

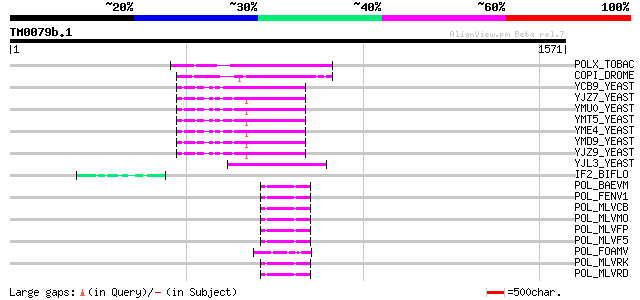
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0079b.1
(1571 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 178 9e-44
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 137 3e-31
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 96 8e-19
YJZ7_YEAST (P47098) Transposon Ty1 protein B 84 3e-15
YMU0_YEAST (Q04670) Transposon Ty1 protein B 84 4e-15
YMT5_YEAST (Q04214) Transposon Ty1 protein B 84 4e-15
YME4_YEAST (Q04711) Transposon Ty1 protein B 84 4e-15
YMD9_YEAST (Q03434) Transposon Ty1 protein B 84 4e-15
YJZ9_YEAST (P47100) Transposon Ty1 protein B 84 4e-15
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 73 7e-12
IF2_BIFLO (Q8G3Y5) Translation initiation factor IF-2 52 1e-05
POL_BAEVM (P10272) Pol polyprotein [Contains: Protease (EC 3.4.2... 51 3e-05
POL_FENV1 (P31792) Pol polyprotein [Contains: Reverse transcript... 50 6e-05
POL_MLVCB (P08361) Pol polyprotein [Contains: Reverse transcript... 49 8e-05
POL_MLVMO (P03355) Pol polyprotein [Contains: Protease (EC 3.4.2... 49 1e-04
POL_MLVFP (P26808) Pol polyprotein [Contains: Protease (EC 3.4.2... 49 1e-04
POL_MLVF5 (P26810) Pol polyprotein [Contains: Protease (EC 3.4.2... 49 1e-04
POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse transcript... 49 1e-04
POL_MLVRK (P31795) Pol polyprotein [Contains: Protease (EC 3.4.2... 48 2e-04
POL_MLVRD (P11227) Pol polyprotein [Contains: Protease (EC 3.4.2... 48 2e-04
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 178 bits (452), Expect = 9e-44
Identities = 138/464 (29%), Positives = 197/464 (41%), Gaps = 45/464 (9%)
Query: 456 QTMLASAPSTLGNWHPDSGATHHVTHDASAISDGMSVTGN-DQVYMGNGQGLPIQSIGSA 514
+ M S P + W D+ A+HH T V G+ V MGN I IG
Sbjct: 283 ECMHLSGPES--EWVVDTAASHHATPVRDLFC--RYVAGDFGTVKMGNTSYSKIAGIGDI 338
Query: 515 SFSSPIHTQTTLLLKNLLLVPNITKNLVSVSKFAQDN-GVYFEFHPFHCTVNSQDTCKTL 573
+ + TL+LK++ VP++ NL+S +D YF + T S K +
Sbjct: 339 CIKTNVGC--TLVLKDVRHVPDLRMNLISGIALDRDGYESYFANQKWRLTKGSLVIAKGV 396
Query: 574 LRGSLGSDGLYSFDGMHSSNFHKDLPQVPVSSAFSLSKPASPAVNTIVSTSNNVPSPTAY 633
RG+L G N +
Sbjct: 397 ARGTLYRTNAEICQG----------------------------------ELNAAQDEISV 422
Query: 634 DLWHSRLGHPHYETLQSALTTCKISVPHKSKYTICHSCCVAKSHRLPSSASSTLYTAPLE 693
DLWH R+GH + LQ IS + C C K HR+ SS L+
Sbjct: 423 DLWHKRMGHMSEKGLQILAKKSLISYAKGTTVKPCDYCLFGKQHRVSFQTSSERKLNILD 482
Query: 694 LIFADLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVELKFGL 753
L+++D+ GP IES G YF+T +D SR W+Y+LK K + VF +F A+VE + G
Sbjct: 483 LVYSDVCGPMEIESMGGNKYFVTFIDDASRKLWVYILKTKDQVFQVFQKFHALVERETGR 542
Query: 754 KIKSVQTDGGTEF--KPLLPHFLKLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLAC 811
K+K +++D G E+ + + GI H T P T NG ER +R IVE S+L
Sbjct: 543 KLKRLRSDNGGEYTSREFEEYCSSHGIRHEKTVPGTPQHNGVAERMNRTIVEKVRSMLRM 602
Query: 812 ANMPTSYWDHAFLTATFLINRLPTPVLGNLSPYHKLFKVPADYKILRVFGSACYPHLRPY 871
A +P S+W A TA +LINR P+ L P Y L+VFG + H+
Sbjct: 603 AKLPKSFWGEAVQTACYLINRSPSVPLAFEIPERVWTNKEVSYSHLKVFGCRAFAHVPKE 662
Query: 872 NSSKLSFRSQECVFLGYSSSHKGYKCL-ASDGRIYISKDVQFNE 914
+KL +S C+F+GY GY+ ++ S+DV F E
