYKSLDHRSFYFKQ 635
+VI+ L++NP A+PV+L RLK K+EEW + FNK+W E Y K + KSLDH++ FKQ
Sbjct: 513 EVIESLKRNPATAVPVVLKRLKAKEEEWREAQQGFNKIWREQYEKAYLKSLDHQAVNFKQ 572
Query: 636 QDSKNLSTKSLVAEIKEIKEKLQKEDHIIQSIAAENRQPLIPHLEFEYSDGGIHEDLYKL 695
D+K L +KSL+ EI+ + ++ Q++ +S + PHL F Y D I ED L
Sbjct: 573 NDTKALRSKSLLNEIESVYDEHQEQHSEGRSAPSSE-----PHLIFVYEDRQILEDAAAL 627
Query: 696 VQY--------SCEEVFSSKELLNKIMRLWSTFLEPMLGVTSQSHGTERVEDRKAGHSSR 747
+ Y E+ + ++LL++ L S F TS ER DR + R
Sbjct: 628 ISYYVKRQPAIQKEDQGTIRQLLHRF--LPSLFFSQQCPGTSDDSADERDRDRDSAEPER 685
>SN3A_HUMAN (Q96ST3) Paired amphipathic helix protein Sin3a
Length = 1273
Score = 278 bits (710), Expect = 8e-74
Identities = 198/622 (31%), Positives = 307/622 (48%), Gaps = 105/622 (16%)
Query: 126 ITLDEDEAPA---KKTVEFEEAISFVNKIKKRFQSDEHVYKSFLDILNMYRKEH---KDI 179
+T+ AP+ + VEF AI++VNKIK RFQ +YK+FL+IL+ Y+KE K+
Sbjct: 286 VTISLGTAPSLQNNQPVEFNHAINYVNKIKNRFQGQPDIYKAFLEILHTYQKEQRNAKEA 345
Query: 180 G----------EVYSEVATLFKDHRDLLEEFTRFLPDTSAAPSTQHAPFGRNSLQRFNER 229
G EVY++VA LFK+ DLL EF +FLPD +++
Sbjct: 346 GGNYTPALTEQEVYAQVARLFKNQEDLLSEFGQFLPDANSS------------------- 386
Query: 230 NSMTPMMRQMQVDKQRYRRDRLPSHDRDRDLSVEHPEMDDDKTMINLHKEQ-RKRDRRIR 288
V + +++ S D +V+ P++++ + + Q R+
Sbjct: 387 -----------VLLSKTTAEKVDSVRNDHGGTVKKPQLNNKPQRPSQNGCQIRRHPTGTT 435
Query: 289 DQDERDPDLDNSRDLTSQRFRDKKKTVKKAEGMYGEAFSFCEKVKEKLSSSDDYQTFLKC 348
++ P L N +D + D K E + F +KV++ L S++ Y+ FL+C
Sbjct: 436 PPVKKKPKLLNLKDSS---MADASKHGGGTESL------FFDKVRKALRSAEAYENFLRC 486
Query: 349 LNIFNNGIIKKNDLQNLVTDLLGKHSDLMDEFKDFLERCENIEGFLAGVMSKKSLSTDAH 408
L IFN +I + +L LV+ LGK +L + FK+FL E++ H
Sbjct: 487 LVIFNQEVISRAELVQLVSPFLGKFPELFNWFKNFLGYKESV-----------------H 529
Query: 409 LSRSSKLEDKDKDQKREMDGAKEKDRYKEKYMGKSIQELDLSDCKRCTPSYRLLPSDYPI 468
L K +R +G E+D + CKR SYR LP Y
Sbjct: 530 LETYPK--------ERATEGI--------------AMEIDYASCKRLGSSYRALPKSYQQ 567
Query: 469 PTASQRSELGAQVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDLLLESVS 528
P + R+ L +VLND WVS S SED +F +K QYEE ++RCED+RFELD++LE+
Sbjct: 568 PKCTGRTPLCKEVLNDTWVSFPSWSEDSTFVSSKKTQYEEHIYRCEDERFELDVVLETNL 627
Query: 529 SASKRAEELYNNINENKISVEALSRIEDHF----TVLNLRCIERLYGDHGLDVIDILRKN 584
+ + E + ++ +A R+++ V++ + ++R+Y D D+ID LRKN
Sbjct: 628 ATIRVLEAIQKKLSRLSAEEQAKFRLDNTLGGTSEVIHRKALQRIYADKAADIIDGLRKN 687
Query: 585 PTHALPVILTRLKQKQEEWNRCRSDFNKVWAEIYAKNHYKSLDHRSFYFKQQDSKNLSTK 644
P+ A+P++L RLK K+EEW + FNKVW E K + KSLDH+ FKQ D+K L +K
Sbjct: 688 PSIAVPIVLKRLKMKEEEWREAQRGFNKVWREQNEKYYLKSLDHQGINFKQNDTKVLRSK 747
Query: 645 SLVAEIKEIKEKLQKEDHIIQSIAAENRQPLIPHLEFEYSDGGIHEDLYKLVQYSCE-EV 703
SL+ EI+ I ++ Q+ Q+ P+ PHL Y D I ED L+ + + +
Sbjct: 748 SLLNEIESIYDERQE-----QATEENAGVPVGPHLSLAYEDKQILEDAAALIIHHVKRQT 802
Query: 704 FSSKELLNKIMRLWSTFLEPML 725
KE KI ++ F+ +L
Sbjct: 803 GIQKEDKYKIKQIMHHFIPDLL 824
Score = 93.2 bits (230), Expect = 4e-18
Identities = 45/80 (56%), Positives = 58/80 (72%)
Query: 53 QKLTTNDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGHNHL 112
Q+L DALSYL QVK F Q + Y+ FL++MK+FK+Q DT GVI+RV +LFKGH L
Sbjct: 119 QRLKVEDALSYLDQVKLQFGSQPQVYNDFLDIMKEFKSQSIDTPGVISRVSQLFKGHPDL 178
Query: 113 IFGFNTFLPKGYEITLDEDE 132
I GFNTFLP GY+I + ++
Sbjct: 179 IMGFNTFLPPGYKIEVQTND 198
Score = 56.2 bits (134), Expect = 5e-07
Identities = 26/67 (38%), Positives = 42/67 (61%)
Query: 139 VEFEEAISFVNKIKKRFQSDEHVYKSFLDILNMYRKEHKDIGEVYSEVATLFKDHRDLLE 198
++ E+A+S+++++K +F S VY FLDI+ ++ + D V S V+ LFK H DL+
Sbjct: 121 LKVEDALSYLDQVKLQFGSQPQVYNDFLDIMKEFKSQSIDTPGVISRVSQLFKGHPDLIM 180
Query: 199 EFTRFLP 205
F FLP
Sbjct: 181 GFNTFLP 187
Score = 42.0 bits (97), Expect = 0.009
Identities = 25/83 (30%), Positives = 43/83 (51%), Gaps = 13/83 (15%)
Query: 52 SQKLTTNDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDT--AG-----------V 98
+Q + N A++Y+ ++K+ FQ Q + Y FLE++ ++ ++ + AG V
Sbjct: 299 NQPVEFNHAINYVNKIKNRFQGQPDIYKAFLEILHTYQKEQRNAKEAGGNYTPALTEQEV 358
Query: 99 IARVKELFKGHNHLIFGFNTFLP 121
A+V LFK L+ F FLP
Sbjct: 359 YAQVARLFKNQEDLLSEFGQFLP 381
>SN3A_MOUSE (Q60520) Paired amphipathic helix protein Sin3a
Length = 1282
Score = 272 bits (696), Expect = 3e-72
Identities = 194/621 (31%), Positives = 304/621 (48%), Gaps = 102/621 (16%)
Query: 126 ITLDEDEAPA---KKTVEFEEAISFVNKIKKRFQSDEHVYKSFLDILNMYRKEH---KDI 179
+T+ AP+ + V F AI++VNKIK RFQ +YK+FL+IL+ Y+KE K+
Sbjct: 286 VTISLGTAPSLQNNQPVHFNHAINYVNKIKNRFQGQPDIYKAFLEILHTYQKEQRNAKEA 345
Query: 180 G----------EVYSEVATLFKDHRDLLEEFTRFLPDTSAAPSTQHAPFGRNSLQRFNER 229
G EVY++VA LFK+ DLL EF +FLPD +++
Sbjct: 346 GGNYTPALTEQEVYAQVARLFKNQEDLLSEFGQFLPDANSS------------------- 386
Query: 230 NSMTPMMRQMQVDKQRYRRDRLPSHDRDRDLSVEHPEMDDDKTMINLHKEQRKRDRRIRD 289
V + +++ S D +V+ P++++ + + Q +R
Sbjct: 387 -----------VLLSKTTAEKVDSVRNDHGGTVKKPQLNNKPQRPSQNGCQIRR----HS 431
Query: 290 QDERDPDLDNSRDLTSQRFRDKKKTVKKAEGMYGEAFSFCEKVKEKLSSSDDYQTFLKCL 349
P + L S ++ G+ E+ F +KV++ L S++ Y+ FL+CL
Sbjct: 432 GTGATPPVKKKPKLMS--LKESSMADASKHGVGTESLFF-DKVRKALRSAEAYENFLRCL 488
Query: 350 NIFNNGIIKKNDLQNLVTDLLGKHSDLMDEFKDFLERCENIEGFLAGVMSKKSLSTDAHL 409
IFN +I + +L LV+ LGK +L + FK+FL E++ HL
Sbjct: 489 VIFNQEVISRAELVQLVSPFLGKFPELFNWFKNFLGYKESV-----------------HL 531
Query: 410 SRSSKLEDKDKDQKREMDGAKEKDRYKEKYMGKSIQELDLSDCKRCTPSYRLLPSDYPIP 469
K +R +G E+D + CKR SYR LP Y P
Sbjct: 532 ESFPK--------ERATEGI--------------AMEIDYASCKRLGSSYRALPKSYQQP 569
Query: 470 TASQRSELGAQVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDLLLESVSS 529
+ R+ L +VLND WVS S SED +F +K QYEE ++RCED+RFELD++LE+ +
Sbjct: 570 KCTGRTPLCKEVLNDTWVSFPSWSEDSTFVSSKKTQYEEHIYRCEDERFELDVVLETNLA 629
Query: 530 ASKRAEELYNNINENKISVEALSRIEDHF----TVLNLRCIERLYGDHGLDVIDILRKNP 585
+ E + ++ +A R+++ V++ + ++R+Y D D+ID LRKNP
Sbjct: 630 TIRVLEAIQKKLSRLSAEEQAKFRLDNTLGGTSEVIHRKALQRIYADKAADIIDGLRKNP 689
Query: 586 THALPVILTRLKQKQEEWNRCRSDFNKVWAEIYAKNHYKSLDHRSFYFKQQDSKNLSTKS 645
+ A+P++L RLK K+EEW + FNKVW + K + KSLDH+ FKQ D+K L +KS
Sbjct: 690 SIAVPIVLKRLKMKEEEWREAQRGFNKVWRDENEKYYLKSLDHQGINFKQNDTKVLRSKS 749
Query: 646 LVAEIKEIKEKLQKEDHIIQSIAAENRQPLIPHLEFEYSDGGIHEDLYKLVQYSCE-EVF 704
L+ EI+ I ++ Q+ Q+ P+ PHL Y D I ED L+ + + +
Sbjct: 750 LLNEIESIYDERQE-----QATEENAGVPVGPHLSLAYEDKQILEDAAALIIHHVKRQTG 804
Query: 705 SSKELLNKIMRLWSTFLEPML 725
KE KI ++ F+ +L
Sbjct: 805 IQKEDKYKIKQIMHHFIPDLL 825
Score = 91.3 bits (225), Expect = 1e-17
Identities = 44/80 (55%), Positives = 58/80 (72%)
Query: 53 QKLTTNDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGHNHL 112
Q+L DALSYL QVK F Q + ++ FL++MK+FK+Q DT GVI+RV +LFKGH L
Sbjct: 119 QRLKVEDALSYLDQVKLQFGSQPQVHNDFLDIMKEFKSQSIDTPGVISRVSQLFKGHPDL 178
Query: 113 IFGFNTFLPKGYEITLDEDE 132
I GFNTFLP GY+I + ++
Sbjct: 179 IMGFNTFLPPGYKIEVQTND 198
Score = 54.3 bits (129), Expect = 2e-06
Identities = 25/67 (37%), Positives = 42/67 (62%)
Query: 139 VEFEEAISFVNKIKKRFQSDEHVYKSFLDILNMYRKEHKDIGEVYSEVATLFKDHRDLLE 198
++ E+A+S+++++K +F S V+ FLDI+ ++ + D V S V+ LFK H DL+
Sbjct: 121 LKVEDALSYLDQVKLQFGSQPQVHNDFLDIMKEFKSQSIDTPGVISRVSQLFKGHPDLIM 180
Query: 199 EFTRFLP 205
F FLP
Sbjct: 181 GFNTFLP 187
Score = 41.6 bits (96), Expect = 0.012
Identities = 25/83 (30%), Positives = 43/83 (51%), Gaps = 13/83 (15%)
