

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0076a.11
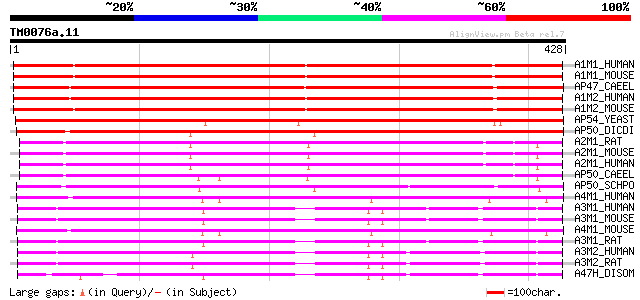
(428 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
A1M1_HUMAN (Q9BXS5) Adaptor-related protein complex 1, mu 1 subu... 546 e-155
A1M1_MOUSE (P35585) Adaptor-related protein complex 1, mu 1 subu... 545 e-155
AP47_CAEEL (P35602) Clathrin coat assembly protein AP47 (Clathri... 539 e-153
A1M2_HUMAN (Q9Y6Q5) Adaptor-related protein complex 1, mu 2 subu... 534 e-151
A1M2_MOUSE (Q9WVP1) Adaptor-related protein complex 1, mu 2 subu... 533 e-151
AP54_YEAST (Q00776) Clathrin coat assembly protein AP54 (Clathri... 432 e-121
AP50_DICDI (P54672) Clathrin coat assembly protein AP50 (Clathri... 356 7e-98
A2M1_RAT (P84092) Clathrin coat assembly protein AP50 (Clathrin ... 319 1e-86
A2M1_MOUSE (P84091) Clathrin coat assembly protein AP50 (Clathri... 319 1e-86
A2M1_HUMAN (Q96CW1) Clathrin coat assembly protein AP50 (Clathri... 319 1e-86
AP50_CAEEL (P35603) Clathrin coat assembly protein AP50 (Clathri... 315 1e-85
AP50_SCHPO (Q09718) Probable clathrin coat assembly protein AP50... 275 2e-73
A4M1_HUMAN (O00189) Adapter-related protein complex 4 mu 1 subun... 205 2e-52
A3M1_HUMAN (Q9Y2T2) Adapter-related protein complex 3 mu 1 subun... 196 1e-49
A3M1_MOUSE (Q9JKC8) Adapter-related protein complex 3 mu 1 subun... 196 1e-49
A4M1_MOUSE (Q9JKC7) Adapter-related protein complex 4 mu 1 subun... 195 2e-49
A3M1_RAT (P53676) Adapter-related protein complex 3 mu 1 subunit... 192 1e-48
A3M2_HUMAN (P53677) Adapter-related protein complex 3 mu 2 subun... 187 3e-47
A3M2_RAT (P53678) Adapter-related protein complex 3 mu 2 subunit... 184 4e-46
A47H_DISOM (P47795) Clathrin coat assembly protein AP47 homolog ... 170 6e-42
>A1M1_HUMAN (Q9BXS5) Adaptor-related protein complex 1, mu 1 subunit
(Mu-adaptin 1) (Adaptor protein complex AP-1 mu-1
subunit) (Golgi adaptor HA1/AP1 adaptin mu-1 subunit)
(Clathrin assembly protein assembly protein complex 1
medium chain 1) (Clathrin co
Length = 423
Score = 546 bits (1408), Expect = e-155
Identities = 259/424 (61%), Positives = 342/424 (80%), Gaps = 5/424 (1%)
Query: 4 AASALFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISYMFV 63
+ASA+++LD+KG+VLI R+YRGDV + E F L+EK+ + P++ G+ +M++
Sbjct: 2 SASAVYVLDLKGKVLICRNYRGDVDMSEVEHFMPILMEKEEEGMLS-PILAHGGVRFMWI 60
Query: 64 QHSNVYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIMDF 123
+H+N+YL+ +++N + + FL++VV VF YF+ELEEES+RDNFV++YELLDE+MDF
Sbjct: 61 KHNNLYLVATSKKNACVSLVFSFLYKVVQVFSEYFKELEEESIRDNFVIIYELLDELMDF 120
Query: 124 GYPQYTEAKILSEFIKTDAYRMEV-TQRPPMAVTNAVSWRSEGISYKKNEVFLDVVESVN 182
GYPQ T++KIL E+I + +++E RPP VTNAVSWRSEGI Y+KNEVFLDV+ESVN
Sbjct: 121 GYPQTTDSKILQEYITQEGHKLETGAPRPPATVTNAVSWRSEGIKYRKNEVFLDVIESVN 180
Query: 183 ILVNSNGQIIRSDVVGALKMRTYLSGMPECKLGLNDRVLLEAQGRTTKGKAIDLEDIKFH 242
+LV++NG ++RS++VG++KMR +LSGMPE +LGLND+VL + GR K K+++LED+KFH
Sbjct: 181 LLVSANGNVLRSEIVGSIKMRVFLSGMPELRLGLNDKVLFDNTGRG-KSKSVELEDVKFH 239
Query: 243 QCVRLARFENDRTISFIPPDGAFDLMTYRLSTQVKPLIWVEATVEKHSKSRIEIMVKARS 302
QCVRL+RFENDRTISFIPPDG F+LM+YRL+T VKPLIW+E+ +EKHS SRIE M+KA+S
Sbjct: 240 QCVRLSRFENDRTISFIPPDGEFELMSYRLNTHVKPLIWIESVIEKHSHSRIEYMIKAKS 299
Query: 303 QFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAPEKDALIWKIRSFPGGKEYMLRAE 362
QFK RSTA NVEI +PVP DA +P +T++GS + PE ++W I+SFPGGKEY++RA
Sbjct: 300 QFKRRSTANNVEIHIPVPNDADSPKFKTTVGSVKWVPENSEIVWSIKSFPGGKEYLMRAH 359
Query: 363 FRLPSITAEEATPERKAPIRVKFEIPYFTVSGIQVRYLKIIEKSGYQALPWVRYITMAGE 422
F LPS+ AE+ E K PI VKFEIPYFT SGIQVRYLKIIEKSGYQALPWVRYIT G+
Sbjct: 360 FGLPSVEAED--KEGKPPISVKFEIPYFTTSGIQVRYLKIIEKSGYQALPWVRYITQNGD 417
Query: 423 YELR 426
Y+LR
Sbjct: 418 YQLR 421
>A1M1_MOUSE (P35585) Adaptor-related protein complex 1, mu 1 subunit
(Mu-adaptin 1) (Adaptor protein complex AP-1 mu-1
subunit) (Golgi adaptor HA1/AP1 adaptin mu-1 subunit)
(Clathrin assembly protein assembly protein complex 1
medium chain 1) (Clathrin co
Length = 423
Score = 545 bits (1405), Expect = e-155
Identities = 258/424 (60%), Positives = 342/424 (79%), Gaps = 5/424 (1%)
Query: 4 AASALFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISYMFV 63
+ASA+++LD+KG+VLI R+YRGDV + E F L+EK+ + P++ G+ +M++
Sbjct: 2 SASAVYVLDLKGKVLICRNYRGDVDMSEVEHFMPILMEKEEEGMLS-PILAHGGVRFMWI 60
Query: 64 QHSNVYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIMDF 123
+H+N+YL+ +++N + + FL++VV VF YF+ELEEES+RDNFV++YELLDE+MDF
Sbjct: 61 KHNNLYLVATSKKNACVSLVFSFLYKVVQVFSEYFKELEEESIRDNFVIIYELLDELMDF 120
Query: 124 GYPQYTEAKILSEFIKTDAYRMEV-TQRPPMAVTNAVSWRSEGISYKKNEVFLDVVESVN 182
GYPQ T++KIL E+I + +++E RPP VTNAVSWRSEGI Y+KNEVFLDV+E+VN
Sbjct: 121 GYPQTTDSKILQEYITQEGHKLETGAPRPPATVTNAVSWRSEGIKYRKNEVFLDVIEAVN 180
Query: 183 ILVNSNGQIIRSDVVGALKMRTYLSGMPECKLGLNDRVLLEAQGRTTKGKAIDLEDIKFH 242
+LV++NG ++RS++VG++KMR +LSGMPE +LGLND+VL + GR K K+++LED+KFH
Sbjct: 181 LLVSANGNVLRSEIVGSIKMRVFLSGMPELRLGLNDKVLFDNTGRG-KSKSVELEDVKFH 239
Query: 243 QCVRLARFENDRTISFIPPDGAFDLMTYRLSTQVKPLIWVEATVEKHSKSRIEIMVKARS 302