Sbjct: 663 QRTKLDDKSIPCIFIGYGDEEFGYRLWDPVKKKVIRSRDVVFRE 706
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 137 bits (344), Expect = 3e-31
Identities = 124/461 (26%), Positives = 198/461 (42%), Gaps = 65/461 (14%)
Query: 472 DSGATHHVTHDASAISDGMSVTGNDQVYMGNGQGLPIQSIGSASFSSPIHTQTTLLLKNL 531
DSGA+ H+ +D S +D + V ++ + QG I + + + L+++
Sbjct: 292 DSGASDHLINDESLYTDSVEVVPPLKIAVAK-QGEFIYATKRGIVR--LRNDHEITLEDV 348
Query: 532 LLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCKTLLRGSLGSDGLYSFDGMHS 591
L NL+SV + Q+ G+ EF T++ G L + + +F
Sbjct: 349 LFCKEAAGNLMSVKRL-QEAGMSIEFDKSGVTISKNGLMVVKNSGMLNNVPVINFQAYSI 407
Query: 592 SNFHKDLPQVPVSSAFSLSKPASPAVNTIVSTSNNVPSPTAYDLWHSRLGHPHYETL--- 648
+ HK NN + LWH R GH L
Sbjct: 408 NAKHK----------------------------NN------FRLWHERFGHISDGKLLEI 433
Query: 649 --------QSALTTCKISVPHKSKYTICHSCCVAKSHRLPSSA--SSTLYTAPLELIFAD 698
QS L ++S IC C K RLP T PL ++ +D
Sbjct: 434 KRKNMFSDQSLLNNLELSCE------ICEPCLNGKQARLPFKQLKDKTHIKRPLFVVHSD 487
Query: 699 LWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVELKFGLKIKSV 758
+ GP + + +YF+ VD F+ + YL+K KS+ ++F F A E F LK+ +
Sbjct: 488 VCGPITPVTLDDKNYFVIFVDQFTHYCVTYLIKYKSDVFSMFQDFVAKSEAHFNLKVVYL 547
Query: 759 QTDGGTEF--KPLLPHFLKLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACANMPT 816
D G E+ + +K GI++ LT PHT NG ER R I E ++++ A +
Sbjct: 548 YIDNGREYLSNEMRQFCVKKGISYHLTVPHTPQLNGVSERMIRTITEKARTMVSGAKLDK 607
Query: 817 SYWDHAFLTATFLINRLPTPVL--GNLSPYHKLFKVPADYKILRVFGSACYPHLRPYNSS 874
S+W A LTAT+LINR+P+ L + +PY K LRVFG+ Y H++
Sbjct: 608 SFWGEAVLTATYLINRIPSRALVDSSKTPYEMWHNKKPYLKHLRVFGATVYVHIK-NKQG 666
Query: 875 KLSFRSQECVFLGYSSSHKGYKCL-ASDGRIYISKDVQFNE 914
K +S + +F+GY + G+K A + + +++DV +E
Sbjct: 667 KFDDKSFKSIFVGYEPN--GFKLWDAVNEKFIVARDVVVDE 705
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 95.9 bits (237), Expect = 8e-19
Identities = 100/383 (26%), Positives = 166/383 (43%), Gaps = 48/383 (12%)
Query: 472 DSGATHHVTHDASAISDGMSVTGNDQVYMGNGQ--GLPIQSIGSASFSSPIHTQTTLLLK 529
DSGA+ + A + T N ++ + + Q +PI +IG+ F+ T+T++
Sbjct: 457 DSGASQTLVRSAHYLHHA---TPNSEINIVDAQKQDIPINAIGNLHFNFQNGTKTSI--- 510
Query: 530 NLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCKTLLRGSLGSDGLYSFDGM 589
L PNI +L+S+S+ A N + + T TL R SDG +
Sbjct: 511 KALHTPNIAYDLLSLSELANQN------------ITACFTRNTLER----SDGTVLAPIV 554
Query: 590 HSSNFHKDLPQVPVSSAFSLSKPASPAVNTIVSTSNNVPSPTAYDLWHSRLGHPHYETLQ 649
+F+ +S + + S V+ S +V + Y L H LGH ++ ++Q
Sbjct: 555 KHGDFYW------LSKKYLIPSHISKLTINNVNKSKSV-NKYPYPLIHRMLGHANFRSIQ 607
Query: 650 -----SALTTCK---ISVPHKSKYTICHSCCVAKSHRLPSSASSTLYTA----PLELIFA 697
+A+T K I + S Y C C + KS + S L P + +
Sbjct: 608 KSLKKNAVTYLKESDIEWSNASTYQ-CPDCLIGKSTKHRHVKGSRLKYQESYEPFQYLHT 666
Query: 698 DLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETS--TVFIQFQAMVELKFGLKI 755
D++GP + SYF++ D +RF W+Y L + E S VF A ++ +F ++
Sbjct: 667 DIFGPVHHLPKSAPSYFISFTDEKTRFQWVYPLHDRREESILNVFTSILAFIKNQFNARV 726
Query: 756 KSVQTDGGTEF--KPLLPHFLKLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACAN 813
+Q D G+E+ K L F GIT T +G ER +R ++ +LL C+
Sbjct: 727 LVIQMDRGSEYTNKTLHKFFTNRGITACYTTTADSRAHGVAERLNRTLLNDCRTLLHCSG 786