Query: 52 SQKLTTNDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDT--AG-----------V 98
+Q + N A++Y+ ++K+ FQ Q + Y FLE++ ++ ++ + AG V
Sbjct: 299 NQPVHFNHAINYVNKIKNRFQGQPDIYKAFLEILHTYQKEQRNAKEAGGNYTPALTEQEV 358
Query: 99 IARVKELFKGHNHLIFGFNTFLP 121
A+V LFK L+ F FLP
Sbjct: 359 YAQVARLFKNQEDLLSEFGQFLP 381
>PST1_SCHPO (Q09750) Paired amphipathic helix protein pst1 (SIN3
homolog 1)
Length = 1522
Score = 192 bits (487), Expect = 6e-48
Identities = 153/573 (26%), Positives = 254/573 (43%), Gaps = 86/573 (15%)
Query: 132 EAPAKKTVEFEEAISFVNKIKKRFQSDEHVYKSFLDILNMYRKEHKDIGEVYSEVATLFK 191
+A K+ + ++AI+FVN +K RF Y SFLDIL Y+ + + I VY +V+ LF
Sbjct: 340 DASVKQAADLDQAINFVNNVKNRFSHKPEAYNSFLDILKSYQHDQRPIQLVYFQVSQLFA 399
Query: 192 DHRDLLEEFTRFLPDTSAAPSTQHAPFGRNSLQRFNERNSMTPMMRQMQVDKQRYRRDRL 251
+ DLLEEF RFLPD S N+ +++++ P K R
Sbjct: 400 EAPDLLEEFKRFLPDVSV-----------NAPAETQDKSTVVPQESATATPK------RS 442
Query: 252 PSHDRDRDLSVEHPEMDDDKTMINLHKEQRKRDRRI--RDQDERDPDLDNSRDLTSQRFR 309
PS L E+R+ + + R+ +R +S + T+ R
Sbjct: 443 PSATPTSALPPIGKFAPPTTAKAQPAPEKRRGEPAVQTRNHSKRTRTATSSVEETTPRAF 502
Query: 310 DKKKTVKKAEGMYGEAFSFCEKVKEKLSSSDDYQTFLKCLNIFNNGIIKKNDLQNLVTDL 369
+ V A+ F E ++ L++ Y F+K L +++ + KN L
Sbjct: 503 N----VPIAQNKNPSELEFLEHARQYLANESKYNEFIKLLELYSQEVFDKNAL------- 551
Query: 370 LGKHSDLMDEFKDFLERCENIEGFLAGVMSKKSLSTDAHLSRSSKLEDKDKDQKREMDGA 429
+ERC G ++ HL M+
Sbjct: 552 --------------VERCYVFFG------------SNEHL----------------MNWL 569
Query: 430 KEKDRYKEKY-MGKSIQELDLSDCKRCTPSYRLLPSDYPIPTASQRSELGAQVLNDHWVS 488
K+ +Y + +DL+ CK C PSYRLLP + S R +L +LND WVS
Sbjct: 570 KDLVKYNPANPIPVPRPRVDLTQCKSCGPSYRLLPKIELLLPCSGRDDLCWTILNDAWVS 629
Query: 489 V-TSGSEDYSFKHMRKNQYEESLFRCEDDRFELDLLLES----VSSASKRAEELYNNINE 543
T SED F RKNQ+EE+L + E++R+E D + + + A+++
Sbjct: 630 FPTLASEDSGFIAHRKNQFEENLHKLEEERYEYDRHIGANMRFIELLQIHADKMLKMSEV 689
Query: 544 NKISVEALSRIEDHFTVLNLRCIERLYG-DHGLDVIDILRKNPTHALPVILTRLKQKQEE 602
K + S + + + I+++YG +H +I+ L+KNP+ +P++L RLK+K E
Sbjct: 690 EKANWTLPSNLGGKSVSIYHKVIKKVYGKEHAQQIIENLQKNPSVTIPIVLERLKKKDRE 749
Query: 603 WNRCRSDFNKVWAEIYAKNHYKSLDHRSFYFKQQDSKNLSTKSLVAEIKEIKEKLQKEDH 662
W ++ +N++W +I KN Y+SLDH+ FK D K+ + K L++E++ + ++ + E
Sbjct: 750 WRSLQNHWNELWHDIEEKNFYRSLDHQGVSFKSVDKKSTTPKFLISELRNLAQQQKVE-- 807
Query: 663 IIQSIAAENRQPLIPHLEFEYSDGGIHEDLYKL 695
+E + F Y D I D+ +L
Sbjct: 808 -----LSEGKVTPSHQFLFSYKDPNIITDIARL 835
Score = 78.6 bits (192), Expect = 9e-14
Identities = 39/76 (51%), Positives = 52/76 (68%)
Query: 53 QKLTTNDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGHNHL 112
++L DALSYL VK F + E Y+ FL++MK+FK+Q +T VI RV +LF G+ +L
Sbjct: 178 RQLNVTDALSYLDLVKLQFHQEPEIYNEFLDIMKEFKSQAIETPEVITRVSKLFAGYPNL 237
Query: 113 IFGFNTFLPKGYEITL 128
I GFNTFLP GY I +
Sbjct: 238 IQGFNTFLPPGYSIEI 253
Score = 50.1 bits (118), Expect = 3e-05
Identities = 52/243 (21%), Positives = 101/243 (41%), Gaps = 15/243 (6%)
Query: 143 EAISFVNKIKKRFQSDEHVYKSFLDILNMYRKEHKDIGEVYSEVATLFKDHRDLLEEFTR 202
+A+S+++ +K +F + +Y FLDI+ ++ + + EV + V+ LF + +L++ F
Sbjct: 184 DALSYLDLVKLQFHQEPEIYNEFLDIMKEFKSQAIETPEVITRVSKLFAGYPNLIQGFNT 243
Query: 203 FLPDTSAAPSTQHAPFGRNSLQRFNERNSMTPMMRQMQVDKQRYRRDRLPSHDRDRDLSV 262
FLP + + P SL + P+M ++ P H L
Sbjct: 244 FLPPGYSIEISSADP---GSLAGIHITTPQGPLM----INDLGKTTAPPPPHGSTTPLPA 296
Query: 263 EHPEMDDDKTMINLHKEQRKRDRRIRDQDERDPDLDNSRDLTSQRFRDKKKTVKKAEGMY 322
T +N+ K+ ++ + S + + +VK+A +
Sbjct: 297 A-----ASYTSMNM-KQSSASHPVLQPPAPSTLQFNPSPSPAAPSYPPVDASVKQAADL- 349
Query: 323 GEAFSFCEKVKEKLSSSDD-YQTFLKCLNIFNNGIIKKNDLQNLVTDLLGKHSDLMDEFK 381
+A +F VK + S + Y +FL L + + + V+ L + DL++EFK
Sbjct: 350 DQAINFVNNVKNRFSHKPEAYNSFLDILKSYQHDQRPIQLVYFQVSQLFAEAPDLLEEFK 409
Query: 382 DFL 384
FL
Sbjct: 410 RFL 412
Score = 40.4 bits (93), Expect = 0.027
Identities = 20/74 (27%), Positives = 39/74 (52%)
Query: 48 EATTSQKLTTNDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFK 107
+A+ Q + A++++ VK+ F + E Y+ FL+++K ++ + V +V +LF
Sbjct: 340 DASVKQAADLDQAINFVNNVKNRFSHKPEAYNSFLDILKSYQHDQRPIQLVYFQVSQLFA 399
Query: 108 GHNHLIFGFNTFLP 121
L+ F FLP
Sbjct: 400 EAPDLLEEFKRFLP 413
>SIN3_YEAST (P22579) Paired amphipathic helix protein SIN3
Length = 1536
Score = 166 bits (421), Expect = 3e-40
Identities = 138/484 (28%), Positives = 216/484 (44%), Gaps = 78/484 (16%)
Query: 324 EAFSFCEKVKEKLSSSDDYQTFLKCLNIFNNGIIKKNDLQNLVTDLLGKHSDLMDEFKDF 383
E +F EK K + + Y FLK LN+++ I+ +DL V LG + +L FK+F
Sbjct: 664 EEVTFFEKAKRYIGNKHLYTEFLKILNLYSQDILDLDDLVEKVDFYLGSNKELFTWFKNF 723
Query: 384 LERCENIEGFLAGVMSKKSLSTDAHLSRSSKLEDKDKDQKREMDGAKEKDRYKEKYMGKS 443
+ G +EK + E + +
Sbjct: 724 V-------------------------------------------GYQEKTKCIENIVHEK 740
Query: 444 IQELDLSDCKRCTPSYRLLPSDYPIPTASQRSELGAQVLNDHWVS-VTSGSEDYSFKHMR 502
LDL C+ PSY+ LP S R ++ +VLND WV SED F R
Sbjct: 741 -HRLDLDLCEAFGPSYKRLPKSDTFMPCSGRDDMCWEVLNDEWVGHPVWASEDSGFIAHR 799
Query: 503 KNQYEESLFRCEDDRFELDLLLESVSSASKRAEELYN---NINENKISVEALSRIEDHFT 559
KNQYEE+LF+ E++R E D +ES + E + N N+ EN+ + L H +
Sbjct: 800 KNQYEETLFKIEEERHEYDFYIESNLRTIQCLETIVNKIENMTENEKANFKLPPGLGHTS 859
Query: 560 V-LNLRCIERLYG-DHGLDVIDILRKNPTHALPVILTRLKQKQEEWNRCRSDFNKVWAEI 617
+ + + I ++Y + G ++ID L ++P PV+L RLKQK EEW R + ++NKVW E+
Sbjct: 860 MTIYKKVIRKVYDKERGFEIIDALHEHPAVTAPVVLKRLKQKDEEWRRAQREWNKVWREL 919
Query: 618 YAKNHYKSLDHRSFYFKQQDSKNLSTKSLVAEIKEIK-EKLQKEDHIIQSIAAENRQPLI 676
K +KSLDH FKQ D K L+TK L++EI IK ++ K+ H +
Sbjct: 920 EQKVFFKSLDHLGLTFKQADKKLLTTKQLISEISSIKVDQTNKKIHWLTPKPKS------ 973
Query: 677 PHLEFEYSDGGIHEDLYKLVQ--------YSCEEVFSSKELLNKIMRLWSTFLEPMLGVT 728
L+F++ D I D+ L YS + K+LL + L+ + + +
Sbjct: 974 -QLDFDFPDKNIFYDILCLADTFITHTTAYSNPDKERLKDLLKYFISLFFSISFEKIEES 1032
Query: 729 SQSHGTERVEDRKAGHSSRNFAASNVGGDGSPHRDSISTNSRLPKS------DKNEVDGR 782
SH + SS + S++ P++ +S L +S N+ DG+
Sbjct: 1033 LYSH------KQNVSESSGSDDGSSIASRKRPYQQEMSLLDILHRSRYQKLKRSNDEDGK 1086
Query: 783 VTEV 786
V ++
Sbjct: 1087 VPQL 1090
Score = 90.9 bits (224), Expect = 2e-17
Identities = 53/137 (38%), Positives = 82/137 (59%), Gaps = 8/137 (5%)
Query: 120 LPKGYEITLDEDEAPAKKTV--EFEEAISFVNKIKKRFQSDEHVYKSFLDILNMYRKEHK 177
+P+ + ED AKK V EF +AIS+VNKIK RF +YK FL+IL Y++E K
Sbjct: 388 IPQSQSLVPQED---AKKNVDVEFSQAISYVNKIKTRFADQPDIYKHFLEILQTYQREQK 444
Query: 178 DIGEVYSEVATLFKDHRDLLEEFTRFLPDTSAAPSTQHAPFGRNSLQRFNER---NSMTP 234
I EVY++V LF++ DLLE+F +FLPD+SA+ + Q +++ Q+ + +
Sbjct: 445 PINEVYAQVTHLFQNAPDLLEDFKKFLPDSSASANQQVQHAQQHAQQQHEAQMHAQAQAQ 504
Query: 235 MMRQMQVDKQRYRRDRL 251
Q QV++Q+ ++ L
Sbjct: 505 AQAQAQVEQQKQQQQFL 521
Score = 87.8 bits (216), Expect = 1e-16
Identities = 43/72 (59%), Positives = 53/72 (72%)
Query: 55 LTTNDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGHNHLIF 114
L DALSYL+QVK F + + Y+LFL++MKDFK+Q DT GVI RV LF+G+ LI
Sbjct: 219 LNVKDALSYLEQVKFQFSSRPDIYNLFLDIMKDFKSQAIDTPGVIERVSTLFRGYPILIQ 278
Query: 115 GFNTFLPKGYEI 126
GFNTFLP+GY I
Sbjct: 279 GFNTFLPQGYRI 290
Score = 62.4 bits (150), Expect = 7e-09
Identities = 66/278 (23%), Positives = 118/278 (41%), Gaps = 41/278 (14%)
Query: 131 DEAPAKKTVEFEEAISFVNKIKKRFQSDEHVYKSFLDILNMYRKEHKDIGEVYSEVATLF 190
DE + + ++A+S++ ++K +F S +Y FLDI+ ++ + D V V+TLF
Sbjct: 211 DEDADYRPLNVKDALSYLEQVKFQFSSRPDIYNLFLDIMKDFKSQAIDTPGVIERVSTLF 270