QCVRL+RFENDRTISFIPPDG F+LM+YRL+T VKPLIW+E+ +EKHS SRIE MVKA+S
Sbjct: 240 QCVRLSRFENDRTISFIPPDGEFELMSYRLNTHVKPLIWIESVIEKHSHSRIEYMVKAKS 299
Query: 303 QFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAPEKDALIWKIRSFPGGKEYMLRAE 362
QFK RSTA NVEI +PVP DA +P +T++GS + PE ++W ++SFPGGKEY++RA
Sbjct: 300 QFKRRSTANNVEIHIPVPNDADSPKFKTTVGSVKWVPENSEIVWSVKSFPGGKEYLMRAH 359
Query: 363 FRLPSITAEEATPERKAPIRVKFEIPYFTVSGIQVRYLKIIEKSGYQALPWVRYITMAGE 422
F LPS+ AE+ E K PI VKFEIPYFT SGIQVRYLKIIEKSGYQALPWVRYIT G+
Sbjct: 360 FGLPSVEAED--KEGKPPISVKFEIPYFTTSGIQVRYLKIIEKSGYQALPWVRYITQNGD 417
Query: 423 YELR 426
Y+LR
Sbjct: 418 YQLR 421
>AP47_CAEEL (P35602) Clathrin coat assembly protein AP47 (Clathrin
coat associated protein AP47) (Golgi adaptor AP-1 47 kDa
protein) (HA1 47 kDa subunit) (Clathrin assembly protein
assembly protein complex 1 medium chain) (Uncoordinated
protein 101)
Length = 422
Score = 539 bits (1389), Expect = e-153
Identities = 254/424 (59%), Positives = 340/424 (79%), Gaps = 4/424 (0%)
Query: 4 AASALFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISYMFV 63
A SA+F+LD+KG+ +I R+YRGD+ ++F L+EK+ + S PV+ +++F+
Sbjct: 2 ATSAMFILDLKGKTIISRNYRGDIDMTAIDKFIHLLMEKEEEG-SAAPVLTYQDTNFVFI 60
Query: 64 QHSNVYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIMDF 123
+H+N+YL+ A R N N +L FL++ V+VF YF+++EEES+RDNFVV+YELLDE+MDF
Sbjct: 61 KHTNIYLVSACRSNVNVTMILSFLYKCVEVFSEYFKDVEEESVRDNFVVIYELLDEMMDF 120
Query: 124 GYPQYTEAKILSEFIKTDAYRMEVTQRPPMAVTNAVSWRSEGISYKKNEVFLDVVESVNI 183
G+PQ TE++IL E+I + ++ RPPMAVTNAVSWRSEGI Y+KNEVFLDV+ESVN+
Sbjct: 121 GFPQTTESRILQEYITQEGQKLISAPRPPMAVTNAVSWRSEGIKYRKNEVFLDVIESVNM 180
Query: 184 LVNSNGQIIRSDVVGALKMRTYLSGMPECKLGLNDRVLLEAQGRTTKGKAIDLEDIKFHQ 243
L ++NG +++S++VG++KMR YL+GMPE +LGLND+VL E GR K K+++LED+KFHQ
Sbjct: 181 LASANGTVLQSEIVGSVKMRVYLTGMPELRLGLNDKVLFEGSGR-GKSKSVELEDVKFHQ 239
Query: 244 CVRLARFENDRTISFIPPDGAFDLMTYRLSTQVKPLIWVEATVEKHSKSRIEIMVKARSQ 303
CVRL+RF+ DRTISFIPPDGAF+LM+YRL+T VKPLIW+E ++E+HS SR+ ++KA+SQ
Sbjct: 240 CVRLSRFDTDRTISFIPPDGAFELMSYRLTTVVKPLIWIETSIERHSHSRVSFIIKAKSQ 299
Query: 304 FKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAPEKDALIWKIRSFPGGKEYMLRAEF 363
FK RSTA NVEI +PVP DA +P +TS+GS Y PE+ A +W I++FPGGKEY+L A
Sbjct: 300 FKRRSTANNVEIIIPVPSDADSPKFKTSIGSVKYTPEQSAFVWTIKNFPGGKEYLLTAHL 359
Query: 364 RLPSITAEEATPERKAPIRVKFEIPYFTVSGIQVRYLKIIEKSGYQALPWVRYITMAGEY 423
LPS+ +EE+ E + PI+VKFEIPYFT SGIQVRYLKIIEKSGYQALPWVRYIT GEY
Sbjct: 360 SLPSVMSEES--EGRPPIKVKFEIPYFTTSGIQVRYLKIIEKSGYQALPWVRYITQNGEY 417
Query: 424 ELRL 427
E+R+
Sbjct: 418 EMRM 421
>A1M2_HUMAN (Q9Y6Q5) Adaptor-related protein complex 1, mu 2 subunit
(Mu-adaptin 2) (Adaptor protein complex AP-1 mu-2
subunit) (Golgi adaptor HA1/AP1 adaptin mu-2 subunit)
(Clathrin assembly protein assembly protein complex 1
medium chain 2) (AP-mu chain
Length = 423
Score = 534 bits (1376), Expect = e-151
Identities = 255/424 (60%), Positives = 342/424 (80%), Gaps = 5/424 (1%)
Query: 4 AASALFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISYMFV 63
+ASA+F+LD+KG+ LI R+Y+GDV E F L++++ + + P++ + ++++
Sbjct: 2 SASAVFILDVKGKPLISRNYKGDVAMSKIEHFMPLLVQREEEG-ALAPLLSHGQVHFLWI 60
Query: 64 QHSNVYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIMDF 123
+HSN+YL+ T +N NA+ + FL++ ++VF YF+ELEEES+RDNFV+VYELLDE+MDF
Sbjct: 61 KHSNLYLVATTSKNANASLVYSFLYKTIEVFCEYFKELEEESIRDNFVIVYELLDELMDF 120
Query: 124 GYPQYTEAKILSEFIKTDAYRMEVTQ-RPPMAVTNAVSWRSEGISYKKNEVFLDVVESVN 182
G+PQ T++KIL E+I + ++E + R P VTNAVSWRSEGI YKKNEVF+DV+ESVN
Sbjct: 121 GFPQTTDSKILQEYITQQSNKLETGKSRVPPTVTNAVSWRSEGIKYKKNEVFIDVIESVN 180
Query: 183 ILVNSNGQIIRSDVVGALKMRTYLSGMPECKLGLNDRVLLEAQGRTTKGKAIDLEDIKFH 242
+LVN+NG ++ S++VG +K++ +LSGMPE +LGLNDRVL E GR+ K K+++LED+KFH
Sbjct: 181 LLVNANGSVLLSEIVGTIKLKVFLSGMPELRLGLNDRVLFELTGRS-KNKSVELEDVKFH 239
Query: 243 QCVRLARFENDRTISFIPPDGAFDLMTYRLSTQVKPLIWVEATVEKHSKSRIEIMVKARS 302
QCVRL+RF+NDRTISFIPPDG F+LM+YRLSTQVKPLIW+E+ +EK S SR+EIMVKA+
Sbjct: 240 QCVRLSRFDNDRTISFIPPDGDFELMSYRLSTQVKPLIWIESVIEKFSHSRVEIMVKAKG 299
Query: 303 QFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAPEKDALIWKIRSFPGGKEYMLRAE 362
QFK++S A VEI +PVP DA +P +TS+GSA Y PE++ +IW I+SFPGGKEY++RA
Sbjct: 300 QFKKQSVANGVEISVPVPSDADSPRFKTSVGSAKYVPERNVVIWSIKSFPGGKEYLMRAH 359
Query: 363 FRLPSITAEEATPERKAPIRVKFEIPYFTVSGIQVRYLKIIEKSGYQALPWVRYITMAGE 422
F LPS+ EE E + PI VKFEIPYFTVSGIQVRY+KIIEKSGYQALPWVRYIT +G+
Sbjct: 360 FGLPSVEKEEV--EGRPPIGVKFEIPYFTVSGIQVRYMKIIEKSGYQALPWVRYITQSGD 417
Query: 423 YELR 426
Y+LR
Sbjct: 418 YQLR 421
>A1M2_MOUSE (Q9WVP1) Adaptor-related protein complex 1, mu 2 subunit
(Mu-adaptin 2) (Adaptor protein complex AP-1 mu-2
subunit) (Golgi adaptor HA1/AP1 adaptin mu-2 subunit)
(Clathrin assembly protein assembly protein complex 1
medium chain 2) (AP-mu chain
Length = 423
Score = 533 bits (1372), Expect = e-151
Identities = 255/424 (60%), Positives = 341/424 (80%), Gaps = 5/424 (1%)
Query: 4 AASALFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISYMFV 63
+ASA+F+LD+KG+ LI R+Y+GDV + + F L++++ + P++ + ++++