Query: 814 MPTSYWDHAFLTATFLINRLPTP 836
+P W A +T + N L +P
Sbjct: 787 LPNHLWFSAVEFSTIIRNSLVSP 809
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 84.0 bits (206), Expect = 3e-15
Identities = 94/383 (24%), Positives = 163/383 (42%), Gaps = 48/383 (12%)
Query: 472 DSGATHHVTHDASAISDGMSVTGNDQVYMGNGQ--GLPIQSIGSASFSSPIHTQTTLLLK 529
DSGA+ + A I S + N + + + Q +PI +IG F +T+T++
Sbjct: 461 DSGASRTLIRSAHHIH---SASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSI--- 514
Query: 530 NLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCKTLLRGSLGSDGLYSFDGM 589
+L PNI +L+S+++ A + + + T L R SDG +
Sbjct: 515 KVLHTPNIAYDLLSLNELAAVD------------ITACFTKNVLER----SDGTVLAPIV 558
Query: 590 HSSNFHKDLPQVPVSSAFSLSKPAS-PAVNTIVSTSNNVPSPTAYDLWHSRLGHPHYETL 648
+F+ VS + L S P +N + ++ + P Y H L H + +T+
Sbjct: 559 QYGDFYW------VSKRYLLPSNISVPTINNVHTSESTRKYP--YPFIHRMLAHANAQTI 610
Query: 649 QSALTTCKISVPHKSKYTI-------CHSCCVAKSHRLPSSASSTLYTA----PLELIFA 697
+ +L I+ ++S C C + KS + S L P + +
Sbjct: 611 RYSLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHT 670
Query: 698 DLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETS--TVFIQFQAMVELKFGLKI 755
D++GP + SYF++ D ++F W+Y L + E S VF A ++ +F +
Sbjct: 671 DIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASV 730
Query: 756 KSVQTDGGTEFKPLLPH-FL-KLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACAN 813
+Q D G+E+ H FL K GIT T +G ER +R +++ + L C+
Sbjct: 731 LVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSG 790
Query: 814 MPTSYWDHAFLTATFLINRLPTP 836
+P W A +T + N L +P
Sbjct: 791 LPNHLWFSAIEFSTIVRNSLASP 813
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 83.6 bits (205), Expect = 4e-15
Identities = 94/383 (24%), Positives = 163/383 (42%), Gaps = 48/383 (12%)
Query: 472 DSGATHHVTHDASAISDGMSVTGNDQVYMGNGQ--GLPIQSIGSASFSSPIHTQTTLLLK 529
DSGA+ + A I S + N + + + Q +PI +IG F +T+T++
Sbjct: 34 DSGASRTLIRSAHHIH---SASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSI--- 87
Query: 530 NLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCKTLLRGSLGSDGLYSFDGM 589
+L PNI +L+S+++ A + + + T L R SDG +
Sbjct: 88 KVLHTPNIAYDLLSLNELAAVD------------ITACFTKNVLER----SDGTVLAPIV 131
Query: 590 HSSNFHKDLPQVPVSSAFSLSKPAS-PAVNTIVSTSNNVPSPTAYDLWHSRLGHPHYETL 648
+F+ VS + L S P +N + ++ + P Y H L H + +T+
Sbjct: 132 KYGDFYW------VSKKYLLPSNISVPTINNVHTSESTRKYP--YPFIHRMLAHANAQTI 183
Query: 649 QSALTTCKISVPHKSKYTI-------CHSCCVAKSHRLPSSASSTLYTA----PLELIFA 697
+ +L I+ ++S C C + KS + S L P + +
Sbjct: 184 RYSLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHT 243
Query: 698 DLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETS--TVFIQFQAMVELKFGLKI 755
D++GP + SYF++ D ++F W+Y L + E S VF A ++ +F +
Sbjct: 244 DIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASV 303
Query: 756 KSVQTDGGTEFKPLLPH-FL-KLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACAN 813
+Q D G+E+ H FL K GIT T +G ER +R +++ + L C+
Sbjct: 304 LVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSG 363
Query: 814 MPTSYWDHAFLTATFLINRLPTP 836
+P W A +T + N L +P
Sbjct: 364 LPNHLWFSAIEFSTIVRNSLASP 386