Query: 191 KDHRDLLEEFTRFLP-----DTSAAPSTQ---HAPFGRNSLQRFNERNSMTPMMRQMQVD 242
+ + L++ F FLP + S+ P P G ++ N+++P R D
Sbjct: 271 RGYPILIQGFNTFLPQGYRIECSSNPDDPIRVTTPMGTTTV-----NNNISPSGRG-TTD 324
Query: 243 KQRYRRDRLPSHDRDRDLSVEHPEMDDDKTMINLHKEQRKRDRRIRDQDERDPDLDNSRD 302
Q L S V+ P + M+ Q +++ +DQ + P L S
Sbjct: 325 AQ-----ELGSFPESDGNGVQQP---SNVPMVPSSVYQSEQN---QDQQQSLPLLATSSG 373
Query: 303 L---------------TSQRFRDKKKTVKKAEGMYGEAFSFCEKVKEKLSSSDD-YQTFL 346
L SQ ++ K + + +A S+ K+K + + D Y+ FL
Sbjct: 374 LPSIQQPEMPAHRQIPQSQSLVPQEDAKKNVDVEFSQAISYVNKIKTRFADQPDIYKHFL 433
Query: 347 KCLNIFNNGIIKKNDLQNLVTDLLGKHSDLMDEFKDFL 384
+ L + N++ VT L DL+++FK FL
Sbjct: 434 EILQTYQREQKPINEVYAQVTHLFQNAPDLLEDFKKFL 471
Score = 43.9 bits (102), Expect = 0.002
Identities = 21/64 (32%), Positives = 37/64 (57%)
Query: 58 NDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGVIARVKELFKGHNHLIFGFN 117
+ A+SY+ ++K F DQ + Y FLE+++ ++ ++ V A+V LF+ L+ F
Sbjct: 409 SQAISYVNKIKTRFADQPDIYKHFLEILQTYQREQKPINEVYAQVTHLFQNAPDLLEDFK 468
Query: 118 TFLP 121
FLP
Sbjct: 469 KFLP 472
Score = 42.4 bits (98), Expect = 0.007
Identities = 25/93 (26%), Positives = 45/93 (47%), Gaps = 9/93 (9%)
Query: 120 LPKGYEITLD--------EDEAPAKKTVEFEEAISFVNKIKKRFQSDEHVYKSFLDILNM 171
+P EI LD E P + + E ++F K K R+ ++H+Y FL ILN+
Sbjct: 633 IPVRPEIDLDPSIVPVVPEPTEPIENNISLNEEVTFFEKAK-RYIGNKHLYTEFLKILNL 691
Query: 172 YRKEHKDIGEVYSEVATLFKDHRDLLEEFTRFL 204
Y ++ D+ ++ +V +++L F F+
Sbjct: 692 YSQDILDLDDLVEKVDFYLGSNKELFTWFKNFV 724
>PST2_SCHPO (O13919) Paired amphipathic helix protein pst2 (SIN3
homolog 2)
Length = 1075
Score = 143 bits (361), Expect = 2e-33
Identities = 136/551 (24%), Positives = 217/551 (38%), Gaps = 101/551 (18%)
Query: 146 SFVNKIKKRFQSDEHVYKSFLDILNMYRKEHKDIGEVYSEVATLFKDHRDLLEEFTRFLP 205
+FV K+ +R +VY +F+D++ E D ++ + +D+ DLLE FLP
Sbjct: 41 AFVQKLGQRLCHRPYVYSAFMDVVKALHNEIVDFPGFIERISVILRDYPDLLEYLNIFLP 100
Query: 206 DT---------------------------------SAAPSTQHAPFGRNSLQRFNERNSM 232
+ + P T H G S R S
Sbjct: 101 SSYKYLLSNSGANFTLQFTTPSGPVSTPSTYVATYNDLPCTYHRAIGFVSRVR-RALLSN 159
Query: 233 TPMMRQMQVDKQRYRRDRLPSHDRDR---DLSVEHPEMDDD-----KTMINLHKEQRKRD 284
++Q ++++ + L EHP + + + I +
Sbjct: 160 PEQFFKLQDSLRKFKNSECSLSELQTIVTSLLAEHPSLAHEFHNFLPSSIFFGSKPPLGS 219
Query: 285 RRIRDQDERDPDLDNSRDLTSQRFRDKKKTVKKAEGMYGEAFSFCEKVKEKLSSSDDYQT 344
+R L N DL SQ D + + E+ F + VK L+ + Y
Sbjct: 220 FPLRGIQSSQFTLSNISDLLSQSRPDN---LSPFSHLSNESSDFFKNVKNVLTDVETYHE 276
Query: 345 FLKCLNIFNNGIIKKNDLQNLVTDLLGKHSDLMDEFKDFLERCENIEGFLAGVMSKKSLS 404
FLK LN++ GII +N L + L +S L F +S SLS
Sbjct: 277 FLKLLNLYVQGIIDRNILVSRGFGFLKSNSGLWRSF-----------------LSLTSLS 319
Query: 405 TDAHLSRSSKLEDKDKDQKREMDGAKEKDRYKEKYMGKSIQELDLSDCKRCTPSYRLLPS 464
+ LS + SD C PSYRLLP
Sbjct: 320 PEEFLS---------------------------------VYNSACSDFPECGPSYRLLPV 346
Query: 465 DYPIPTASQRSELGAQVLNDHWVS-VTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDLL 523
+ + S R + +LND WVS T SE+ F RK YEE++ + E++R+E D
Sbjct: 347 EERNISCSGRDDFAWGILNDDWVSHPTWASEESGFIVQRKTPYEEAMTKLEEERYEFDRH 406
Query: 524 LESVSSASKRAEELYNNINENKISVEALSRIEDHFTV----LNLRCIERLY-GDHGLDVI 578
+E+ S K +++ N INE +E+ + + + I+ +Y +H ++
Sbjct: 407 IEATSWTIKSLKKIQNRINELPEEERETYTLEEGLGLPSKSIYKKTIKLVYTSEHAEEMF 466
Query: 579 DILRKNPTHALPVILTRLKQKQEEWNRCRSDFNKVWAEIYAKNHYKSLDHRSFYFKQQDS 638
L + P LP++++RL++K EEW + W I KN+ KSLD + YFK +D
Sbjct: 467 KALERMPCLTLPLVISRLEEKNEEWKSVKRSLQPGWRSIEFKNYDKSLDSQCVYFKARDK 526
Query: 639 KNLSTKSLVAE 649
KN+S+K L+AE
Sbjct: 527 KNVSSKFLLAE 537
Score = 52.0 bits (123), Expect = 9e-06
Identities = 34/152 (22%), Positives = 62/152 (40%), Gaps = 22/152 (14%)
Query: 78 YDLFLEVMKDFKAQRTDTAGVIARVKELFKGHNHLIFGFNTFLPKGYEITLDEDEA---- 133
Y F++V+K + D G I R+ + + + L+ N FLP Y+ L A
Sbjct: 57 YSAFMDVVKALHNEIVDFPGFIERISVILRDYPDLLEYLNIFLPSSYKYLLSNSGANFTL 116
Query: 134 -----------PAKKTVEFEE-------AISFVNKIKKRFQSDEHVYKSFLDILNMYRKE 175
P+ + + AI FV+++++ S+ + D L ++
Sbjct: 117 QFTTPSGPVSTPSTYVATYNDLPCTYHRAIGFVSRVRRALLSNPEQFFKLQDSLRKFKNS 176
Query: 176 HKDIGEVYSEVATLFKDHRDLLEEFTRFLPDT 207
+ E+ + V +L +H L EF FLP +
Sbjct: 177 ECSLSELQTIVTSLLAEHPSLAHEFHNFLPSS 208
>WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY
DNA-binding protein 19)
Length = 1895
Score = 103 bits (257), Expect = 3e-21
Identities = 59/119 (49%), Positives = 77/119 (64%), Gaps = 21/119 (17%)
Query: 39 GGGGAGGGGEATTSQKLTTNDALSYLKQVKDMFQDQREKYDLFLEVMKDFKAQRTDTAGV 98
GGG GG +TT DAL+YLK VKD F+D EKYD FLEV+ D K Q DT+GV
Sbjct: 297 GGGNMGG---------VTTGDALNYLKAVKDKFEDS-EKYDTFLEVLNDCKHQGVDTSGV 346
Query: 99 IARVKELFKGHNHLIFGFNTFLPKGYEIT-LDEDEAPAKKTVEFEEAISFVNKIKKRFQ 156
IAR+K+LFKGH+ L+ GFNT+L K Y+IT L ED+ P I F++K++ ++
Sbjct: 347 IARLKDLFKGHDDLLLGFNTYLSKEYQITILPEDDFP----------IDFLDKVEGPYE 395
Score = 41.2 bits (95), Expect = 0.016
Identities = 30/126 (23%), Positives = 54/126 (42%), Gaps = 2/126 (1%)
Query: 143 EAISFVNKIKKRFQSDEHVYKSFLDILNMYRKEHKDIGEVYSEVATLFKDHRDLLEEFTR 202
+A++++ +K +F+ E Y +FL++LN + + D V + + LFK H DLL F
Sbjct: 308 DALNYLKAVKDKFEDSEK-YDTFLEVLNDCKHQGVDTSGVIARLKDLFKGHDDLLLGFNT 366
Query: 203 FLPDTSAAPSTQHAPFGRNSLQRFNERNSMTPMMRQ-MQVDKQRYRRDRLPSHDRDRDLS 261
+L F + L + MT Q +Q + + PS + S
Sbjct: 367 YLSKEYQITILPEDDFPIDFLDKVEGPYEMTYQQAQTVQANANMQPQTEYPSSSAVQSFS 426
Query: 262 VEHPEM 267
P++
Sbjct: 427 SGQPQI 432
Score = 38.1 bits (87), Expect = 0.14
Identities = 19/64 (29%), Positives = 34/64 (52%)
Query: 323 GEAFSFCEKVKEKLSSSDDYQTFLKCLNIFNNGIIKKNDLQNLVTDLLGKHSDLMDEFKD 382
G+A ++ + VK+K S+ Y TFL+ LN + + + + + DL H DL+ F
Sbjct: 307 GDALNYLKAVKDKFEDSEKYDTFLEVLNDCKHQGVDTSGVIARLKDLFKGHDDLLLGFNT 366
Query: 383 FLER 386
+L +
Sbjct: 367 YLSK 370
>RA50_PYRHO (O58687) DNA double-strand break repair rad50 ATPase
Length = 879
Score = 63.2 bits (152), Expect = 4e-09
Identities = 139/653 (21%), Positives = 258/653 (39%), Gaps = 82/653 (12%)
Query: 113 IFGFNTFLPKGY------EITLDEDEAPAKKTVEF------EEAISFVNKIKKRFQSDEH 160
I +N FL Y + L+ DE K E E+A + KI+K +
Sbjct: 123 IIPYNVFLNAIYVRQGQIDAILESDETRDKIVKEILNLDKLEKAYDNLGKIRKYIKYSIE 182
Query: 161 VYKSFL----DILNMYRKEHKDIGEVYSEVATLFKDHRDLLEEFTRFLPDTSAAPSTQHA 216
+ F+ +I ++ R + K EV +E+ + + L R L T A
Sbjct: 183 EKEKFIMKTENIEDLIRTQEKSFTEVLNEIRNISSN----LPRLRRELEGIKEEVKTLEA 238
Query: 217 PFGRNSLQRFNERNSMTPMMRQMQVDKQRYRRDRLPSHDRDRDLSVEHPEMDDD------ 270
F NS+T + ++++ + ++ RL R + +E
Sbjct: 239 TF-----------NSITEL--KLRLGELNGKKGRLEERIRQLESGIEEKRKKSKELEEVV 285
Query: 271 KTMINLHKEQRKRDRRIRDQDE---RDPDLDNSRDLTSQRFRDKKKTVKKAEGMYGEAFS 327
K + L K++ + R I +DE + +L+ + S R ++ K+ +K AE
Sbjct: 286 KELPELEKKETEYRRLIEFKDEYLVKKNELEKRLGILSNRLQEVKRKIKDAESKVARIRW 345
Query: 328 FCEKVKEKLSSSDDYQTFLKCLNIFNNGIIKKNDLQNLVTDLLGKHSDLMDEFKDFLE-R 386
E++KE + + F + + K +++L + L G + ++E +LE R
Sbjct: 346 IEERLKE---IQEKIMKLEPRVREFEDAMRLKAQMESLKSKLGGLEPEKINEKLLYLENR 402
Query: 387 CENIEGFLAGVMSK-KSLSTDAHLSRSSKLEDKDKDQKREMDGAKEKDRYKEKYMGKSIQ 445
+ +E + + K L+ + R + +E K K + G + + +K + K
Sbjct: 403 KKELEEEIDKITRKIGELNQRSKDRRLAIIELKKARGKCPVCGRELTEEHKADLLRK--Y 460
Query: 446 ELDLSDCKRCTPSYRLLPSDYPIPTASQRSELGA----QVLNDHWVSVTSGSEDYSFKHM 501
L+LS ++ + L +EL + + D + + + + +