Sbjct: 2 SASAVFILDVKGKPLISRNYKGDVPMTEIDHFMPLLMQREEEGVLA-PLLSHGRVHFLWI 60
Query: 64 QHSNVYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIMDF 123
+HSN+YL+ T +N NA+ + FL++ V+VF YF+ELEEES+RDNFV+VY+LLDE+MDF
Sbjct: 61 KHSNLYLVATTLKNANASLVYSFLYKTVEVFCEYFKELEEESIRDNFVIVYDLLDELMDF 120
Query: 124 GYPQYTEAKILSEFIKTDAYRMEVTQ-RPPMAVTNAVSWRSEGISYKKNEVFLDVVESVN 182
G+PQ T++KIL E+I ++E + R P VTNAVSWRSEGI YKKNEVF+DV+ESVN
Sbjct: 121 GFPQTTDSKILQEYITQQGNKLETGKSRVPPTVTNAVSWRSEGIKYKKNEVFIDVIESVN 180
Query: 183 ILVNSNGQIIRSDVVGALKMRTYLSGMPECKLGLNDRVLLEAQGRTTKGKAIDLEDIKFH 242
+LVN+NG ++ S++VG +K++ +LSGMPE +LGLNDRVL E GR+ K K+++LED+KFH
Sbjct: 181 LLVNANGSVLLSEIVGTIKLKVFLSGMPELRLGLNDRVLFELTGRS-KNKSVELEDVKFH 239
Query: 243 QCVRLARFENDRTISFIPPDGAFDLMTYRLSTQVKPLIWVEATVEKHSKSRIEIMVKARS 302
QCVRL+RF+NDRTISFIPPDG F+LM+YRLSTQVKPLIW+E+ +EK S SR+EIMVKA+
Sbjct: 240 QCVRLSRFDNDRTISFIPPDGDFELMSYRLSTQVKPLIWIESVIEKFSHSRVEIMVKAKG 299
Query: 303 QFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAPEKDALIWKIRSFPGGKEYMLRAE 362
QFK++S A VEI +PVP DA +P +TS+GSA Y PEK+ +IW I+SFPGGKEY++RA
Sbjct: 300 QFKKQSVANGVEISVPVPSDADSPRFKTSVGSAKYVPEKNVVIWSIKSFPGGKEYLMRAH 359
Query: 363 FRLPSITAEEATPERKAPIRVKFEIPYFTVSGIQVRYLKIIEKSGYQALPWVRYITMAGE 422
F LPS+ EE E + PI VKFEIPYFTVSGIQVRY+KIIEKSGYQALPWVRYIT +G+
Sbjct: 360 FGLPSVETEEV--EGRPPIGVKFEIPYFTVSGIQVRYMKIIEKSGYQALPWVRYITQSGD 417
Query: 423 YELR 426
Y+LR
Sbjct: 418 YQLR 421
>AP54_YEAST (Q00776) Clathrin coat assembly protein AP54 (Clathrin
coat associated protein AP54) (Golgi adaptor AP-1 54 kDa
protein) (HA1 54 kDa subunit) (Clathrin assembly protein
complex 1 medium chain)
Length = 475
Score = 432 bits (1111), Expect = e-121
Identities = 223/473 (47%), Positives = 316/473 (66%), Gaps = 50/473 (10%)
Query: 5 ASALFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISYMFVQ 64
ASA++ D G+ L+ R YR D+ ++F L + + + P ++ NG+ Y+F+Q
Sbjct: 2 ASAVYFCDHNGKPLLSRRYRDDIPLSAIDKFPILLSDLEEQSNLIPPCLNHNGLEYLFIQ 61
Query: 65 HSNVYLM-IATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIMDF 123
H+++Y++ I T + NAA++ FLH++V+V Y + +EEES+RDNFV++YELLDE+MD+
Sbjct: 62 HNDLYVVAIVTSLSANAAAIFTFLHKLVEVLSDYLKTVEEESIRDNFVIIYELLDEVMDY 121
Query: 124 GYPQYTEAKILSEFIKTDAYRMEVTQ-------RPPMAVTNAVSWRSEGISYKKNEVFLD 176
G PQ TE K+L ++I ++++ + RPP+A+TN+VSWR EGI++KKNE FLD
Sbjct: 122 GIPQITETKMLKQYITQKSFKLVKSAKKKRNATRPPVALTNSVSWRPEGITHKKNEAFLD 181
Query: 177 VVESVNILVNSNGQIIRSDVVGALKMRTYLSGMPECKLGLNDRVL--------------- 221
+VES+N+L+ GQ++RS+++G +K+ + LSGMP+ KLG+ND+ +
Sbjct: 182 IVESINMLMTQKGQVLRSEIIGDVKVNSKLSGMPDLKLGINDKGIFSKYLDDDTNIPSAS 241
Query: 222 ---------------LEAQGRTTKGKA-IDLEDIKFHQCVRLARFENDRTISFIPPDGAF 265
+ + T K K I+LED+KFHQCVRL++FEN++ I+FIPPDG F
Sbjct: 242 ATTSDNNTETDKKPSITSSSATNKKKVNIELEDLKFHQCVRLSKFENEKIITFIPPDGKF 301
Query: 266 DLMTYRLSTQVKPLIWVEATVEKHSKSRIEIMVKARSQFKERSTATNVEIELPVPVDAMN 325
DLM YRLST +KPLIW + V+ HS SRIEI KA++Q K +STATNVEI +PVP DA
Sbjct: 302 DLMNYRLSTTIKPLIWCDVNVQVHSNSRIEIHCKAKAQIKRKSTATNVEILIPVPDDADT 361
Query: 326 PNVRTSMGSAAYAPEKDALIWKIRSFPGGKEYMLRAEFRLPSITAEE----ATPER---- 377
P + S GS Y PEK A++WKIRSFPGGKEY + AE LPSI+ E P+
Sbjct: 362 PTFKYSHGSLKYVPEKSAILWKIRSFPGGKEYSMSAELGLPSISNNEDGNRTMPKSNAEI 421
Query: 378 -KAPIRVKFEIPYFTVSGIQVRYLKIIE-KSGYQALPWVRYITMAG-EYELRL 427
K P+++KF+IPYFT SGIQVRYLKI E K Y++ PWVRYIT +G +Y +RL
Sbjct: 422 LKGPVQIKFQIPYFTTSGIQVRYLKINEPKLQYKSYPWVRYITQSGDDYTIRL 474
>AP50_DICDI (P54672) Clathrin coat assembly protein AP50 (Clathrin
coat associated protein AP50) (Plasma membrane adaptor
AP-2 50 kDa protein) (Clathrin assembly protein complex
2 medium chain)
Length = 439
Score = 356 bits (913), Expect = 7e-98
Identities = 179/440 (40%), Positives = 273/440 (61%), Gaps = 21/440 (4%)
Query: 6 SALFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISYMFVQH 65
SALFL++ KG VLI R YR D++ F ++I Q +++ PV S+M+++
Sbjct: 3 SALFLMNGKGEVLISRIYRDDISRGVGNAFRLEVIGVQ---ETRSPVKLIGSTSFMYIKV 59
Query: 66 SNVYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIMDFGY 125
N+Y++ +RQN NA + LH++VD+FK YF+ L+E+S+R+NFV+VYELLDEI+DFGY
Sbjct: 60 GNIYIVGVSRQNVNACMVFEVLHQLVDIFKSYFDNLDEDSIRNNFVLVYELLDEILDFGY 119
Query: 126 PQYTEAKILSEFI--------KTDAYRMEVTQRPPMAVTNAVSWRSEGISYKKNEVFLDV 177
PQ +L +I D + + + + T WR+ I YK+NE+++DV
Sbjct: 120 PQNCSTDVLKLYITQGQGKLKSLDKLKQDKISKITIHATGTTPWRTPDIKYKRNELYIDV 179
Query: 178 VESVNILVNSNGQIIRSDVVGALKMRTYLSGMPECKLGLNDRVLLEAQGRTTKGKA---- 233
VESVN+L+++ G I+R+DV G + M+ +LSGMPECK G+ND+V+++ + T G A
Sbjct: 180 VESVNLLMSAEGNILRADVSGQVMMKCFLSGMPECKFGMNDKVIMDREKSTNGGSAARSG 239
Query: 234 ------IDLEDIKFHQCVRLARFENDRTISFIPPDGAFDLMTYRLSTQVKPLIWVEATVE 287
I+++DI FHQCVRL +F++DRT+SFIPPDG F+LM YR + + V V
Sbjct: 240 RRRANGIEIDDITFHQCVRLGKFDSDRTVSFIPPDGEFELMRYRTTEHINLPFKVIPIVR 299
Query: 288 KHSKSRIEIMVKARSQFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAPEKDALIWK 347
+ ++R+E V +S F + NV++ +P P + + + G A Y PE+DA+IW+
Sbjct: 300 EMGRTRLECSVTVKSNFSSKMFGANVKVIIPTPKNTAVCKIVVAAGKAKYMPEQDAIIWR 359