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 83.6 bits (205), Expect = 4e-15
Identities = 94/383 (24%), Positives = 163/383 (42%), Gaps = 48/383 (12%)
Query: 472 DSGATHHVTHDASAISDGMSVTGNDQVYMGNGQ--GLPIQSIGSASFSSPIHTQTTLLLK 529
DSGA+ + A I S + N + + + Q +PI +IG F +T+T++
Sbjct: 34 DSGASRTLIRSAHHIH---SASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSI--- 87
Query: 530 NLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCKTLLRGSLGSDGLYSFDGM 589
+L PNI +L+S+++ A + + + T L R SDG +
Sbjct: 88 KVLHTPNIAYDLLSLNELAAVD------------ITACFTKNVLER----SDGTVLAPIV 131
Query: 590 HSSNFHKDLPQVPVSSAFSLSKPAS-PAVNTIVSTSNNVPSPTAYDLWHSRLGHPHYETL 648
+F+ VS + L S P +N + ++ + P Y H L H + +T+
Sbjct: 132 KYGDFYW------VSKKYLLPSNISVPTINNVHTSESTRKYP--YPFIHRMLAHANAQTI 183
Query: 649 QSALTTCKISVPHKSKYTI-------CHSCCVAKSHRLPSSASSTLYTA----PLELIFA 697
+ +L I+ ++S C C + KS + S L P + +
Sbjct: 184 RYSLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHT 243
Query: 698 DLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETS--TVFIQFQAMVELKFGLKI 755
D++GP + SYF++ D ++F W+Y L + E S VF A ++ +F +
Sbjct: 244 DIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASV 303
Query: 756 KSVQTDGGTEFKPLLPH-FL-KLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACAN 813
+Q D G+E+ H FL K GIT T +G ER +R +++ + L C+
Sbjct: 304 LVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSG 363
Query: 814 MPTSYWDHAFLTATFLINRLPTP 836
+P W A +T + N L +P
Sbjct: 364 LPNHLWFSAIEFSTIVRNSLASP 386
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 83.6 bits (205), Expect = 4e-15
Identities = 94/383 (24%), Positives = 163/383 (42%), Gaps = 48/383 (12%)
Query: 472 DSGATHHVTHDASAISDGMSVTGNDQVYMGNGQ--GLPIQSIGSASFSSPIHTQTTLLLK 529
DSGA+ + A I S + N + + + Q +PI +IG F +T+T++
Sbjct: 34 DSGASRTLIRSAHHIH---SASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSI--- 87
Query: 530 NLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCKTLLRGSLGSDGLYSFDGM 589
+L PNI +L+S+++ A + + + T L R SDG +
Sbjct: 88 KVLHTPNIAYDLLSLNELAAVD------------ITACFTKNVLER----SDGTVLAPIV 131
Query: 590 HSSNFHKDLPQVPVSSAFSLSKPAS-PAVNTIVSTSNNVPSPTAYDLWHSRLGHPHYETL 648
+F+ VS + L S P +N + ++ + P Y H L H + +T+
Sbjct: 132 KYGDFYW------VSKKYLLPSNISVPTINNVHTSESTRKYP--YPFIHRMLAHANAQTI 183
Query: 649 QSALTTCKISVPHKSKYTI-------CHSCCVAKSHRLPSSASSTLYTA----PLELIFA 697
+ +L I+ ++S C C + KS + S L P + +
Sbjct: 184 RYSLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHT 243
Query: 698 DLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETS--TVFIQFQAMVELKFGLKI 755
D++GP + SYF++ D ++F W+Y L + E S VF A ++ +F +
Sbjct: 244 DIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASV 303
Query: 756 KSVQTDGGTEFKPLLPH-FL-KLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACAN 813
+Q D G+E+ H FL K GIT T +G ER +R +++ + L C+
Sbjct: 304 LVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSG 363
Query: 814 MPTSYWDHAFLTATFLINRLPTP 836
+P W A +T + N L +P
Sbjct: 364 LPNHLWFSAIEFSTIVRNSLASP 386
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 83.6 bits (205), Expect = 4e-15
Identities = 94/383 (24%), Positives = 163/383 (42%), Gaps = 48/383 (12%)