Sbjct: 461 SLELSSIEKEIQEAKALERQLRAEFRKVENELSRLSSLKTIADQIIEIRERLSKINLEDL 520
Query: 502 RKNQYEESLFRCEDDRFELDLLLESVSSASKRAEELYNNINEN-KISVEA------LSRI 554
++++ E L + E ++ L V S K EL + NE+ K+ +E LS I
Sbjct: 521 KRDKEEYELLKSESNK-----LKGEVESLKKEVNELNDYKNESTKLEIEIDKAKKELSEI 575
Query: 555 EDHFTVLNLRCIERLYGD-HGLDVID---ILRKNPTHALPVILTRLKQKQEEWNRCRSDF 610
ED L + I+ L G L+ I KN L IL LK ++EE ++ +
Sbjct: 576 EDRLLRLGFKTIDELSGRIRELEKFHNKYIEAKNAEKELRDILESLKDEREELDKAFEEL 635
Query: 611 NKVWAEIYAKNHYKSLDHRSFYFKQQDSKNLSTKSLVAEIKEIKEKLQ----KEDHI--- 663
K+ +I + R F K+ + K L EIK ++ KL+ + D I
Sbjct: 636 AKIETDIEKVTSQLNELQRKFDQKKYEEKREKMMKLSMEIKGLETKLEELERRRDEIKST 695
Query: 664 IQSIAAENRQPLIPHLEFEYSDGGIH--EDLY-KLVQYSCEEVFSSKELLNKI 713
I+ + E ++ +E E + I E+L K+ +Y + +E LNKI
Sbjct: 696 IEKLKEERKERESAKMELEKLNIAIKRIEELRGKIKEY---KALIKEEALNKI 745
>UBF1_HUMAN (P17480) Nucleolar transcription factor 1 (Upstream
binding factor 1) (UBF-1) (Autoantigen NOR-90)
Length = 764
Score = 58.5 bits (140), Expect = 1e-07
Identities = 44/126 (34%), Positives = 59/126 (45%), Gaps = 13/126 (10%)
Query: 939 SEEDFVAYRD---SNAQSMAKSKH---NIERRKYESRDREEECGPETGGDNDADADDEDS 992
S +D AY++ + +SM K + R +S+ EE E D D D ++ED
Sbjct: 638 SPQDRAAYKEYISNKRKSMTKLRGPNPKSSRTTLQSKSESEEDDEEDEDDEDEDEEEEDD 697
Query: 993 EN--VSEAGEDVSGSESA-----GDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQG 1045
EN SE G D S S S GDE ++D +EDD E DD D ESEG + +
Sbjct: 698 ENGDSSEDGGDSSESSSEDESEDGDENEEDDEDEDDDEDDDEDEDNESEGSSSSSSSSGD 757
Query: 1046 GGDSSS 1051
DS S
Sbjct: 758 SSDSDS 763
Score = 39.7 bits (91), Expect = 0.046
Identities = 33/103 (32%), Positives = 46/103 (44%), Gaps = 16/103 (15%)
Query: 908 LVENSKVKSHEESSGPCKVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYE 967
L S+ + +E + E EE E NGDS ED +S+++ E
Sbjct: 671 LQSKSESEEDDEEDEDDEDEDEEEEDDENGDSSEDGGDSSESSSED-------------E 717
Query: 968 SRDREEECGPETGGDNDADADDEDSENVSEAGEDVSGSESAGD 1010
S D +E + D+D D DDED +N SE S S S+GD
Sbjct: 718 SEDGDENEEDDEDEDDDED-DDEDEDNESEGSS--SSSSSSGD 757
Score = 37.4 bits (85), Expect = 0.23
Identities = 25/92 (27%), Positives = 36/92 (38%), Gaps = 3/92 (3%)
Query: 925 KVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDND 984
K E EE + D +ED D N S S+ + + S D E+ D D
Sbjct: 674 KSESEEDDEEDEDDEDEDEEEEDDENGDS---SEDGGDSSESSSEDESEDGDENEEDDED 730
Query: 985 ADADDEDSENVSEAGEDVSGSESAGDECFQED 1016
D D++D E+ E S S S+ + D
Sbjct: 731 EDDDEDDDEDEDNESEGSSSSSSSSGDSSDSD 762
>UBF1_XENLA (P25979) Nucleolar transcription factor 1 (Upstream
binding factor-1) (UBF-1) (xUBF-1)
Length = 677
Score = 57.4 bits (137), Expect = 2e-07
Identities = 37/118 (31%), Positives = 62/118 (52%), Gaps = 9/118 (7%)
Query: 937 GDSEEDFVAYRDSNA---QSMAKSKHNIERRKYESRDREEECGPETGGDNDADADDEDSE 993
G S +D AY++ N+ +S AK + + + K ++ + ++ + D D D DD++ E
Sbjct: 554 GLSSQDRAAYKEQNSNKRKSTAKIQVPVAKPKLVAQSKSDDDDEDDDDDEDEDDDDDEDE 613
Query: 994 NVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQGGGDSSS 1051
+ ++ ED SES+ DE ED E++D +G+ E E E E D + G SSS
Sbjct: 614 DKEDSSEDGDSSESSSDE-----DSEDGEENEDEEGE-EDEDEEEDDDDNESGSSSSS 665
Score = 35.8 bits (81), Expect = 0.67
Identities = 28/98 (28%), Positives = 40/98 (40%), Gaps = 6/98 (6%)
Query: 901 VPVANGVLVENSKVKSHEESSGPCKVEKEEG-ELSPNGDSEEDFVAYRDSNAQSMAKSKH 959
VPVA LV SK +E + E ++ E DS ED + S+ + +
Sbjct: 579 VPVAKPKLVAQSKSDDDDEDDDDDEDEDDDDDEDEDKEDSSEDGDSSESSSDEDSEDGEE 638
Query: 960 NIERRKYESRDREE-----ECGPETGGDNDADADDEDS 992
N + E D EE E G + + AD+ D DS
Sbjct: 639 NEDEEGEEDEDEEEDDDDNESGSSSSSSSSADSSDSDS 676
>UBF1_RAT (P25977) Nucleolar transcription factor 1 (Upstream binding
factor 1) (UBF-1)
Length = 764
Score = 56.2 bits (134), Expect = 5e-07
Identities = 37/118 (31%), Positives = 54/118 (45%), Gaps = 7/118 (5%)
Query: 941 EDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDADADDEDSEN--VSEA 998
+++++ + N + R +S+ EE E D+D + ++ED EN SE
Sbjct: 646 KEYISNKRKNMTKLRGPNPKSSRTTLQSKSESEEDDDEEDDDDDDEEEEEDDENGDSSED 705
Query: 999 GEDVSGSESA-----GDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQGGGDSSS 1051
G D S S S GDE +D +EDD E DD D ESEG + + DS S
Sbjct: 706 GGDSSESSSEDESEDGDENEDDDDDEDDDEDDDEDEDNESEGSSSSSSSSGDSSDSDS 763
Score = 35.4 bits (80), Expect = 0.88
Identities = 23/92 (25%), Positives = 38/92 (41%), Gaps = 3/92 (3%)
Query: 925 KVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDND 984
K E EE + + D +++ D N S S+ + + S D E+ D+D
Sbjct: 674 KSESEEDDDEEDDDDDDEEEEEDDENGDS---SEDGGDSSESSSEDESEDGDENEDDDDD 730
Query: 985 ADADDEDSENVSEAGEDVSGSESAGDECFQED 1016
D D++D E+ E S S S+ + D
Sbjct: 731 EDDDEDDDEDEDNESEGSSSSSSSSGDSSDSD 762
>PLS_STAAU (P80544) Surface protein precursor (Plasmin-sensitive
surface protein) (230 kDa cell-wall protein)
Length = 1637
Score = 55.1 bits (131), Expect = 1e-06
Identities = 33/126 (26%), Positives = 62/126 (49%), Gaps = 2/126 (1%)
Query: 928 KEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDADA 987
K++ + + D++ D A DS+A S + + + + D + + ++ D+D+DA
Sbjct: 1299 KDDSDADSDSDADSDSDADSDSDADSDSDADSDSDSDSDSDSDSDSDADSDSDSDSDSDA 1358
Query: 988 D-DEDSENVSEAGEDV-SGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQG 1045
D D D+++ S+A D S ++S D D + D D D ++S+ +A+ DA
Sbjct: 1359 DSDSDADSDSDADSDSDSDADSDSDSDADSDSDSDSDSDADSDSDSDSDSDADSDSDADS 1418
Query: 1046 GGDSSS 1051
DS S
Sbjct: 1419 DSDSDS 1424
Score = 53.9 bits (128), Expect = 2e-06
Identities = 37/143 (25%), Positives = 67/143 (45%), Gaps = 3/143 (2%)
Query: 911 NSKVKSHEESSGPCKVEKE-EGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESR 969
+S S +S + + + + + DS+ D + DS++ S A S + +
Sbjct: 1425 DSDADSDSDSDSDSDADSDSDADSDSDADSDSDADSDSDSDSDSDADSDSDADSDSDADS 1484
Query: 970 DREEECGPETGGDNDADAD-DEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVD 1028
D + + ++ D+DAD+D D DS++ S+A D S ++S D D + D D D
Sbjct: 1485 DSDADSDSDSDSDSDADSDSDSDSDSDSDADSD-SDADSDSDSDADSDSDADSDSDADGD 1543
Query: 1029 GKAESEGEAEGMCDAQGGGDSSS 1051
A+S+ +A+ D+ DS S
Sbjct: 1544 SDADSDSDADSDSDSDSDSDSDS 1566
Score = 53.5 bits (127), Expect = 3e-06
Identities = 36/146 (24%), Positives = 68/146 (45%), Gaps = 3/146 (2%)
Query: 907 VLVENSKVKSHEESSGPCKVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKY 966
V++ +K S +S + + + + DS+ D + DS++ S + S + +
Sbjct: 1293 VIIHGAKDDSDADSDSDADSDSD-ADSDSDADSDSDADSDSDSDSDSDSDSDSDADSDSD 1351
Query: 967 ESRDREEECGPETGGDNDADAD-DEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHD 1025
D + + + D+DAD+D D D+++ S++ D S S+S D D + D
Sbjct: 1352 SDSDSDADSDSDADSDSDADSDSDSDADSDSDSDAD-SDSDSDSDSDADSDSDSDSDSDA 1410
Query: 1026 DVDGKAESEGEAEGMCDAQGGGDSSS 1051
D D A+S+ +++ DA DS S
Sbjct: 1411 DSDSDADSDSDSDSDSDADSDSDSDS 1436
Score = 53.5 bits (127), Expect = 3e-06
Identities = 34/126 (26%), Positives = 60/126 (46%), Gaps = 4/126 (3%)
Query: 930 EGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDADAD- 988
+ + + DS+ D A DS++ S + + + + D + + ++ D+D+DAD
Sbjct: 1371 DSDSDSDADSDSDSDADSDSDSDSDSDADSDSDSDSDSDADSDSDADSDSDSDSDSDADS 1430
Query: 989 DEDSENVSEAGEDV---SGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQG 1045
D DS++ S+A D S S++ D D + D D D A+S+ +A+ DA
Sbjct: 1431 DSDSDSDSDADSDSDADSDSDADSDSDADSDSDSDSDSDADSDSDADSDSDADSDSDADS 1490