Query: 348 IRSFPGGKEYMLRAEFRLPSITAEEATPERKAPIRVKFEIPYFTVSGIQVRYLKIIEKSG 407
IR FPG E+ LRAE L + + + PI ++F++ FT SG VR+LK++EKS
Sbjct: 360 IRRFPGDTEFTLRAEVELMASVNLDKKAWSRPPISMEFQVTMFTASGFSVRFLKVVEKSN 419
Query: 408 YQALPWVRYITMAGEYELRL 427
Y + WVRY+T AG Y+ R+
Sbjct: 420 YTPIKWVRYLTKAGTYQNRI 439
>A2M1_RAT (P84092) Clathrin coat assembly protein AP50 (Clathrin
coat associated protein AP50) (Plasma membrane adaptor
AP-2 50 kDa protein) (Clathrin assembly protein complex
2 medium chain) (AP-2 mu 2 chain)
Length = 435
Score = 319 bits (817), Expect = 1e-86
Identities = 173/435 (39%), Positives = 262/435 (59%), Gaps = 21/435 (4%)
Query: 8 LFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISYMFVQHSN 67
LF+ + KG VLI R YR D+ + F +I A Q + PV + S+ V+ SN
Sbjct: 5 LFIYNHKGEVLISRVYRDDIGRNAVDAFRVNVIH--ARQQVRSPVTNIARTSFFHVKRSN 62
Query: 68 VYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIMDFGYPQ 127
++L T+QN NAA + FL+++ DV YF ++ EE++++NFV++YELLDEI+DFGYPQ
Sbjct: 63 IWLAAVTKQNVNAAMVFEFLYKMCDVMAAYFGKISEENIKNNFVLIYELLDEILDFGYPQ 122
Query: 128 YTEAKILSEFI-----KTDAYRMEVTQRPPMAVTNAVSWRSEGISYKKNEVFLDVVESVN 182
+E L FI K+ E + VT + WR EGI Y++NE+FLDV+ESVN
Sbjct: 123 NSETGALKTFITQQGIKSQHQTKEEQSQITSQVTGQIGWRREGIKYRRNELFLDVLESVN 182
Query: 183 ILVNSNGQIIRSDVVGALKMRTYLSGMPECKLGLNDRVLLEAQGRTT------KGK-AID 235
+L++ GQ++ + V G + M++YLSGMPECK G+ND++++E QG+ T GK +I
Sbjct: 183 LLMSPQGQVLSAHVSGRVVMKSYLSGMPECKFGMNDKIVIEKQGKGTADETSKSGKQSIA 242
Query: 236 LEDIKFHQCVRLARFENDRTISFIPPDGAFDLMTYRLSTQVKPLIWVEATVEKHSKSRIE 295
++D FHQCVRL++F+++R+ISFIPPDG F+LM YR + + V V + ++++E
Sbjct: 243 IDDCTFHQCVRLSKFDSERSISFIPPDGEFELMRYRTTKDIILPFRVIPLVREVGRTKLE 302
Query: 296 IMVKARSQFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAPEKDALIWKIRSFPGGK 355
+ V +S FK A +E+ +P P++ V G A Y ++A++WKI+ G K
Sbjct: 303 VKVVIKSNFKPSLLAQKIEVRIPTPLNTSGVQVICMKGKAKYKASENAIVWKIKRMAGMK 362
Query: 356 EYMLRAEFRLPSITAEEATPERKAPIRVKFEIPYFTVSGIQVRYLKIIEK----SGYQAL 411
E + AE L + + + PI + FE+P F SG++VRYLK+ E S + +
Sbjct: 363 ESQISAEIEL--LPTNDKKKWARPPISMNFEVP-FAPSGLKVRYLKVFEPKLNYSDHDVI 419
Query: 412 PWVRYITMAGEYELR 426
WVRYI +G YE R
Sbjct: 420 KWVRYIGRSGIYETR 434
>A2M1_MOUSE (P84091) Clathrin coat assembly protein AP50 (Clathrin
coat associated protein AP50) (Plasma membrane adaptor
AP-2 50 kDa protein) (Clathrin assembly protein complex
2 medium chain) (AP-2 mu 2 chain)
Length = 435
Score = 319 bits (817), Expect = 1e-86
Identities = 173/435 (39%), Positives = 262/435 (59%), Gaps = 21/435 (4%)
Query: 8 LFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISYMFVQHSN 67
LF+ + KG VLI R YR D+ + F +I A Q + PV + S+ V+ SN
Sbjct: 5 LFIYNHKGEVLISRVYRDDIGRNAVDAFRVNVIH--ARQQVRSPVTNIARTSFFHVKRSN 62
Query: 68 VYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIMDFGYPQ 127
++L T+QN NAA + FL+++ DV YF ++ EE++++NFV++YELLDEI+DFGYPQ
Sbjct: 63 IWLAAVTKQNVNAAMVFEFLYKMCDVMAAYFGKISEENIKNNFVLIYELLDEILDFGYPQ 122
Query: 128 YTEAKILSEFI-----KTDAYRMEVTQRPPMAVTNAVSWRSEGISYKKNEVFLDVVESVN 182
+E L FI K+ E + VT + WR EGI Y++NE+FLDV+ESVN
Sbjct: 123 NSETGALKTFITQQGIKSQHQTKEEQSQITSQVTGQIGWRREGIKYRRNELFLDVLESVN 182
Query: 183 ILVNSNGQIIRSDVVGALKMRTYLSGMPECKLGLNDRVLLEAQGRTT------KGK-AID 235
+L++ GQ++ + V G + M++YLSGMPECK G+ND++++E QG+ T GK +I
Sbjct: 183 LLMSPQGQVLSAHVSGRVVMKSYLSGMPECKFGMNDKIVIEKQGKGTADETSKSGKQSIA 242
Query: 236 LEDIKFHQCVRLARFENDRTISFIPPDGAFDLMTYRLSTQVKPLIWVEATVEKHSKSRIE 295
++D FHQCVRL++F+++R+ISFIPPDG F+LM YR + + V V + ++++E
Sbjct: 243 IDDCTFHQCVRLSKFDSERSISFIPPDGEFELMRYRTTKDIILPFRVIPLVREVGRTKLE 302
Query: 296 IMVKARSQFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAPEKDALIWKIRSFPGGK 355
+ V +S FK A +E+ +P P++ V G A Y ++A++WKI+ G K
Sbjct: 303 VKVVIKSNFKPSLLAQKIEVRIPTPLNTSGVQVICMKGKAKYKASENAIVWKIKRMAGMK 362
Query: 356 EYMLRAEFRLPSITAEEATPERKAPIRVKFEIPYFTVSGIQVRYLKIIEK----SGYQAL 411
E + AE L + + + PI + FE+P F SG++VRYLK+ E S + +
Sbjct: 363 ESQISAEIEL--LPTNDKKKWARPPISMNFEVP-FAPSGLKVRYLKVFEPKLNYSDHDVI 419
Query: 412 PWVRYITMAGEYELR 426
WVRYI +G YE R
Sbjct: 420 KWVRYIGRSGIYETR 434
>A2M1_HUMAN (Q96CW1) Clathrin coat assembly protein AP50 (Clathrin
coat associated protein AP50) (Plasma membrane adaptor
AP-2 50 kDa protein) (HA2 50 kDa subunit) (Clathrin
assembly protein complex 2 medium chain) (AP-2 mu 2
chain)
Length = 435
Score = 319 bits (817), Expect = 1e-86
Identities = 173/435 (39%), Positives = 262/435 (59%), Gaps = 21/435 (4%)
Query: 8 LFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISYMFVQHSN 67
LF+ + KG VLI R YR D+ + F +I A Q + PV + S+ V+ SN
Sbjct: 5 LFIYNHKGEVLISRVYRDDIGRNAVDAFRVNVIH--ARQQVRSPVTNIARTSFFHVKRSN 62
Query: 68 VYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIMDFGYPQ 127
++L T+QN NAA + FL+++ DV YF ++ EE++++NFV++YELLDEI+DFGYPQ
Sbjct: 63 IWLAAVTKQNVNAAMVFEFLYKMCDVMAAYFGKISEENIKNNFVLIYELLDEILDFGYPQ 122
Query: 128 YTEAKILSEFI-----KTDAYRMEVTQRPPMAVTNAVSWRSEGISYKKNEVFLDVVESVN 182
+E L FI K+ E + VT + WR EGI Y++NE+FLDV+ESVN
Sbjct: 123 NSETGALKTFITQQGIKSQHQTKEEQSQITSQVTGQIGWRREGIKYRRNELFLDVLESVN 182