Query: 472 DSGATHHVTHDASAISDGMSVTGNDQVYMGNGQ--GLPIQSIGSASFSSPIHTQTTLLLK 529
DSGA+ + A I S + N + + + Q +PI +IG F +T+T++
Sbjct: 34 DSGASRTLIRSAHHIH---SASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSI--- 87
Query: 530 NLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCKTLLRGSLGSDGLYSFDGM 589
+L PNI +L+S+++ A + + + T L R SDG +
Sbjct: 88 KVLHTPNIAYDLLSLNELAAVD------------ITACFTKNVLER----SDGTVLAPIV 131
Query: 590 HSSNFHKDLPQVPVSSAFSLSKPAS-PAVNTIVSTSNNVPSPTAYDLWHSRLGHPHYETL 648
+F+ VS + L S P +N + ++ + P Y H L H + +T+
Sbjct: 132 KYGDFYW------VSKKYLLPSNISVPTINNVHTSESTRKYP--YPFIHRMLAHANAQTI 183
Query: 649 QSALTTCKISVPHKSKYTI-------CHSCCVAKSHRLPSSASSTLYTA----PLELIFA 697
+ +L I+ ++S C C + KS + S L P + +
Sbjct: 184 RYSLKNNTITYFNESDVDRSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHT 243
Query: 698 DLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETS--TVFIQFQAMVELKFGLKI 755
D++GP + SYF++ D ++F W+Y L + E S VF A ++ +F +
Sbjct: 244 DIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASV 303
Query: 756 KSVQTDGGTEFKPLLPH-FL-KLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACAN 813
+Q D G+E+ H FL K GIT T +G ER +R +++ + L C+
Sbjct: 304 LVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSG 363
Query: 814 MPTSYWDHAFLTATFLINRLPTP 836
+P W A +T + N L +P
Sbjct: 364 LPNHLWFSAIEFSTIVRNSLASP 386
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 83.6 bits (205), Expect = 4e-15
Identities = 94/383 (24%), Positives = 163/383 (42%), Gaps = 48/383 (12%)
Query: 472 DSGATHHVTHDASAISDGMSVTGNDQVYMGNGQ--GLPIQSIGSASFSSPIHTQTTLLLK 529
DSGA+ + A I S + N + + + Q +PI +IG F +T+T++
Sbjct: 461 DSGASRTLIRSAHHIH---SASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSI--- 514
Query: 530 NLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCKTLLRGSLGSDGLYSFDGM 589
+L PNI +L+S+++ A + + + T L R SDG +
Sbjct: 515 KVLHTPNIAYDLLSLNELAAVD------------ITACFTKNVLER----SDGTVLAPIV 558
Query: 590 HSSNFHKDLPQVPVSSAFSLSKPAS-PAVNTIVSTSNNVPSPTAYDLWHSRLGHPHYETL 648
+F+ VS + L S P +N + ++ + P Y H L H + +T+
Sbjct: 559 KYGDFYW------VSKKYLLPSNISVPTINNVHTSESTRKYP--YPFIHRMLAHANAQTI 610
Query: 649 QSALTTCKISVPHKSKYTI-------CHSCCVAKSHRLPSSASSTLYTA----PLELIFA 697
+ +L I+ ++S C C + KS + S L P + +
Sbjct: 611 RYSLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHT 670
Query: 698 DLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETS--TVFIQFQAMVELKFGLKI 755
D++GP + SYF++ D ++F W+Y L + E S VF A ++ +F +
Sbjct: 671 DIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASV 730
Query: 756 KSVQTDGGTEFKPLLPH-FL-KLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACAN 813
+Q D G+E+ H FL K GIT T +G ER +R +++ + L C+
Sbjct: 731 LVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSG 790
Query: 814 MPTSYWDHAFLTATFLINRLPTP 836
+P W A +T + N L +P
Sbjct: 791 LPNHLWFSAIEFSTIVRNSLASP 813
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 72.8 bits (177), Expect = 7e-12
Identities = 72/298 (24%), Positives = 125/298 (41%), Gaps = 21/298 (7%)
Query: 617 VNTIVSTSN-----NVPSPTAYDLWHSRLGHPHYETLQSALTTC----KISVPHKSKYTI 667
+N I TS+ N S T D H R+GH + +++++ + + +