Query: 1046 GGDSSS 1051
DS S
Sbjct: 1491 DSDSDS 1496
Score = 53.1 bits (126), Expect = 4e-06
Identities = 36/143 (25%), Positives = 67/143 (46%), Gaps = 3/143 (2%)
Query: 911 NSKVKSHEESSGPCKVEKE-EGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESR 969
+S S +S + + + + + DS+ D + D+++ S A S + +
Sbjct: 1413 DSDADSDSDSDSDSDADSDSDSDSDSDADSDSDADSDSDADSDSDADSDSDSDSDSDADS 1472
Query: 970 DREEECGPETGGDNDADAD-DEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVD 1028
D + + + D+DAD+D D DS++ +++ D S S+S D D + D D D
Sbjct: 1473 DSDADSDSDADSDSDADSDSDSDSDSDADSDSD-SDSDSDSDADSDSDADSDSDSDADSD 1531
Query: 1029 GKAESEGEAEGMCDAQGGGDSSS 1051
A+S+ +A+G DA D+ S
Sbjct: 1532 SDADSDSDADGDSDADSDSDADS 1554
Score = 52.8 bits (125), Expect = 5e-06
Identities = 33/130 (25%), Positives = 60/130 (45%), Gaps = 8/130 (6%)
Query: 930 EGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDADAD- 988
+ + + DS+ D A DS+A S + S + + D + + ++ D+D+DAD
Sbjct: 1395 DSDADSDSDSDSDSDADSDSDADSDSDSDSDSDADSDSDSDSDSDADSDSDADSDSDADS 1454
Query: 989 ------DEDSENVSEAGEDV-SGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMC 1041
D DS++ S+A D + S+S D D + D D D ++S+ +++
Sbjct: 1455 DSDADSDSDSDSDSDADSDSDADSDSDADSDSDADSDSDSDSDSDADSDSDSDSDSDSDA 1514
Query: 1042 DAQGGGDSSS 1051
D+ DS S
Sbjct: 1515 DSDSDADSDS 1524
Score = 52.4 bits (124), Expect = 7e-06
Identities = 33/126 (26%), Positives = 60/126 (47%), Gaps = 4/126 (3%)
Query: 930 EGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDADAD- 988
+ + + DS+ D A DS++ S + + + + D + + ++ D+D+DAD
Sbjct: 1383 DSDADSDSDSDSDSDADSDSDSDSDSDADSDSDADSDSDSDSDSDADSDSDSDSDSDADS 1442
Query: 989 DEDSENVSEAGEDV---SGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQG 1045
D D+++ S+A D S S+S D D + D D D A+S+ +++ DA
Sbjct: 1443 DSDADSDSDADSDSDADSDSDSDSDSDADSDSDADSDSDADSDSDADSDSDSDSDSDADS 1502
Query: 1046 GGDSSS 1051
DS S
Sbjct: 1503 DSDSDS 1508
Score = 52.0 bits (123), Expect = 9e-06
Identities = 33/126 (26%), Positives = 60/126 (47%), Gaps = 4/126 (3%)
Query: 930 EGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDADAD- 988
+ + + DS+ D + DS++ S A S + + D + + ++ D+D+DAD
Sbjct: 1353 DSDSDADSDSDADSDSDADSDSDSDADSDSDSDADSDSDSDSDSDADSDSDSDSDSDADS 1412
Query: 989 --DEDSENVSEAGEDV-SGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQG 1045
D DS++ S++ D S S+S D D + D D D A+S+ +++ DA
Sbjct: 1413 DSDADSDSDSDSDSDADSDSDSDSDSDADSDSDADSDSDADSDSDADSDSDSDSDSDADS 1472
Query: 1046 GGDSSS 1051
D+ S
Sbjct: 1473 DSDADS 1478
Score = 52.0 bits (123), Expect = 9e-06
Identities = 32/126 (25%), Positives = 60/126 (47%), Gaps = 4/126 (3%)
Query: 930 EGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDADAD- 988
+ + + DS+ D + DS+A S + S + + D + + ++ D+D+D+D
Sbjct: 1365 DSDSDADSDSDSDADSDSDSDADSDSDSDSDSDADSDSDSDSDSDADSDSDADSDSDSDS 1424
Query: 989 --DEDSENVSEAGEDV-SGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQG 1045
D DS++ S++ D S S++ D D + D D D A+S+ +A+ DA
Sbjct: 1425 DSDADSDSDSDSDSDADSDSDADSDSDADSDSDADSDSDSDSDSDADSDSDADSDSDADS 1484
Query: 1046 GGDSSS 1051
D+ S
Sbjct: 1485 DSDADS 1490
Score = 51.6 bits (122), Expect = 1e-05
Identities = 32/123 (26%), Positives = 59/123 (47%), Gaps = 2/123 (1%)
Query: 930 EGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDADAD- 988
+ + + DS+ D + DS++ S + S + + D + + + D+D+DAD
Sbjct: 1329 DSDSDSDSDSDSDSDSDADSDSDSDSDSDADSDSDADSDSDADSDSDSDADSDSDSDADS 1388
Query: 989 DEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQGGGD 1048
D DS++ S+A D S S+S D D + D D D ++S+ +++ D+ D
Sbjct: 1389 DSDSDSDSDADSD-SDSDSDSDADSDSDADSDSDSDSDSDADSDSDSDSDSDADSDSDAD 1447
Query: 1049 SSS 1051
S S
Sbjct: 1448 SDS 1450
Score = 50.8 bits (120), Expect = 2e-05
Identities = 30/123 (24%), Positives = 60/123 (48%), Gaps = 2/123 (1%)
Query: 930 EGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDADAD- 988
+ + + D++ D + DS+A S + S + + D + + ++ D+D+D+D
Sbjct: 1407 DSDADSDSDADSDSDSDSDSDADSDSDSDSDSDADSDSDADSDSDADSDSDADSDSDSDS 1466
Query: 989 DEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQGGGD 1048
D D+++ S+A D S ++S D D + D D D ++S+ +A+ DA D
Sbjct: 1467 DSDADSDSDADSD-SDADSDSDADSDSDSDSDSDADSDSDSDSDSDSDADSDSDADSDSD 1525
Query: 1049 SSS 1051
S +
Sbjct: 1526 SDA 1528
Score = 50.4 bits (119), Expect = 3e-05
Identities = 35/134 (26%), Positives = 59/134 (43%), Gaps = 12/134 (8%)
Query: 930 EGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDADAD- 988
+ + + DS+ D + DS++ S A S + + D + + ++ D D+D+D
Sbjct: 1333 DSDSDSDSDSDSDADSDSDSDSDSDADSDSDADSDSDADSDSDSDADSDSDSDADSDSDS 1392
Query: 989 --------DEDSENVSEAGEDV---SGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEA 1037
D DS++ S+A D S S+S D D + D D D A+S+ +A
Sbjct: 1393 DSDSDADSDSDSDSDSDADSDSDADSDSDSDSDSDADSDSDSDSDSDADSDSDADSDSDA 1452
Query: 1038 EGMCDAQGGGDSSS 1051
+ DA DS S
Sbjct: 1453 DSDSDADSDSDSDS 1466
Score = 49.7 bits (117), Expect = 4e-05
Identities = 31/122 (25%), Positives = 58/122 (47%), Gaps = 2/122 (1%)
Query: 930 EGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDADADD 989
+ + + D++ D + DS++ S A S + + D + + + D+DAD+ D
Sbjct: 1361 DSDADSDSDADSDSDSDADSDSDSDADSDSDSDSDSDADSDSDSDSDSDADSDSDADS-D 1419
Query: 990 EDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQGGGDS 1049
DS++ S+A D S S+S D D + D D D ++S+ +++ D+ DS
Sbjct: 1420 SDSDSDSDADSD-SDSDSDSDADSDSDADSDSDADSDSDADSDSDSDSDSDADSDSDADS 1478
Query: 1050 SS 1051
S
Sbjct: 1479 DS 1480
Score = 49.7 bits (117), Expect = 4e-05
Identities = 31/128 (24%), Positives = 60/128 (46%), Gaps = 8/128 (6%)
Query: 930 EGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDADAD- 988
+ + + DS+ D A DS+A S + + + + D + + ++ D+D+D+D
Sbjct: 1455 DSDADSDSDSDSDSDADSDSDADSDSDADSDSDADSDSDSDSDSDADSDSDSDSDSDSDA 1514
Query: 989 ----DEDSENVSEAGEDV---SGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMC 1041
D DS++ S+A D S S++ GD D + D D D ++S+ +A+
Sbjct: 1515 DSDSDADSDSDSDADSDSDADSDSDADGDSDADSDSDADSDSDSDSDSDSDSDSDADSDS 1574
Query: 1042 DAQGGGDS 1049
D+ D+
Sbjct: 1575 DSDSDSDA 1582
Score = 49.3 bits (116), Expect = 6e-05
Identities = 30/123 (24%), Positives = 59/123 (47%), Gaps = 2/123 (1%)
Query: 930 EGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDADAD- 988
+ + + DS+ D + D+++ S A S + + D + + + D+DAD+D
Sbjct: 1391 DSDSDSDADSDSDSDSDSDADSDSDADSDSDSDSDSDADSDSDSDSDSDADSDSDADSDS 1450
Query: 989 DEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQGGGD 1048
D DS++ +++ D S S+S D D + D D D ++S+ +++ D+ D
Sbjct: 1451 DADSDSDADSDSD-SDSDSDADSDSDADSDSDADSDSDADSDSDSDSDSDADSDSDSDSD 1509
Query: 1049 SSS 1051
S S
Sbjct: 1510 SDS 1512
Score = 48.9 bits (115), Expect = 8e-05
Identities = 30/125 (24%), Positives = 60/125 (48%), Gaps = 4/125 (3%)
Query: 930 EGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDADAD- 988
+ + + DS+ D + DS++ S + S + + D + + ++ D+D+D+D
Sbjct: 1411 DSDSDADSDSDSDSDSDADSDSDSDSDSDADSDSDADSDSDADSDSDADSDSDSDSDSDA 1470
Query: 989 --DEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQGG 1046
D D+++ S+A D S ++S D D + D D D A+S+ +A+ D+
Sbjct: 1471 DSDSDADSDSDADSD-SDADSDSDSDSDSDADSDSDSDSDSDSDADSDSDADSDSDSDAD 1529
Query: 1047 GDSSS 1051
DS +
Sbjct: 1530 SDSDA 1534