Query: 183 ILVNSNGQIIRSDVVGALKMRTYLSGMPECKLGLNDRVLLEAQGRTT------KGK-AID 235
+L++ GQ++ + V G + M++YLSGMPECK G+ND++++E QG+ T GK +I
Sbjct: 183 LLMSPQGQVLSAHVSGRVVMKSYLSGMPECKFGMNDKIVIEKQGKGTADETSKSGKQSIA 242
Query: 236 LEDIKFHQCVRLARFENDRTISFIPPDGAFDLMTYRLSTQVKPLIWVEATVEKHSKSRIE 295
++D FHQCVRL++F+++R+ISFIPPDG F+LM YR + + V V + ++++E
Sbjct: 243 IDDCTFHQCVRLSKFDSERSISFIPPDGEFELMRYRTTKDIILPFRVIPLVREVGRTKLE 302
Query: 296 IMVKARSQFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAPEKDALIWKIRSFPGGK 355
+ V +S FK A +E+ +P P++ V G A Y ++A++WKI+ G K
Sbjct: 303 VKVVIKSNFKPSLLAQKIEVRIPTPLNTSGVQVICMKGKAKYKASENAIVWKIKRMAGMK 362
Query: 356 EYMLRAEFRLPSITAEEATPERKAPIRVKFEIPYFTVSGIQVRYLKIIEK----SGYQAL 411
E + AE L + + + PI + FE+P F SG++VRYLK+ E S + +
Sbjct: 363 ESQISAEIEL--LPTNDKKKWARPPISMNFEVP-FAPSGLKVRYLKVFEPKLNYSDHDVI 419
Query: 412 PWVRYITMAGEYELR 426
WVRYI +G YE R
Sbjct: 420 KWVRYIGRSGIYETR 434
>AP50_CAEEL (P35603) Clathrin coat assembly protein AP50 (Clathrin
coat associated protein AP50) (Plasma membrane adaptor
AP-2 50 kDa protein) (Clathrin assembly protein complex
2 medium chain) (Dumpy protein 23)
Length = 441
Score = 315 bits (807), Expect = 1e-85
Identities = 171/439 (38%), Positives = 256/439 (57%), Gaps = 23/439 (5%)
Query: 8 LFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISYMFVQHSN 67
LF+ + KG VLI R YR DVT + F +I A Q + PV + S+ V+ N
Sbjct: 5 LFVYNHKGEVLISRIYRDDVTRNAVDAFRVNVIH--ARQQVRSPVTNMARTSFFHVKRGN 62
Query: 68 VYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIMDFGYPQ 127
V++ TRQN NAA + FL R D + YF +L EE++++NFV++YELLDEI+DFGYPQ
Sbjct: 63 VWICAVTRQNVNAAMVFEFLKRFADTMQSYFGKLNEENVKNNFVLIYELLDEILDFGYPQ 122
Query: 128 YTEAKILSEFIKTDAYR-----MEVTQRPPMAVTNAVS----WRSEGISYKKNEVFLDVV 178
T+ +L FI R + VT+ +T+ V+ WR EGI Y++NE+FLDV+
Sbjct: 123 NTDPGVLKTFITQQGVRTADAPVPVTKEEQSQITSQVTGQIGWRREGIKYRRNELFLDVI 182
Query: 179 ESVNILVNSNGQIIRSDVVGALKMRTYLSGMPECKLGLNDRVLLEAQGRT-------TKG 231
E VN+L+N GQ++ + V G + M++YLSGMPECK G+ND++ +E + +
Sbjct: 183 EYVNLLMNQQGQVLSAHVAGKVAMKSYLSGMPECKFGINDKITIEGKSKPGSDDPNKASR 242
Query: 232 KAIDLEDIKFHQCVRLARFENDRTISFIPPDGAFDLMTYRLSTQVKPLIWVEATVEKHSK 291
A+ ++D +FHQCV+L +FE + ISFIPPDG ++LM YR + ++ V V + S+
Sbjct: 243 AAVAIDDCQFHQCVKLTKFETEHAISFIPPDGEYELMRYRTTKDIQLPFRVIPLVREVSR 302
Query: 292 SRIEIMVKARSQFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAPEKDALIWKIRSF 351
+++E+ V +S FK A +E+ +P P + + G A Y ++A++WKI+
Sbjct: 303 NKMEVKVVVKSNFKPSLLAQKLEVRIPTPPNTSGVQLICMKGKAKYKAGENAIVWKIKRM 362
Query: 352 PGGKEYMLRAEFRLPSITAEEATPERKAPIRVKFEIPYFTVSGIQVRYLKIIEK----SG 407
G KE + AE L S E + P+ + FE+P F SG++VRYLK+ E S
Sbjct: 363 AGMKESQISAEIDLLSTGNVEKKKWNRPPVSMNFEVP-FAPSGLKVRYLKVFEPKLNYSD 421
Query: 408 YQALPWVRYITMAGEYELR 426
+ + WVRYI +G YE R
Sbjct: 422 HDVIKWVRYIGRSGLYETR 440
>AP50_SCHPO (Q09718) Probable clathrin coat assembly protein AP50
(Clathrin coat associated protein AP50) (Plasma membrane
adaptor AP-2 50 kDa protein) (Clathrin assembly protein
complex 2 medium chain)
Length = 446
Score = 275 bits (702), Expect = 2e-73
Identities = 150/450 (33%), Positives = 247/450 (54%), Gaps = 34/450 (7%)
Query: 6 SALFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISYMFVQH 65
S LF+ ++KG LI + +R D+ E F ++ + + P+V +Y++ +H
Sbjct: 3 SGLFIFNLKGDTLICKTFRHDLKKSVTEIFRVAIL---TNTDYRHPIVSIGSSTYIYTKH 59
Query: 66 SNVYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIMDFGY 125
++Y++ T+ N N +L FL ++ HYF +L E +++DN ++ELLDE++D+G
Sbjct: 60 EDLYVVAITKGNPNVMIVLEFLESLIQDLTHYFGKLNENTVKDNVSFIFELLDEMIDYGI 119
Query: 126 PQYTEAKILSEFIKTDAYRM-------------EVTQRPPMAVTNAVSWRSEGISYKKNE 172
Q TE L+ + A + ++ + +V WR GI Y+KN
Sbjct: 120 IQTTEPDALARSVSITAVKKKGNALSLKRSHSSQLAHTTSSEIPGSVPWRRAGIKYRKNS 179
Query: 173 VFLDVVESVNILVNSNGQIIRSDVVGALKMRTYLSGMPECKLGLNDRVLLEAQGRTTKGK 232
+++D+VE +N+L++S G ++RSDV G +KMR LSGMPEC+ GLND++ + + +K K
Sbjct: 180 IYIDIVERMNLLISSTGNVLRSDVSGVVKMRAMLSGMPECQFGLNDKLDFKLKQSESKSK 239
Query: 233 A-------------IDLEDIKFHQCVRLARFENDRTISFIPPDGAFDLMTYRLSTQVKPL 279
+ + LED +FHQCVRL FEN+ I+FIPPDG +LM+YR +
Sbjct: 240 SNNSRNPSSVNGGFVILEDCQFHQCVRLPEFENEHRITFIPPDGEVELMSYRSHENINIP 299
Query: 280 IWVEATVEKHSKSRIEIMVKARSQFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAP 339
+ VE+ SK +I + R+ + + ++++ +PVP + + N R + G A Y P
Sbjct: 300 FRIVPIVEQLSKQKIIYRISIRADYPHK-LSSSLNFRIPVPTNVVKANPRVNRGKAGYEP 358
Query: 340 EKDALIWKIRSFPGGKEYMLRAEFRLPSITAEEATPERKAPIRVKFEIPYFTVSGIQVRY 399
++ + WKI F G E + AE L + T ++ K PI + F I FT SG+ V+Y
Sbjct: 359 SENIINWKIPRFLGETELIFYAEVELSNTTNQQIW--AKPPISLDFNILMFTSSGLHVQY 416
Query: 400 LKIIEKSG--YQALPWVRYITMAGEYELRL 427
L++ E S Y+++ WVRY T AG E+R+
Sbjct: 417 LRVSEPSNSKYKSIKWVRYSTRAGTCEIRI 446
>A4M1_HUMAN (O00189) Adapter-related protein complex 4 mu 1 subunit
(Mu subunit of AP-4) (AP-4 adapter complex mu subunit)
(Mu-adaptin-related protein 2) (mu-ARP2) (mu4)
Length = 453
Score = 205 bits (521), Expect = 2e-52
Identities = 136/454 (29%), Positives = 227/454 (49%), Gaps = 35/454 (7%)