Sbjct: 538 INAIKPTSSPGFKLNKRSITLEDA-HKRMGHTGIQQIENSIKHNHYEESLDLIKEPNEFW 596
Query: 668 CHSCCVAKS----HRLPSSASSTLYTAPLELIFADLWGPASIESSAGYSYFLTCVDAFSR 723
C +C ++K+ H S + + P D++GP S ++ Y L VD +R
Sbjct: 597 CQTCKISKATKRNHYTGSMNNHSTDHEPGSSWCMDIFGPVSSSNADTKRYMLIMVDNNTR 656
Query: 724 FTWIYLLKRKSETSTVFIQFQA---MVELKFGLKIKSVQTDGGTEFK--PLLPHFLKLGI 778
+ K+ T+ Q + VE +F K++ + +D GTEF + +F+ GI
Sbjct: 657 YCMTSTHFNKN-AETILAQVRKNIQYVETQFDRKVREINSDRGTEFTNDQIEEYFISKGI 715
Query: 779 THRLTCPHTHHQNGSVERKHRHIVETGLSLLACANMPTSYWDHAFLTATFLINRLPTPVL 838
H LT H NG ER R I+ +LL +N+ +W++A +AT + N L
Sbjct: 716 HHILTSTQDHAANGRAERYIRTIITDATTLLRQSNLRVKFWEYAVTSATNIRNYLEHKST 775
Query: 839 GNLSPYHKLFKVPADYKILRVFGSACYPHLRPYNSSKLSFRSQECVFLGYSSSHKGYK 896
G L P + + P +++ + +N KL + L + GYK
Sbjct: 776 GKL-PLKAISRQPVTVRLMSFLPFGEKGIIWNHNHKKLKPSGLPSIILCKDPNSYGYK 832
>IF2_BIFLO (Q8G3Y5) Translation initiation factor IF-2
Length = 954
Score = 52.0 bits (123), Expect = 1e-05
Identities = 64/252 (25%), Positives = 95/252 (37%), Gaps = 36/252 (14%)
Query: 190 EDYSHLMTIVTSREFPCSISQAEAMVIAHEARLDRLRLRQHAASSPSAFLAQATAPPQST 249
+D + +S P + +A A+ D ++Q ++PSA + +++ P T
Sbjct: 25 KDMGEFVKSASSTIEPPVARRLKAEFAKDNAKGDSKPVQQRRPAAPSAPASTSSSAP--T 82
Query: 250 PQAPPLVPSQSPAPPQAPPPVAIAPPSSEVNYISRTGSDDRAYEDRDDDRSRSDDNRDRG 309
P AP S + A QAP P A P+ +R G ++D+R DNR+
Sbjct: 83 PAAPARQASPASAHQQAPTPGA---PTPRPQGGARPGMPTPGRHGQNDNRENGRDNRE-- 137
Query: 310 GYNNRGRGGRGSYNNRGRGGDRSSVQCQICHKYGHDASVCYYRGNSTSPATNSQPNMTNQ 369
G N GR S N R DR + Q R N++ P +
Sbjct: 138 GREN----GRQSRPNDRRNNDRRNNQG---------------RPNNSQPVQHQNNRGNAS 178
Query: 370 APAPVAPTFGFGSSFGMMPQFGYSPFRGFQPYGAPSPFGMSFPNSGFGSGSPYASGGMFT 429
AP P A P+ G +PF Q P+P + P+ + GG
Sbjct: 179 APRPHAQG----------PRPGNNPFSRKQGMHTPTPGDIPRPHPMARPTADNGRGGRPG 228
Query: 430 PPGYGFGSGYGF 441
PG G G G GF
Sbjct: 229 RPGQGQGQGRGF 240
>POL_BAEVM (P10272) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1189
Score = 50.8 bits (120), Expect = 3e-05
Identities = 46/147 (31%), Positives = 67/147 (45%), Gaps = 12/147 (8%)
Query: 709 AGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVEL--KFGLKIKSVQTDGGTEF 766
AGY Y L VD FS W+ + ET+ + + + + E+ +FGL K + +D G F
Sbjct: 920 AGYKYLLVFVDTFSG--WVEAFPTRQETAHIVAK-KILEEIFPRFGLP-KVIGSDNGPAF 975
Query: 767 KPLLPHFLK--LGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACANMPTSYWDHAFL 824
+ L LGI +L C + +G VER +R I ET L + T D L
Sbjct: 976 VSQVSQGLARILGINWKLHCAYRPQSSGQVERMNRTIKET----LTKLTLETGLKDWRRL 1031
Query: 825 TATFLINRLPTPVLGNLSPYHKLFKVP 851
+ L+ TP L+PY L+ P
Sbjct: 1032 LSLALLRARNTPNRFGLTPYEILYGGP 1058
>POL_FENV1 (P31792) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 1046
Score = 49.7 bits (117), Expect = 6e-05
Identities = 46/147 (31%), Positives = 67/147 (45%), Gaps = 12/147 (8%)
Query: 709 AGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVEL--KFGLKIKSVQTDGGTEF 766
AGY Y L VD FS W+ + ET+ + + + + E+ +FGL K + +D G F
Sbjct: 777 AGYKYLLVFVDTFSG--WVEAYPTRQETAHMVAK-KILEEIFPRFGLP-KVIGSDNGPAF 832
Query: 767 KPLLPHFLK--LGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACANMPTSYWDHAFL 824
+ L LGI +L C + +G VER +R I ET L + T D L
Sbjct: 833 VSQVSQGLARTLGINWKLHCAYRPQSSGQVERMNRTIKET----LTKLTLETGLKDWRRL 888
Query: 825 TATFLINRLPTPVLGNLSPYHKLFKVP 851
+ L+ TP L+PY L+ P
Sbjct: 889 LSLALLRARNTPNRFGLTPYEILYGGP 915
>POL_MLVCB (P08361) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 282
Score = 49.3 bits (116), Expect = 8e-05
Identities = 46/146 (31%), Positives = 64/146 (43%), Gaps = 12/146 (8%)
Query: 710 GYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVEL--KFGLKIKSVQTDGGTEFK 767
GY Y L VD FS WI K ET+ V + + + E+ +FG+ + + TD G F
Sbjct: 12 GYKYLLVFVDTFSG--WIEAFPTKKETAKVVTK-KLLEEIFPRFGMP-QVLGTDNGPAFV 67
Query: 768 PLLPHFLK--LGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACANMPTSYWDHAFLT 825
+ + LGI +L C + +G VER +R I ET L + T D L
Sbjct: 68 SKVSQTVADLLGIDWKLHCAYRPQSSGQVERMNRTIKET----LTKLTLATGSRDWVLLL 123
Query: 826 ATFLINRLPTPVLGNLSPYHKLFKVP 851
L TP L+PY L+ P
Sbjct: 124 PLALYRARNTPGPHGLTPYEILYGAP 149
>POL_MLVMO (P03355) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1199
Score = 48.9 bits (115), Expect = 1e-04
Identities = 45/146 (30%), Positives = 64/146 (43%), Gaps = 12/146 (8%)
Query: 710 GYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVEL--KFGLKIKSVQTDGGTEFK 767
GY Y L +D FS WI K ET+ V + + + E+ +FG+ + + TD G F
Sbjct: 926 GYKYLLVFIDTFSG--WIEAFPTKKETAKVVTK-KLLEEIFPRFGMP-QVLGTDNGPAFV 981
Query: 768 PLLPHFLK--LGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACANMPTSYWDHAFLT 825
+ + LGI +L C + +G VER +R I ET L + T D L
Sbjct: 982 SKVSQTVADLLGIDWKLHCAYRPQSSGQVERMNRTIKET----LTKLTLATGSRDWVLLL 1037
Query: 826 ATFLINRLPTPVLGNLSPYHKLFKVP 851
L TP L+PY L+ P
Sbjct: 1038 PLALYRARNTPGPHGLTPYEILYGAP 1063
>POL_MLVFP (P26808) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1204
Score = 48.5 bits (114), Expect = 1e-04
Identities = 44/146 (30%), Positives = 64/146 (43%), Gaps = 12/146 (8%)
Query: 710 GYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVEL--KFGLKIKSVQTDGGTEFK 767
GY Y L VD FS W+ K ET+ V + + + E+ +FG+ + + TD G F
Sbjct: 931 GYKYLLVFVDTFSG--WVEAFPTKKETAKVVTK-KLLEEIFPRFGMP-QVLGTDNGPAFV 986
Query: 768 PLLPHFLK--LGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACANMPTSYWDHAFLT 825
+ + LG+ +L C + +G VER +R I ET L + T D L
Sbjct: 987 SKVSQTVADLLGVDWKLHCAYRPQSSGQVERMNRTIKET----LTKLTLATGSRDWVLLL 1042
Query: 826 ATFLINRLPTPVLGNLSPYHKLFKVP 851
L TP L+PY L+ P
Sbjct: 1043 PLALYRARNTPGPHGLTPYEILYGAP 1068
>POL_MLVF5 (P26810) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1204
Score = 48.5 bits (114), Expect = 1e-04
Identities = 44/146 (30%), Positives = 64/146 (43%), Gaps = 12/146 (8%)
Query: 710 GYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVEL--KFGLKIKSVQTDGGTEFK 767
GY Y L VD FS W+ K ET+ V + + + E+ +FG+ + + TD G F
Sbjct: 931 GYKYLLVFVDTFSG--WVEAFPTKKETAKVVTK-KLLEEIFPRFGMP-QVLGTDNGPAFV 986
Query: 768 PLLPHFLK--LGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACANMPTSYWDHAFLT 825
+ + LG+ +L C + +G VER +R I ET L + T D L
Sbjct: 987 SKVSQTVADLLGVDWKLHCAYRPQSSGQVERMNRTIKET----LTKLTLATGSRDWVLLL 1042
Query: 826 ATFLINRLPTPVLGNLSPYHKLFKVP 851
L TP L+PY L+ P