Score = 47.8 bits (112), Expect = 2e-04
Identities = 29/123 (23%), Positives = 59/123 (47%), Gaps = 2/123 (1%)
Query: 930 EGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDADAD- 988
+ + + DS+ D + D+++ S A S + + D + + + D+D+D+D
Sbjct: 1451 DADSDSDADSDSDSDSDSDADSDSDADSDSDADSDSDADSDSDSDSDSDADSDSDSDSDS 1510
Query: 989 DEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQGGGD 1048
D D+++ S+A D S S++ D D + D D D A+S+ +++ D+ D
Sbjct: 1511 DSDADSDSDADSD-SDSDADSDSDADSDSDADGDSDADSDSDADSDSDSDSDSDSDSDSD 1569
Query: 1049 SSS 1051
+ S
Sbjct: 1570 ADS 1572
Score = 45.8 bits (107), Expect = 6e-04
Identities = 33/122 (27%), Positives = 58/122 (47%), Gaps = 5/122 (4%)
Query: 930 EGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDADAD- 988
+ + + DS+ D + DS++ S A S + + D + + ++ D D+DAD
Sbjct: 1489 DSDSDSDSDSDADSDSDSDSDSDSDADSDSDADSDSDSDADSDSDADSDSDADGDSDADS 1548
Query: 989 --DEDSENVSEAGEDVSGSESAGDECFQEDHEED-DIEHDDVDGKAESEGEAEGMCDAQG 1045
D DS++ S++ D S S+S D D + D D +H+D K ++ + DAQ
Sbjct: 1549 DSDADSDSDSDSDSD-SDSDSDADSDSDSDSDSDADRDHNDKTDKPNNKELPDTGNDAQN 1607
Query: 1046 GG 1047
G
Sbjct: 1608 NG 1609
Score = 42.7 bits (99), Expect = 0.005
Identities = 31/125 (24%), Positives = 56/125 (44%), Gaps = 5/125 (4%)
Query: 927 EKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDAD 986
+ E GE+ + +D V + S A S + + D + + + D+D+D
Sbjct: 1279 DPETGEII---EEPQDEVIIHGAKDDSDADSDSDADSDSDADSDSDADSDSDADSDSDSD 1335
Query: 987 ADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQGG 1046
+ D DS++ S+A D S S+S D D + D D D A+S+ +++ D+
Sbjct: 1336 S-DSDSDSDSDADSD-SDSDSDSDADSDSDADSDSDADSDSDSDADSDSDSDADSDSDSD 1393
Query: 1047 GDSSS 1051
DS +
Sbjct: 1394 SDSDA 1398
Score = 42.0 bits (97), Expect = 0.009
Identities = 29/132 (21%), Positives = 59/132 (43%), Gaps = 5/132 (3%)
Query: 911 NSKVKSHEESSGPCKVEKE-EGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESR 969
+S S +S + + + + + DS+ D + DS++ S A S + +
Sbjct: 1455 DSDADSDSDSDSDSDADSDSDADSDSDADSDSDADSDSDSDSDSDADSDSDSDSDSDSDA 1514
Query: 970 DREEECGPETGGDNDADAD---DEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDD 1026
D + + ++ D D+D+D D D++ S+A D S ++S D D + D D
Sbjct: 1515 DSDSDADSDSDSDADSDSDADSDSDADGDSDADSD-SDADSDSDSDSDSDSDSDSDADSD 1573
Query: 1027 VDGKAESEGEAE 1038
D ++S+ + +
Sbjct: 1574 SDSDSDSDADRD 1585
>GARP_PLAFF (P13816) Glutamic acid-rich protein precursor
Length = 678
Score = 54.7 bits (130), Expect = 1e-06
Identities = 31/113 (27%), Positives = 56/113 (49%), Gaps = 2/113 (1%)
Query: 926 VEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDA 985
V+KEE + + + +E+ ++ + + E + E + EEE E + +
Sbjct: 545 VDKEEDKKEESKEVQEESKEVQEDEEEVEEDEEEEEEEEEEEEEEEEEE--EEEEEEEEE 602
Query: 986 DADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAE 1038
+ +DED E+ +A ED +E D+ ++D EEDD E DD + + E E + E
Sbjct: 603 EEEDEDEEDEDDAEEDEDDAEEDEDDAEEDDDEEDDDEEDDDEDEDEDEEDEE 655
Score = 52.0 bits (123), Expect = 9e-06
Identities = 36/129 (27%), Positives = 63/129 (47%), Gaps = 17/129 (13%)
Query: 910 ENSKVKSHEESSGPCKVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESR 969
E+ K +S E +V+++E E+ + + EE+ + + + E + E
Sbjct: 549 EDKKEESKEVQEESKEVQEDEEEVEEDEEEEEE---------EEEEEEEEEEEEEEEEEE 599
Query: 970 DREEECGPETGGDNDADADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDG 1029
+ EEE + ++DA+ D++D+E ED +E DE ++D EEDD E +D D
Sbjct: 600 EEEEEEDEDEEDEDDAEEDEDDAE------EDEDDAEEDDDE--EDDDEEDDDEDEDEDE 651
Query: 1030 KAESEGEAE 1038
+ E E E E
Sbjct: 652 EDEEEEEEE 660
Score = 45.4 bits (106), Expect = 8e-04
Identities = 34/115 (29%), Positives = 54/115 (46%), Gaps = 11/115 (9%)
Query: 944 VAYRDSNAQSMAK--------SKH--NIERRKYESRDREEECGPETGGDNDADADDEDSE 993
V RD++ + MAK KH E +K ES++ +EE E D + +DE+ E
Sbjct: 521 VPRRDNHKKKMAKIEEAELQKQKHVDKEEDKKEESKEVQEE-SKEVQEDEEEVEEDEEEE 579
Query: 994 NVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQGGGD 1048
E E+ E +E +E+ EED+ E D+ D + + + E DA+ D
Sbjct: 580 EEEEEEEEEEEEEEEEEEEEEEEEEEDEDEEDEDDAEEDEDDAEEDEDDAEEDDD 634
Score = 35.8 bits (81), Expect = 0.67
Identities = 46/271 (16%), Positives = 119/271 (42%), Gaps = 51/271 (18%)
Query: 220 RNSLQRFNERNSMTPMMRQMQVDKQRYRRD----RLPSHDRDRDLSVEHPEMDDDKT--- 272
R ++ + M +Q +++K+R +++ + HD++ + +++ P+ ++T
Sbjct: 297 RKKQEKKERKQKEKEMKKQKKIEKERKKKEEKEKKKKKHDKENEETMQQPDQTSEETNNE 356
Query: 273 -MINL----------------------HKEQRKRDRRIRDQDERDPDLDNSRDLTSQ-RF 308
M+ L HKE ++ ++++ ++ + + + +
Sbjct: 357 IMVPLPSPLTDVTTPEEHKEGEHKEEEHKEGEHKEGEHKEEEHKEEEHKKEEHKSKEHKS 416
Query: 309 RDKKKTVKKAEGMYGEAFSFCEKVKEKLSSS---DDYQTFLKCLNIFNNGIIKKNDLQNL 365
+ KK KK +G + +A EKVK+ + + D+ + ++ +N+ + ++ +
Sbjct: 417 KGKKDKGKKDKGKHKKAKK--EKVKKHVVKNVIEDEDKDGVEIINLEDKEACEEQHITVE 474
Query: 366 VTDLLGKHSDLMDEFKDFL--------ERCENIEGFLAGVMSKKSL--STDAHLSRSSKL 415
L L+DE + E+ +I+ L G + + ++ D H + +K+
Sbjct: 475 SRPLSQPQCKLIDEPEQLTLMDKSKVEEKNLSIQEQLIGTIGRVNVVPRRDNHKKKMAKI 534
Query: 416 EDKDKDQKREMDGAKEKDRYKEKYMGKSIQE 446
E+ + +++ +D KE+D+ +E K +QE
Sbjct: 535 EEAELQKQKHVD--KEEDKKEE---SKEVQE 560
Score = 34.7 bits (78), Expect = 1.5
Identities = 37/213 (17%), Positives = 85/213 (39%), Gaps = 16/213 (7%)
Query: 254 HDRDRDLSVEHPEMDDDKTMINLHKEQRKRDRRIRDQDERDPDLDNSRDLTSQRFRDKKK 313
+D+D + SV+ + +K HK+ +K + +D+ E+ + + +DKKK
Sbjct: 109 NDKDNENSVDKKKDKKEKK----HKKDKKEKKEKKDKKEKKDKKEKKHKKEKKHKKDKKK 164
Query: 314 TVK-KAEGMYGEAFSFCEKVKEKLSSSDDYQTFLKCLNIFNNGIIKKNDLQ-------NL 365
+ +Y + E + D + + N G++ + Q +
Sbjct: 165 KENSEVMSLYKTGQHKPKNATEHGEENLDEEMVSEINNNAQGGLLLSSPYQYREQGGCGI 224
Query: 366 VTDLLGKHSDLMDEFKDFLERCENI----EGFLAGVMSKKSLSTDAHLSRSSKLEDKDKD 421
++ + +D D K+ + + E L + K+ + + K+E K K
Sbjct: 225 ISSVHETSNDTKDNDKENISEDKKEDHQQEEMLKTLDKKERKQKEKEMKEQEKIEKKKKK 284
Query: 422 QKREMDGAKEKDRYKEKYMGKSIQELDLSDCKR 454
Q+ + +EK+R K++ + +E ++ K+
Sbjct: 285 QEEKEKKKQEKERKKQEKKERKQKEKEMKKQKK 317
>DMP1_HUMAN (Q13316) Dentin matrix acidic phosphoprotein 1 precursor
(Dentin matrix protein-1) (DMP-1)
Length = 513
Score = 53.1 bits (126), Expect = 4e-06
Identities = 72/340 (21%), Positives = 129/340 (37%), Gaps = 33/340 (9%)
Query: 735 ERVEDRKAGHSSRNFAASN---VGG--DG-SPHRDSISTNSRLPKSDKNEVDGRVTEVKN 788
+RV+ + G S + S VGG DG S H D + +SD E +E N
Sbjct: 172 DRVDSKPEGGDSTQESESEEHWVGGGSDGESSHGDGSELDDEGMQSDDPE--SIRSERGN 229
Query: 789 IHRTSVAANDKENGSVGGELVCRDD---QLMDKGLKKVECSDKAGFSKQFASDEQGVK-- 843
S KE+G + +D QL++ +K+ F K S+E
Sbjct: 230 SRMNSAGMKSKESGENSEQANTQDSGGSQLLEHPSRKI-------FRKSRISEEDDRSEL 282
Query: 844 --NNPSIAIRGENSLNRTNLDVSPGCVSAPSRPTDADDSVAKSQTVNLPLVEGGDIAAPV 901
NN ++ +++ N + D +S P R + D + NL E ++ P
Sbjct: 283 DDNNTMEEVKSDSTENSNSRDTG---LSQPRRDSKGDSQEDSKE--NLSQEESQNVDGPS 337
Query: 902 PVANGVLVENSKVKSHEESSGPCKVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNI 961
++ + + + S E SS + E N D +V ++ + S S H +
Sbjct: 338 SESS----QEANLSSQENSSESQEEVVSESR-GDNPDPTTSYVEDQEDSDSSEEDSSHTL 392
Query: 962 ERRKYESRDREEECGPETGGDNDADADDEDSENVSEAGEDVSGSESAGDECFQEDH-EED 1020
K ESR+ + + + ++ + + S + E + S+ + +E H EED
Sbjct: 393 SHSKSESREEQADSESSESLNFSEESPESPEDENSSSQEGLQSHSSSAESQSEESHSEED 452
Query: 1021 DIEHDDVDGKAESEGEAEGMCDAQGGGDSSSLPLSERFLS 1060
D + D E E ++ G ++ + R L+