Query: 6 SALFLLDIKGRVLIWRDYRGDVTALD-AERFFTKLIEKQADAQSQDPVV-HDNGISYMFV 63
S F+L KG LI++D+RGD D AE F+ KL D + PVV H +G ++ +
Sbjct: 3 SQFFILSSKGDPLIYKDFRGDSGGRDVAELFYRKLTGLPGD---ESPVVMHHHGRHFIHI 59
Query: 64 QHSNVYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIMDF 123
+HS +YL++ T +N + SLL L R+ + Y L E ++ N +VYELLDE++D+
Sbjct: 60 RHSGLYLVVTTSENVSPFSLLELLSRLATLLGDYCGSLGEGTISRNVALVYELLDEVLDY 119
Query: 124 GYPQYTEAKILSEFIKTDAYRMEV---------------TQRPPMAVTNAVS---WRSEG 165
GY Q T ++L FI+T+A + TQ+ +A ++A S S
Sbjct: 120 GYVQTTSTEMLRNFIQTEAVVSKPFSLFDLSSVGLFGAETQQSKVAPSSAASRPVLSSRS 179
Query: 166 ISYKKNEVFLDVVESVNILVNSNGQIIRSDVVGALKMRTYLSGMPECKLGLNDRVLLEAQ 225
+KNEVFLDVVE +++L+ SNG +++ DV G ++++++L E ++GL + +
Sbjct: 180 DQSQKNEVFLDVVERLSVLIASNGSLLKVDVQGEIRLKSFLPSGSEMRIGLTEEFCVGKS 239
Query: 226 GRTTKGKAIDLEDIKFHQCVRLARFENDRTISFIPPDGAFDLMTYRLSTQVK---PLIWV 282
G I ++++ FH V L FE+ R + PP G +M Y+LS + P
Sbjct: 240 ELRGYGPGIRVDEVSFHSSVNLDEFESHRILRLQPPQGELTVMRYQLSDDLPSPLPFRLF 299
Query: 283 EATVEKHSKSRIEIMVKARSQFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAPEKD 342
+ R+++ +K R +S A NV + LP+P ++ + S +
Sbjct: 300 PSVQWDRGSGRLQVYLKLRCDLLSKSQALNVRLHLPLPRGVVSLSRELSSPEQKAELAEG 359
Query: 343 ALIWKIRSFPGGKEYMLRAEFRLPSI-------TAEEATPERKAPIRVKFEIPYFTVSGI 395
AL W + GG + + +P + A+P P + FE+P T SG+
Sbjct: 360 ALRWDLPRVQGGSQLSGLFQMDVPGPPGPPSHGLSTSASPLGLGPASLSFELPRHTCSGL 419
Query: 396 QVRYLKIIEKSGYQALP--WVRYITMAGEYELRL 427
QVR+L++ + A P WVR+++ + Y +R+
Sbjct: 420 QVRFLRLAFRPCGNANPHKWVRHLSHSDAYVIRI 453
>A3M1_HUMAN (Q9Y2T2) Adapter-related protein complex 3 mu 1 subunit
(Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit)
Length = 418
Score = 196 bits (498), Expect = 1e-49
Identities = 121/436 (27%), Positives = 218/436 (49%), Gaps = 38/436 (8%)
Query: 7 ALFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISYMFVQHS 66
+LFL++ G + + + ++ V+ + FF + EK AD ++ PV+ + +
Sbjct: 4 SLFLINCSGDIFLEKHWKSVVSQSVCDYFF-EAQEKAADVENVPPVISTPHHYLISIYRD 62
Query: 67 NVYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIMDFGYP 126
++ + + ++ FLHRV D F+ YF E E +++DN V+VYELL+E++D G+P
Sbjct: 63 KLFFVSVIQTEVPPLFVIEFLHRVADTFQDYFGECSEAAIKDNVVIVYELLEEMLDNGFP 122
Query: 127 QYTEAKILSEFIKTDAYRMEVT----------QRPPMAVTNAVSWRSEGISYKKNEVFLD 176
TE+ IL E IK V P + + WR G+ Y NE + D
Sbjct: 123 LATESNILKELIKPPTILRSVVNSITGSSNVGDTLPTGQLSNIPWRRAGVKYTNNEAYFD 182
Query: 177 VVESVNILVNSNGQIIRSDVVGALKMRTYLSGMPECKLG-LNDRVLLEAQGRTTKGKAID 235
VVE ++ +++ +G + +++ G + LSGMP+ L +N R+
Sbjct: 183 VVEEIDAIIDKSGSTVFAEIQGVIDACIKLSGMPDLSLSFMNPRL--------------- 227
Query: 236 LEDIKFHQCVRLARFENDRTISFIPPDGAFDLMTYRLSTQ--VKPLIWVEATV---EKHS 290
L+D+ FH C+R R+E++R +SFIPPDG F L++YR+S+Q V ++V+ ++ E S
Sbjct: 228 LDDVSFHPCIRFKRWESERVLSFIPPDGNFRLISYRVSSQNLVAIPVYVKHSISFKENSS 287
Query: 291 KSRIEIMVKARSQFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAPEKDALIWKIRS 350
R +I + + + V + +P V +N N+ + GS + P L W +
Sbjct: 288 CGRFDITIGPKQNMGKTIEGITVTVHMPKVV--LNMNLTPTQGSYTFDPVTKVLTWDVGK 345
Query: 351 FPGGKEYMLRAEFRLPSITAEEATPERKAPIRVKFEIPYFTVSGIQVRYLKIIEKSGYQA 410
K L+ L ++ + PE + ++F+I +SG++V L + + Y+
Sbjct: 346 ITPQKLPSLKG---LVNLQSGAPKPEENPSLNIQFKIQQLAISGLKVNRLDMYGEK-YKP 401
Query: 411 LPWVRYITMAGEYELR 426
V+Y+T AG++++R
Sbjct: 402 FKGVKYVTKAGKFQVR 417
>A3M1_MOUSE (Q9JKC8) Adapter-related protein complex 3 mu 1 subunit
(Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit)
Length = 418
Score = 196 bits (497), Expect = 1e-49
Identities = 121/436 (27%), Positives = 218/436 (49%), Gaps = 38/436 (8%)
Query: 7 ALFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISYMFVQHS 66
+LFL++ G + + + ++ V+ + FF + EK AD ++ PV+ + +
Sbjct: 4 SLFLINCSGDIFLEKHWKSVVSQSVCDYFF-EAQEKAADVENVPPVISTPHHYLISIYRD 62
Query: 67 NVYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIMDFGYP 126
++ + + ++ FLHRV D F+ YF E E +++DN V+VYELL+E++D G+P
Sbjct: 63 KLFFVSVIQTEVPPLFVIEFLHRVADTFQDYFGECSEAAIKDNVVIVYELLEEMLDNGFP 122
Query: 127 QYTEAKILSEFIKTDAYRMEVT----------QRPPMAVTNAVSWRSEGISYKKNEVFLD 176
TE+ IL E IK V P + + WR G+ Y NE + D
Sbjct: 123 LATESNILKELIKPPTILRSVVNSITGSSNVGDTLPTGQLSNIPWRRAGVKYTNNEAYFD 182
Query: 177 VVESVNILVNSNGQIIRSDVVGALKMRTYLSGMPECKLG-LNDRVLLEAQGRTTKGKAID 235
VVE ++ +++ +G + +++ G + LSGMP+ L +N R+
Sbjct: 183 VVEEIDAIIDKSGSTVFAEIQGVIDACIKLSGMPDLSLSFMNPRL--------------- 227
Query: 236 LEDIKFHQCVRLARFENDRTISFIPPDGAFDLMTYRLSTQ--VKPLIWVEATV---EKHS 290
L+D+ FH C+R R+E++R +SFIPPDG F L++YR+S+Q V ++V+ ++ E S
Sbjct: 228 LDDVSFHPCIRFKRWESERVLSFIPPDGNFRLISYRVSSQNLVAIPVYVKHSISFKENSS 287
Query: 291 KSRIEIMVKARSQFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAPEKDALIWKIRS 350
R +I + + + V + +P V +N N+ + GS + P L W +
Sbjct: 288 CGRFDITIGPKQNMGKTIEGITVTVHMPKVV--LNMNLTPTQGSYTFDPVTKVLAWDVGK 345
Query: 351 FPGGKEYMLRAEFRLPSITAEEATPERKAPIRVKFEIPYFTVSGIQVRYLKIIEKSGYQA 410
K L+ L ++ + PE + ++F+I +SG++V L + + Y+
Sbjct: 346 ITPQKLPSLKG---LVNLQSGAPKPEENPNLNIQFKIQQLAISGLKVNRLDMYGEK-YKP 401
Query: 411 LPWVRYITMAGEYELR 426
V+Y+T AG++++R
Sbjct: 402 FKGVKYVTKAGKFQVR 417