Sbjct: 1043 PLALYRARNTPGPHGLTPYEILYGAP 1068
>POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)]
Length = 886
Score = 48.5 bits (114), Expect = 1e-04
Identities = 49/166 (29%), Positives = 74/166 (44%), Gaps = 14/166 (8%)
Query: 691 PLELIFADLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVELK 750
P + F D GP + S GY Y L VD + FTW+Y K S ++TV +++ L
Sbjct: 675 PFDKFFIDYIGP--LPPSQGYLYVLVVVDGMTGFTWLYPTKAPSTSATV----KSLNVLT 728
Query: 751 FGLKIKSVQTDGGTEF--KPLLPHFLKLGITHRLTCPHTHHQNGS-VERKHRHIVETGLS 807
K + +D G F + GI + P+ H Q+GS VERK+ I
Sbjct: 729 SIAIPKVIHSDQGAAFTSSTFAEWAKERGIHLEFSTPY-HPQSGSKVERKNSDIKRLLTK 787
Query: 808 LLACANMPTSYWDHAFLTATFLINRLPTPVLGNLSPYHKLFKVPAD 853
LL PT ++D +N +PVL +P+ LF + ++
Sbjct: 788 LL--VGRPTKWYD-LLPVVQLALNNTYSPVL-KYTPHQLLFGIDSN 829
>POL_MLVRK (P31795) Pol polyprotein [Contains: Protease (EC
3.4.23.-); Reverse transcriptase/ribonuclease H (EC
2.7.7.49) (EC 3.1.26.4) (RT); Integrase (IN)] (Fragment)
Length = 581
Score = 48.1 bits (113), Expect = 2e-04
Identities = 44/146 (30%), Positives = 64/146 (43%), Gaps = 12/146 (8%)
Query: 710 GYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVEL--KFGLKIKSVQTDGGTEFK 767
GY Y L VD FS W+ K ET+ + + + + E+ +FG+ + + TD G F
Sbjct: 311 GYKYLLVFVDTFSG--WVEAFPTKHETAKIVTK-KLLEEIFPRFGMP-QVLGTDNGPAFV 366
Query: 768 PLLPHFLK--LGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACANMPTSYWDHAFLT 825
+ + LGI +L C + +G VER +R I ET L + T D L
Sbjct: 367 SQVSQSVAKLLGIDWKLHCAYRPQSSGQVERMNRTIKET----LTKLTLATGTRDWVLLL 422
Query: 826 ATFLINRLPTPVLGNLSPYHKLFKVP 851
L TP L+PY L+ P
Sbjct: 423 PLALYRARNTPGPHGLTPYEILYGAP 448
>POL_MLVRD (P11227) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1196
Score = 48.1 bits (113), Expect = 2e-04
Identities = 44/146 (30%), Positives = 64/146 (43%), Gaps = 12/146 (8%)
Query: 710 GYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVEL--KFGLKIKSVQTDGGTEFK 767
GY Y L VD FS W+ K ET+ + + + + E+ +FG+ + + TD G F
Sbjct: 926 GYKYLLVFVDTFSG--WVEAFPTKHETAKIVTK-KLLEEIFPRFGMP-QVLGTDNGPAFV 981
Query: 768 PLLPHFLK--LGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACANMPTSYWDHAFLT 825
+ + LGI +L C + +G VER +R I ET L + T D L
Sbjct: 982 SQVSQSVAKLLGIDWKLHCAYRPQSSGQVERMNRTIKET----LTKLTLATGTRDWVLLL 1037
Query: 826 ATFLINRLPTPVLGNLSPYHKLFKVP 851
L TP L+PY L+ P
Sbjct: 1038 PLALYRARNTPGPHGLTPYEILYGAP 1063
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.331 0.142 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 182,728,080
Number of Sequences: 164201
Number of extensions: 8148544
Number of successful extensions: 46350
Number of sequences better than 10.0: 415
Number of HSP's better than 10.0 without gapping: 98
Number of HSP's successfully gapped in prelim test: 338
Number of HSP's that attempted gapping in prelim test: 40749
Number of HSP's gapped (non-prelim): 3360
length of query: 1571
length of database: 59,974,054
effective HSP length: 124
effective length of query: 1447
effective length of database: 39,613,130
effective search space: 57320199110
effective search space used: 57320199110
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0079b.1