Sbjct: 453 DSDSQDSSRSKEDSNSTESKSSSEEDGQLKNIEIESRKLT 492
Score = 35.0 bits (79), Expect = 1.1
Identities = 40/174 (22%), Positives = 68/174 (38%), Gaps = 19/174 (10%)
Query: 919 ESSGPCKVEKEEGELSPNG---DSEEDFVAYRDSNAQSMAKSK---HNIERRKYESRDRE 972
+S P + E G N S+E +N Q S+ H + +SR E
Sbjct: 216 QSDDPESIRSERGNSRMNSAGMKSKESGENSEQANTQDSGGSQLLEHPSRKIFRKSRISE 275
Query: 973 EECGPETGGDN---DADADDEDSENVSEAGEDVSGSESAGDECFQEDHEED--DIEHDDV 1027
E+ E +N + +D ++ N + G +S GD QED +E+ E +V
Sbjct: 276 EDDRSELDDNNTMEEVKSDSTENSNSRDTGLSQPRRDSKGDS--QEDSKENLSQEESQNV 333
Query: 1028 DGKAESEGEAEGMCDAQGGGDSSSLPLSERFLSSVKPLTKHVSAVSFAEEMKDS 1081
DG + + + + +S +SE + P T S+ E+ +DS
Sbjct: 334 DGPSSESSQEANLSSQENSSESQEEVVSESRGDNPDPTT------SYVEDQEDS 381
Score = 32.7 bits (73), Expect = 5.7
Identities = 41/169 (24%), Positives = 64/169 (37%), Gaps = 29/169 (17%)
Query: 902 PVANGVLVENSKVKSHEESSGPCK--VEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKH 959
P+ + E SKV S E+++ + EEG G + ++ YR + S + K
Sbjct: 44 PLESSESSEGSKVSSEEQANEDPSDSTQSEEGL----GSDDHQYI-YRLAGGFSRSTGKG 98
Query: 960 NIERRKYES----------------RDREEECGPETGGDNDADADDEDSENVSEAGED-V 1002
++ E +DR+E G D D+D + SE + G+D
Sbjct: 99 GDDKDDDEDDSGDDTFGDDDSGPGPKDRQEGGNSRLGSDEDSDDTIQASEESAPQGQDSA 158
Query: 1003 SGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQGGGDSSS 1051
+ S E ED + E D ++ESE G GG D S
Sbjct: 159 QDTTSESRELDNEDRVDSKPEGGDSTQESESEEHWVG-----GGSDGES 202
Score = 32.7 bits (73), Expect = 5.7
Identities = 38/155 (24%), Positives = 59/155 (37%), Gaps = 30/155 (19%)
Query: 918 EESSGPCKVEKEEGELSPNGD----------SEEDFVAYRDSNAQSMAKSKHNIERRKYE 967
++ SGP +++EG S G SEE +DS + ++S R+ +
Sbjct: 116 DDDSGPGPKDRQEGGNSRLGSDEDSDDTIQASEESAPQGQDSAQDTTSES------RELD 169
Query: 968 SRDREEECGPETGGDNDADADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEH--- 1024
+ DR + PE G D+ + E E+ G D S G E E + DD E
Sbjct: 170 NEDRVDS-KPEGG---DSTQESESEEHWVGGGSDGESSHGDGSELDDEGMQSDDPESIRS 225
Query: 1025 -------DDVDGKAESEGEAEGMCDAQGGGDSSSL 1052
+ K++ GE + Q G S L
Sbjct: 226 ERGNSRMNSAGMKSKESGENSEQANTQDSGGSQLL 260
>UBF2_XENLA (P25980) Nucleolar transcription factor 2 (Upstream
binding factor-2) (UBF-2) (xUBF-2)
Length = 701
Score = 52.0 bits (123), Expect = 9e-06
Identities = 34/122 (27%), Positives = 55/122 (44%), Gaps = 8/122 (6%)
Query: 937 GDSEEDFVAYRDSNAQSMAKSKHNIERRKYESR-------DREEECGPETGGDNDADADD 989
G S +D AY++ N KS I+ +S+ D +E+ + ++D D DD
Sbjct: 576 GLSTQDRAAYKEQNTNKR-KSTTKIQAPSSKSKLVIQSKSDDDEDDEDDEDEEDDDDDDD 634
Query: 990 EDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQGGGDS 1049
ED E+ SE G+ S E +E+ +E+D E DD D + + + G +
Sbjct: 635 EDKEDSSEDGDSSDSSSDEDSEEGEENEDEEDEEDDDEDNEEDDDDNESGSSSSSSSSAD 694
Query: 1050 SS 1051
SS
Sbjct: 695 SS 696
Score = 33.5 bits (75), Expect = 3.3
Identities = 27/106 (25%), Positives = 43/106 (40%), Gaps = 9/106 (8%)
Query: 902 PVANGVLVENSKVKSHEESSGPCKVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNI 961
P + LV SK E+ E +E E + D +ED + S + S +
Sbjct: 602 PSSKSKLVIQSKSDDDEDD------EDDEDEEDDDDDDDEDKEDSSEDGDSSDSSSDEDS 655
Query: 962 ERRKYESRDREEECGPETGGDNDADADDEDSENVSEAGEDVSGSES 1007
E + + +EE E DN+ D DD +S + S + S+S
Sbjct: 656 EEGEENEDEEDEEDDDE---DNEEDDDDNESGSSSSSSSSADSSDS 698
>NKX1_BISBI (O46383) Sodium/potassium/calcium exchanger 1
(Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod
Na-Ca+K exchanger) (Fragment)
Length = 300
Score = 52.0 bits (123), Expect = 9e-06
Identities = 50/221 (22%), Positives = 85/221 (37%), Gaps = 14/221 (6%)
Query: 824 ECSDKAGFSKQFASDEQGVKNNPSIAIRGENSLNRTNLDVSPGCVSAPSRPTDADDSVAK 883
E +++G + +G N S G+ R D G + A D+ +
Sbjct: 29 ENGERSGGDAALGGESEGKAENES---EGDIPAERRGDDEDEGEIQAEGGEVKGDEDEGE 85
Query: 884 SQTVNLPLVEGGDIAAPVPVANGVLVENSKVKSHEESS--GPCKVEKEEGELSPNGDSEE 941
Q VEG + + G VE + + ++ G + +++EGE+ E
Sbjct: 86 IQAGEGGEVEGDEDEGEIQAGEGGEVEGDEDEGEIQAGEGGEVEGDEDEGEIQAGEGGE- 144
Query: 942 DFVAYRDSNAQSMAKSKHNIERR--KYESRDREEECGPETGGDNDADADDEDSENVSEAG 999
+D + A +E + E + E E GG+ + + ++E EA
Sbjct: 145 ----VKDDEGEIQAGEAGEVEGEDGEVEGGEDEGEIQAGEGGEGETGEQELNAEIQGEAK 200
Query: 1000 EDVSG--SESAGDECFQEDHEEDDIEHDDVDGKAESEGEAE 1038
+D G E GD ED EE+D E D+ + + E E E E
Sbjct: 201 DDEEGVDGEGGGDGGDSEDEEEEDEEEDEEEEEEEEEEEEE 241
>VG48_SHV21 (Q01033) Hypothetical gene 48 protein
Length = 797
Score = 51.2 bits (121), Expect = 2e-05
Identities = 33/114 (28%), Positives = 53/114 (45%), Gaps = 8/114 (7%)
Query: 927 EKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDAD 986
EKE ++ D +D RD + + ++N + + E D E+E + G+++ D
Sbjct: 384 EKEYKKIIDKSDDRDD----RDKDEYELENEEYNRDEEEDEGEDEEDEKDEKEEGEDEGD 439
Query: 987 ADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVD-GKAESEGEAEG 1039
+++ E + GED GDE E +EDD E + D G EGE EG
Sbjct: 440 DGEDEGE---DEGEDEGDEGDEGDEGEDEGEDEDDEEDEGEDEGDEGDEGEDEG 490
Score = 40.4 bits (93), Expect = 0.027
Identities = 24/115 (20%), Positives = 49/115 (41%)
Query: 925 KVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDND 984
++E EE D ED +D + + + + E D +E G+++
Sbjct: 406 ELENEEYNRDEEEDEGEDEEDEKDEKEEGEDEGDDGEDEGEDEGEDEGDEGDEGDEGEDE 465
Query: 985 ADADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEG 1039
+ +D++ + + G++ E GDE + + E D+ + +G EG+ EG
Sbjct: 466 GEDEDDEEDEGEDEGDEGDEGEDEGDEGDEGEDEGDEGDEGKDEGDEGDEGKDEG 520
Score = 40.0 bits (92), Expect = 0.036
Identities = 36/139 (25%), Positives = 58/139 (40%), Gaps = 16/139 (11%)
Query: 910 ENSKVKSHEESSGPCKVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESR 969
E + K + K E +EG+ GD ED + D + E E
Sbjct: 502 EGDEGKDEGDEGDEGKDEGDEGDEGDEGDEGED--EWEDEGDEG--------EDEGDEGE 551
Query: 970 DREEECGPETGGDNDADADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDG 1029
D +E E G+++ D +++ E+ + GED E GDE ED ED+ + + +G
Sbjct: 552 DEGDE--GEDEGEDEGDEGEDEGEDEGDEGEDEG--EDEGDE--GEDEGEDEGDEGEDEG 605
Query: 1030 KAESEGEAEGMCDAQGGGD 1048
+ E + EG + G D
Sbjct: 606 EDEGDEGDEGEDEGDEGED 624
Score = 39.7 bits (91), Expect = 0.046
Identities = 29/125 (23%), Positives = 52/125 (41%), Gaps = 3/125 (2%)
Query: 927 EKEEGELSP---NGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDN 983
+K+E EL N D EED + + + + + E D E+ G E +
Sbjct: 401 DKDEYELENEEYNRDEEEDEGEDEEDEKDEKEEGEDEGDDGEDEGEDEGEDEGDEGDEGD 460
Query: 984 DADADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDA 1043
+ + + ED ++ + GED G++ E E +D + +GK E + EG +
Sbjct: 461 EGEDEGEDEDDEEDEGEDEGDEGDEGEDEGDEGDEGEDEGDEGDEGKDEGDEGDEGKDEG 520
Query: 1044 QGGGD 1048
G +
Sbjct: 521 DEGDE 525
Score = 39.3 bits (90), Expect = 0.061
Identities = 39/125 (31%), Positives = 51/125 (40%), Gaps = 8/125 (6%)
Query: 928 KEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETG--GDNDA 985
++EG+ GD ED D + + E + E E + G + G GD
Sbjct: 450 EDEGDEGDEGDEGED--EGEDEDDEEDEGEDEGDEGDEGEDEGDEGDEGEDEGDEGDEGK 507
Query: 986 DADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVD-GKAE-SEGEAEGMCDA 1043
D DE E E E G E GDE E +E D D+ D G+ E EGE EG +
Sbjct: 508 DEGDEGDEGKDEGDEGDEGDE--GDEGEDEWEDEGDEGEDEGDEGEDEGDEGEDEGEDEG 565
Query: 1044 QGGGD 1048
G D
Sbjct: 566 DEGED 570
Score = 39.3 bits (90), Expect = 0.061
Identities = 30/128 (23%), Positives = 54/128 (41%), Gaps = 3/128 (2%)