>A4M1_MOUSE (Q9JKC7) Adapter-related protein complex 4 mu 1 subunit
(Mu subunit of AP-4) (AP-4 adapter complex mu subunit)
(Mu-adaptin-related protein 2) (mu-ARP2) (mu4)
Length = 449
Score = 195 bits (495), Expect = 2e-49
Identities = 132/450 (29%), Positives = 222/450 (49%), Gaps = 31/450 (6%)
Query: 6 SALFLLDIKGRVLIWRDYRGDVTALD-AERFFTKLIEKQADAQSQDPVVHDNGIS-YMFV 63
S F+L KG LI++D+RGD D AE F+ KL + PVV +G ++ +
Sbjct: 3 SQFFILSSKGDPLIYKDFRGDSGGRDVAELFYRKLTGLPG---GESPVVMYHGDRHFIHI 59
Query: 64 QHSNVYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIMDF 123
+HS +YL+ T +N + SLL L R+ + Y L E ++ N +VYELLDE++D+
Sbjct: 60 RHSGLYLVATTLENVSPFSLLELLSRLATLLGDYCGSLNEGTISRNVALVYELLDEVLDY 119
Query: 124 GYPQYTEAKILSEFIKTDAYRMEV---------------TQRPPMAVTNAVS---WRSEG 165
GY Q T ++L FI+T+A + TQ+ +A ++A S S
Sbjct: 120 GYVQTTSTEMLRNFIQTEAVVSKPFSLFDLSSVGLFGAETQQNKVAPSSAASRPVLSSRS 179
Query: 166 ISYKKNEVFLDVVESVNILVNSNGQIIRSDVVGALKMRTYLSGMPECKLGLNDRVLLEAQ 225
+KNEVFLDVVE +++L+ SNG +++ DV G ++++++L E +GL + +
Sbjct: 180 DQSQKNEVFLDVVERLSVLIASNGSLLKVDVQGEIRLKSFLPSGSEICIGLTEEFCVGKS 239
Query: 226 GRTTKGKAIDLEDIKFHQCVRLARFENDRTISFIPPDGAFDLMTYRLSTQVK---PLIWV 282
G I ++++ FH V L FE+ R + PP G +M Y+LS + P
Sbjct: 240 ELRGYGPGIRVDEVSFHSSVNLDEFESHRILRLQPPQGELTVMRYQLSDDLPSPLPFRLF 299
Query: 283 EATVEKHSKSRIEIMVKARSQFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAPEKD 342
+ R+++ +K R +S A N+ + LP+P ++ + S +
Sbjct: 300 PSVQWDQGSGRLQVYLKLRCDLPPKSQALNIHLHLPLPRGVISLSQELSSPDQKAELGEG 359
Query: 343 ALIWKIRSFPGGKEYMLRAEFRLPSITA---EEATPERKAPIRVKFEIPYFTVSGIQVRY 399
AL W + GG + + +P + +P P + FE+P T SG+QVR+
Sbjct: 360 ALHWDLPRVQGGSQLSGLFQMDVPGLQGLPNHGPSPLGLGPASLSFELPRHTCSGLQVRF 419
Query: 400 LKIIEKSGYQALP--WVRYITMAGEYELRL 427
L++ + A P WVR+++ + Y +R+
Sbjct: 420 LRLSFSACGNANPHKWVRHLSHSNAYVIRI 449
>A3M1_RAT (P53676) Adapter-related protein complex 3 mu 1 subunit
(Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit)
(Clathrin coat assembly protein AP47 homolog 1)
(Clathrin coat associated protein AP47 homolog 1) (Golgi
adaptor AP-1 47 kDa protein homo
Length = 418
Score = 192 bits (489), Expect = 1e-48
Identities = 121/436 (27%), Positives = 216/436 (48%), Gaps = 38/436 (8%)
Query: 7 ALFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISYMFVQHS 66
+LFL++ G + + + ++ V+ + FF + EK AD ++ V+ + +
Sbjct: 4 SLFLINCSGDIFLEKHWKSVVSQSVCDYFF-EAQEKAADVENVPTVISTPHHYLISIYRD 62
Query: 67 NVYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIMDFGYP 126
++ + + ++ FLHRV D F+ YF E E +++DN V+VYELL+E++D G+P
Sbjct: 63 KLFFVSVIQTEVPPLFVIEFLHRVADTFQDYFGECSEAAIKDNVVIVYELLEEMLDNGFP 122
Query: 127 QYTEAKILSEFIKTDAYRMEVT----------QRPPMAVTNAVSWRSEGISYKKNEVFLD 176
TE+ IL E IK V P + + WR G+ Y NE + D
Sbjct: 123 LATESNILKELIKPPTILRSVVNSITGSSNVGDTLPTGQLSNIPWRRAGVKYTNNEAYFD 182
Query: 177 VVESVNILVNSNGQIIRSDVVGALKMRTYLSGMPECKLG-LNDRVLLEAQGRTTKGKAID 235
VVE ++ +++ +G + +++ G + LSGMP+ L +N R+
Sbjct: 183 VVEEIDAIIDKSGSTVFAEIQGVIDACIKLSGMPDLSLSFMNPRL--------------- 227
Query: 236 LEDIKFHQCVRLARFENDRTISFIPPDGAFDLMTYRLSTQ--VKPLIWVEATV---EKHS 290
L+D+ FH C+R R+E++R +SFIPPDG F L++YR+S+Q V ++V+ + E S
Sbjct: 228 LDDVSFHPCIRFKRWESERVLSFIPPDGNFRLISYRVSSQNLVAIPVYVKHNISFKENSS 287
Query: 291 KSRIEIMVKARSQFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAPEKDALIWKIRS 350
R +I + + + V + +P V +N N+ + GS + P L W +
Sbjct: 288 CGRFDITIGPKQNMGKTIEGITVTVHMPKVV--LNMNLTPTQGSYTFDPVTKVLAWDVGK 345
Query: 351 FPGGKEYMLRAEFRLPSITAEEATPERKAPIRVKFEIPYFTVSGIQVRYLKIIEKSGYQA 410
K L+ L ++ + PE + ++F+I +SG++V L + + Y+
Sbjct: 346 ITPQKLPSLKG---LVNLQSGAPKPEENPNLNIQFKIQQLAISGLKVNRLDMYGEK-YKP 401
Query: 411 LPWVRYITMAGEYELR 426
V+YIT AG++++R
Sbjct: 402 FKGVKYITKAGKFQVR 417
>A3M2_HUMAN (P53677) Adapter-related protein complex 3 mu 2 subunit
(Clathrin coat assembly protein AP47 homolog 2)
(Clathrin coat associated protein AP47 homolog 2) (Golgi
adaptor AP-1 47 kDa protein homolog 2) (HA1 47 kDa
subunit homolog 2) (Clathrin as
Length = 418
Score = 187 bits (476), Expect = 3e-47
Identities = 121/436 (27%), Positives = 216/436 (48%), Gaps = 38/436 (8%)
Query: 7 ALFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISYMFVQHS 66
+LFL++ G + + + ++ V+ + FF + E+ +A++ PV+ + V
Sbjct: 4 SLFLINSSGDIFLEKHWKSVVSRSVCDYFF-EAQERATEAENVPPVIPTPHHYLLSVYRH 62
Query: 67 NVYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIMDFGYP 126
++ + + ++ FLHRVVD F+ YF E ++DN VVVYE+L+E++D G+P
Sbjct: 63 KIFFVAVIQTEVPPLFVIEFLHRVVDTFQDYFGVCSEPVIKDNVVVVYEVLEEMLDNGFP 122
Query: 127 QYTEAKILSEFIK----------TDAYRMEVTQRPPMAVTNAVSWRSEGISYKKNEVFLD 176
TE+ IL E IK T V + P + V WR G+ Y NE + D
Sbjct: 123 LATESNILKELIKPPTILRTVVNTITGSTNVGDQLPTGQLSVVPWRRTGVKYTNNEAYFD 182
Query: 177 VVESVNILVNSNGQIIRSDVVGALKMRTYLSGMPECKLG-LNDRVLLEAQGRTTKGKAID 235
V+E ++ +++ +G I +++ G + L+GMP+ L +N R+
Sbjct: 183 VIEEIDAIIDKSGSTITAEIQGVIDACVKLTGMPDLTLSFMNPRL--------------- 227
Query: 236 LEDIKFHQCVRLARFENDRTISFIPPDGAFDLMTYRLSTQ--VKPLIWVEATV---EKHS 290
L+D+ FH CVR R+E++R +SFIPPDG F L++Y +S Q V ++V+ + + S
Sbjct: 228 LDDVSFHPCVRFKRWESERILSFIPPDGNFRLLSYHVSAQNLVAIPVYVKHNISFRDSSS 287
Query: 291 KSRIEIMVKARSQFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAPEKDALIWKIRS 350
R EI V + + T V + +P +N ++ S G+ + P L W +
Sbjct: 288 LGRFEITVGPKQTMGK--TIEGVTVTSQMPKGVLNMSLTPSQGTHTFDPVTKMLSWDVGK 345
Query: 351 FPGGKEYMLRAEFRLPSITAEEATPERKAPIRVKFEIPYFTVSGIQVRYLKIIEKSGYQA 410
K L+ S+ A + P+ I ++F+I +SG++V L + + Y+
Sbjct: 346 INPQKLPSLKGTM---SLQAGASKPDENPTINLQFKIQQLAISGLKVNRLDMYGEK-YKP 401
Query: 411 LPWVRYITMAGEYELR 426
++Y+T AG++++R
Sbjct: 402 FKGIKYMTKAGKFQVR 417
>A3M2_RAT (P53678) Adapter-related protein complex 3 mu 2 subunit
(Clathrin coat assembly protein AP47 homolog 2)
(Clathrin coat associated protein AP47 homolog 2) (Golgi
adaptor AP-1 47 kDa protein homolog 2) (HA1 47 kDa
subunit homolog 2) (Clathrin asse
Length = 418
Score = 184 bits (467), Expect = 4e-46
Identities = 119/436 (27%), Positives = 215/436 (49%), Gaps = 38/436 (8%)
Query: 7 ALFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVVHDNGISYMFVQHS 66
+LFL++ G + + + ++ V+ + FF + E+ +A++ PV+ + V
Sbjct: 4 SLFLINSAGDIFLEKHWKSVVSRSVCDYFF-EAQERATEAENVPPVIPTPHHYLLSVYRH 62
Query: 67 NVYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVVYELLDEIMDFGYP 126
++ + + ++ FLHRVVD F+ YF E ++DN VVVYE+L+E++D G+P
Sbjct: 63 KIFFVAVIQTEVPPLFVIEFLHRVVDTFQDYFGVCSEPVIKDNVVVVYEVLEEMLDNGFP 122
Query: 127 QYTEAKILSEFIK----------TDAYRMEVTQRPPMAVTNAVSWRSEGISYKKNEVFLD 176
TE+ IL E IK T V + P + V WR G+ Y NE + D
Sbjct: 123 LATESNILKELIKPPTILRTVVNTITGSTNVGDQLPTGQLSVVPWRRTGVKYTNNEAYFD 182
Query: 177 VVESVNILVNSNGQIIRSDVVGALKMRTYLSGMPECKLG-LNDRVLLEAQGRTTKGKAID 235
VVE ++ +++ +G + +++ G + L+GMP+ L +N R+
Sbjct: 183 VVEEIDAIIDKSGSTVTAEIQGVIDACVKLTGMPDLTLSFMNPRL--------------- 227
Query: 236 LEDIKFHQCVRLARFENDRTISFIPPDGAFDLMTYRLSTQ--VKPLIWVEATV---EKHS 290
L+D+ FH CVR R+E++R +SFIPPDG F L++Y +S Q V ++V+ ++ + S
Sbjct: 228 LDDVSFHPCVRFKRWESERILSFIPPDGNFRLLSYHVSAQNLVAIPVYVKHSISFRDSGS 287
Query: 291 KSRIEIMVKARSQFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAYAPEKDALIWKIRS 350
R EI V + + T V + +P +N ++ S G+ + P L W +
Sbjct: 288 LGRFEITVGPKQTMGK--TIEGVTVTSQMPKGVLNMSLTPSQGTHTFDPVTKMLSWDVGK 345
Query: 351 FPGGKEYMLRAEFRLPSITAEEATPERKAPIRVKFEIPYFTVSGIQVRYLKIIEKSGYQA 410
K L+ + + P+ I ++F+I +SG++V L + + Y+
Sbjct: 346 INPQKLPSLKGTM---GLQVGASKPDENPTINLQFKIQQLAISGLKVNRLDMYGEK-YKP 401
Query: 411 LPWVRYITMAGEYELR 426
++Y+T AG++++R
Sbjct: 402 FKGIKYMTKAGKFQVR 417
>A47H_DISOM (P47795) Clathrin coat assembly protein AP47 homolog
(Clathrin coat associated protein AP47 homolog) (Golgi
adaptor AP-1 47 kDa protein homolog) (HA1 47 kDa subunit
homolog) (Clathrin assembly protein assembly protein
complex 1 medium chain ho
Length = 418
Score = 170 bits (431), Expect = 6e-42
Identities = 123/449 (27%), Positives = 214/449 (47%), Gaps = 64/449 (14%)
Query: 7 ALFLLDIKGRVLIWRDYRGDVTALDAERFFTKLIEKQADAQSQDPVV------------- 53
+LFL++ G V + + +R V+ L+E Q A + V
Sbjct: 4 SLFLMNGGGAVFLEKHWRSVVS----RSVCAYLLEAQLKAGQPENVAPVLATPHHYLVST 59
Query: 54 HDNGISYMFVQHSNVYLMIATRQNCNAASLLFFLHRVVDVFKHYFEELEEESLRDNFVVV 113
H +GIS++ V + V + ++ FLHRV + + YF E E S++DN V+V
Sbjct: 60 HRHGISFVAVIQAEVPPLF----------VIEFLHRVAETLQDYFGECSEASIKDNVVIV 109
Query: 114 YELLDEIMDFGYPQYTEAKILSEFIKTDAYRMEVT----------QRPPMAVTNAVSWRS 163
YELL+E++D G+P TE+ IL E IK V + P + + WR
Sbjct: 110 YELLEEMLDNGFPLATESNILKELIKPPTILRSVVNSITGSSNVGDQLPTGQLSNIPWRR 169
Query: 164 EGISYKKNEVFLDVVESVNILVNSNGQIIRSDVVGALKMRTYLSGMPECKLG-LNDRVLL 222
G+ Y NE + DV E ++ +++ +G + +++ G + L+GMP+ L LN R+
Sbjct: 170 VGVKYTNNEAYFDVTEEIDAIIDKSGSTVFAEIQGVIDACIKLTGMPDLTLSFLNPRL-- 227
Query: 223 EAQGRTTKGKAIDLEDIKFHQCVRLARFENDRTISFIPPDGAFDLMTYRLSTQ--VKPLI 280
L+D+ FH CVR R+E++R +SFIPP G F LM+Y +++Q V +
Sbjct: 228 -------------LDDVSFHPCVRFKRWESERVLSFIPPVGNFRLMSYHVNSQNLVAIPV 274
Query: 281 WVEATV---EKHSKSRIEIMVKARSQFKERSTATNVEIELPVPVDAMNPNVRTSMGSAAY 337
+V+ + + S +I + + + N+ + + +P +N + + G+ +
Sbjct: 275 YVKHNINFRDDGSTGWFDITIGPKQTMGK--VVENILVIIHMPKVVLNMTLTAAQGNFTF 332
Query: 338 APEKDALIWKIRSFPGGKEYMLRAEFRLPSITAEEATPERKAPIRVKFEIPYFTVSGIQV 397
P LIW I K L+ L ++ + EA PE + ++F I VSG++V
Sbjct: 333 DPVTKVLIWDIGKIILPKLPTLKG---LINLQSGEAKPEENPTLNIQFRIQQLAVSGLKV 389
Query: 398 RYLKIIEKSGYQALPWVRYITMAGEYELR 426
L + + Y+ V+Y+T AG++++R
Sbjct: 390 NRLDMYGER-YKPFKGVKYVTKAGKFQVR 417
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.137 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 47,848,157
Number of Sequences: 164201
Number of extensions: 1942858
Number of successful extensions: 4861
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 44
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 4708
Number of HSP's gapped (non-prelim): 66
length of query: 428
length of database: 59,974,054
effective HSP length: 113
effective length of query: 315
effective length of database: 41,419,341
effective search space: 13047092415
effective search space used: 13047092415
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0076a.11