Query: 919 ESSGPCKVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPE 978
E G + ++ E E GD ++ D + + + + E + E+E G E
Sbjct: 591 EDEGEDEGDEGEDEGEDEGDEGDEGEDEGDEGEDEGDEGEDEGDEGEDEGDEGEDE-GDE 649
Query: 979 TGGDNDADADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAE 1038
G+++ D +++ + E ++ E GDE E E D+ + D + E EGE E
Sbjct: 650 --GEDEGDEGEDEGDEGDEGEDEGDEGEDEGDEGEDEGDEGDEGDEGDEGDEGEDEGEDE 707
Query: 1039 GMCDAQGG 1046
G + G
Sbjct: 708 GEDEGDEG 715
Score = 38.5 bits (88), Expect = 0.10
Identities = 30/129 (23%), Positives = 57/129 (43%), Gaps = 5/129 (3%)
Query: 925 KVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKY--ESRDREEECGPETGGD 982
+V+ E+G L D E + ++ K + E +K +S DR++ E +
Sbjct: 349 EVDSEDGNLCVLDDESESVNSVALRQVLTVDKQANEKEYKKIIDKSDDRDDRDKDEYELE 408
Query: 983 NDA---DADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEG 1039
N+ D ++++ E+ + ++ E GD+ E +E + E D+ D E E E E
Sbjct: 409 NEEYNRDEEEDEGEDEEDEKDEKEEGEDEGDDGEDEGEDEGEDEGDEGDEGDEGEDEGED 468
Query: 1040 MCDAQGGGD 1048
D + G+
Sbjct: 469 EDDEEDEGE 477
Score = 38.1 bits (87), Expect = 0.14
Identities = 35/139 (25%), Positives = 55/139 (39%), Gaps = 8/139 (5%)
Query: 919 ESSGPCKVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPE 978
E G + ++ E E GD ED E E D +E G +
Sbjct: 580 EDEGEDEGDEGEDEGEDEGDEGEDEGEDEGDEGDEGEDEGDEGEDEGDEGEDEGDE-GED 638
Query: 979 TG--GDNDADADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVD----GKAE 1032
G G+++ D +++ + + G++ E GDE E E +D E D+ D G
Sbjct: 639 EGDEGEDEGDEGEDEGDEGEDEGDEGDEGEDEGDEGEDEGDEGED-EGDEGDEGDEGDEG 697
Query: 1033 SEGEAEGMCDAQGGGDSSS 1051
EGE EG + + GD +
Sbjct: 698 DEGEDEGEDEGEDEGDEGT 716
Score = 37.4 bits (85), Expect = 0.23
Identities = 34/126 (26%), Positives = 51/126 (39%), Gaps = 4/126 (3%)
Query: 918 EESSGPCKVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGP 977
E G + ++ E E GD ED + + + + + E E E
Sbjct: 546 EGDEGEDEGDEGEDEGEDEGDEGEDEGEDEGDEGEDEGEDEGDEGEDEGEDEGDEGEDEG 605
Query: 978 ETGGDNDADADDE--DSENVSEAGEDVSG-SESAGDECFQE-DHEEDDIEHDDVDGKAES 1033
E GD + +DE + E+ + GED E GDE E D ED+ + + +G
Sbjct: 606 EDEGDEGDEGEDEGDEGEDEGDEGEDEGDEGEDEGDEGEDEGDEGEDEGDEGEDEGDEGD 665
Query: 1034 EGEAEG 1039
EGE EG
Sbjct: 666 EGEDEG 671
Score = 35.8 bits (81), Expect = 0.67
Identities = 33/125 (26%), Positives = 57/125 (45%), Gaps = 7/125 (5%)
Query: 927 EKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERR-KYESRDREEECGPETGGDNDA 985
E++EGE GD ++ D + + E + + + D ++ G E ++
Sbjct: 472 EEDEGE--DEGDEGDEGEDEGDEGDEGEDEGDEGDEGKDEGDEGDEGKDEGDEGDEGDEG 529
Query: 986 DADDEDSENVSEAGEDVSGS-ESAGDECFQEDHEEDDIEHDDVDGKAES-EGEAEGMCDA 1043
D +++ E+ + GED E GDE ED ED+ + + +G+ E EGE EG +
Sbjct: 530 DEGEDEWEDEGDEGEDEGDEGEDEGDE--GEDEGEDEGDEGEDEGEDEGDEGEDEGEDEG 587
Query: 1044 QGGGD 1048
G D
Sbjct: 588 DEGED 592
Score = 35.8 bits (81), Expect = 0.67
Identities = 33/116 (28%), Positives = 46/116 (39%), Gaps = 8/116 (6%)
Query: 927 EKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDAD 986
E +EGE GD ED + E E D +E G + G + D
Sbjct: 611 EGDEGE--DEGDEGEDEGDEGEDEGDEGEDEGDEGEDEGDEGEDEGDE-GEDEGDEGDEG 667
Query: 987 ADDEDS-ENVSEAGEDVSGSESAGDECFQ----EDHEEDDIEHDDVDGKAESEGEA 1037
D+ D E+ + GED GDE + ED ED+ E + +G + EG A
Sbjct: 668 EDEGDEGEDEGDEGEDEGDEGDEGDEGDEGDEGEDEGEDEGEDEGDEGTKDKEGNA 723
>UBF1_MOUSE (P25976) Nucleolar transcription factor 1 (Upstream
binding factor 1) (UBF-1)
Length = 765
Score = 50.8 bits (120), Expect = 2e-05
Identities = 29/100 (29%), Positives = 46/100 (46%), Gaps = 7/100 (7%)
Query: 952 QSMAKSKHNIERRKYESRDREEECGPETGGDNDADADDEDSENVSEAGEDVSGSESAGDE 1011
QS ++S+ + + + + + EEE + GD+ D D + + ED GDE
Sbjct: 672 QSKSESEEDDDEEEEDDEEEEEEEEDDENGDSSEDGGDSSESSSEDESED-------GDE 724
Query: 1012 CFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQGGGDSSS 1051
+D +EDD + DD D ESEG + + DS S
Sbjct: 725 NDDDDDDEDDEDDDDEDEDNESEGSSSSSSSSGDSSDSGS 764
Score = 45.1 bits (105), Expect = 0.001
Identities = 22/111 (19%), Positives = 50/111 (44%)
Query: 941 EDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDADADDEDSENVSEAGE 1000
+++++ + N + R +S+ EE E D++ + ++E+ + ++ E
Sbjct: 646 KEYISNKRKNMTKLRGPNPKSSRTTLQSKSESEEDDDEEEEDDEEEEEEEEDDENGDSSE 705
Query: 1001 DVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQGGGDSSS 1051
D S + E ED +E+D + DD D + + + + + + SSS
Sbjct: 706 DGGDSSESSSEDESEDGDENDDDDDDEDDEDDDDEDEDNESEGSSSSSSSS 756
Score = 44.7 bits (104), Expect = 0.001
Identities = 25/96 (26%), Positives = 46/96 (47%), Gaps = 1/96 (1%)
Query: 956 KSKHNIERRKYESRDREEECGPETGGDNDADADDEDSENVSEAGEDVSGSESAGDECFQE 1015
KS + K ES + ++E + + + + DDE+ ++ + G+ S E E
Sbjct: 665 KSSRTTLQSKSESEEDDDEEEEDDEEEEEEEEDDENGDSSEDGGDSSESSSEDESEDGDE 724
Query: 1016 DHEEDDIEHDDVDGKAESEGEAEG-MCDAQGGGDSS 1050
+ ++DD E D+ D + + E+EG + GDSS
Sbjct: 725 NDDDDDDEDDEDDDDEDEDNESEGSSSSSSSSGDSS 760
Score = 39.3 bits (90), Expect = 0.061
Identities = 34/102 (33%), Positives = 47/102 (45%), Gaps = 19/102 (18%)
Query: 912 SKVKSHE---ESSGPCKVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYES 968
SK +S E E + E+EE E NGDS ED +S+++ ES
Sbjct: 673 SKSESEEDDDEEEEDDEEEEEEEEDDENGDSSEDGGDSSESSSED-------------ES 719
Query: 969 RDREEECGPETGGDNDADADDEDSENVSEAGEDVSGSESAGD 1010
D +E + ++D D DDED +N SE S S S+GD
Sbjct: 720 EDGDEN-DDDDDDEDDEDDDDEDEDNESEGSS--SSSSSSGD 758
>ECX1_METMA (Q8PTT8) Probable exosome complex exonuclease 1 (EC
3.1.13.-)
Length = 493
Score = 50.4 bits (119), Expect = 3e-05
Identities = 58/251 (23%), Positives = 109/251 (43%), Gaps = 32/251 (12%)
Query: 814 QLMDKGLKKVECSDKAGFSKQFASDEQGVKNNPSIAIRGENSLNRTNLDVSPGCVSAPSR 873
+L+ KG K++ +A K+F + + V + E + L+ SP
Sbjct: 216 ELVKKGCKEILEIQQAVLRKKFETPVEEVSEETA----PEKGAEKEVLEPSPVAAIVEET 271
Query: 874 PTDADD-SVAKSQTVNLPLVEGGDIAAPVPVANGVLVENSKVKSHEESSGPCKVEKEE-- 930
P +A++ V S+ V ++ + +P L E + + EES + E+EE
Sbjct: 272 PEEAEEPEVEISEEVEAEILA----SEVIPDFEDELEEEIE-EELEESEEDLETEEEEFE 326
Query: 931 -----GELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEE--------CGP 977
E P D EED + + + + + +E ++E EEE C P
Sbjct: 327 EEALEEEAEPEEDLEEDL---EEDLGEELEEEEEELEEEEFEEEALEEETELEASLECAP 383
Query: 978 ETGGDNDADADDEDSENVSEAGEDVSG-SESAGDECFQEDHEEDDI---EHDDVDGKAES 1033
E ++ +A E + EA E++ +E A +E +E+ E ++ E ++++ +AE+
Sbjct: 384 ELKEFDEIEARLEKEDASIEAEEEIEPEAEEATEEGLEEEAEIEETAASEEENIEAEAEA 443
Query: 1034 EGEAEGMCDAQ 1044
E EAE +A+
Sbjct: 444 EEEAEPEVEAE 454
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.312 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 137,237,968
Number of Sequences: 164201
Number of extensions: 6365587
Number of successful extensions: 48111
Number of sequences better than 10.0: 804
Number of HSP's better than 10.0 without gapping: 307
Number of HSP's successfully gapped in prelim test: 518
Number of HSP's that attempted gapping in prelim test: 36110
Number of HSP's gapped (non-prelim): 5431
length of query: 1129
length of database: 59,974,054
effective HSP length: 121
effective length of query: 1008
effective length of database: 40,105,733
effective search space: 40426578864
effective search space used: 40426578864
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0081b.6


