

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
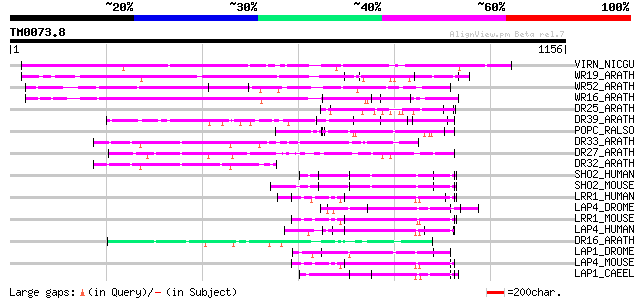
Query= TM0073.8
(1156 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
VIRN_NICGU (Q40392) TMV resistance protein N 528 e-149
WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY ... 385 e-106
WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY ... 360 2e-98
WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY ... 358 6e-98
DR25_ARATH (O50052) Putative disease resistance protein At4g19050 115 7e-25
DR39_ARATH (Q9LVT1) Putative disease resistance protein At5g47280 114 2e-24
POPC_RALSO (Q9RBS2) PopC protein 104 1e-21
DR33_ARATH (Q9FG90) Probable disease resistance protein At5g43740 94 3e-18
DR27_ARATH (Q9T048) Putative disease resistance protein At4g27190 94 3e-18
DR32_ARATH (Q9FG91) Probable disease resistance protein At5g43730 91 1e-17
SHO2_HUMAN (Q9UQ13) Leucine-rich repeat protein SHOC-2 (Ras-bind... 89 7e-17
SHO2_MOUSE (O88520) Leucine-rich repeat protein SHOC-2 (Ras-bind... 87 3e-16
LRR1_HUMAN (Q9BTT6) Leucine-rich repeat-containing protein 1 (LA... 87 3e-16
LAP4_DROME (Q7KRY7) LAP4 protein (Scribble protein) (Smell-impai... 86 4e-16
LRR1_MOUSE (Q80VQ1) Leucine-rich repeat-containing protein 1 85 1e-15
LAP4_HUMAN (Q14160) LAP4 protein (Scribble homolog protein) (hSc... 84 2e-15
DR16_ARATH (O22727) Putative disease resistance protein At1g61190 84 2e-15
LAP1_DROME (Q9V780) Lap1 protein 83 5e-15
LAP4_MOUSE (Q80U72) LAP4 protein (Scribble homolog protein) 82 8e-15
LAP1_CAEEL (O61967) Lap1 protein (Lethal protein 413) 82 1e-14
>VIRN_NICGU (Q40392) TMV resistance protein N
Length = 1144
Score = 528 bits (1359), Expect = e-149
Identities = 372/1080 (34%), Positives = 570/1080 (52%), Gaps = 129/1080 (11%)
Query: 24 YDVFLSFSGEDTGKTFTSHLHAALRRKNV--FIDDRSLETG----DEICKAIEASTIYVI 77
YDVFLSF GEDT KTFTSHL+ L K + F DD+ LE G E+CKAIE S ++
Sbjct: 12 YDVFLSFRGEDTRKTFTSHLYEVLNDKGIKTFQDDKRLEYGATIPGELCKAIEESQFAIV 71
Query: 78 VFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNRL 137
VFSENYA+SR CL+ELVKIMECK + + VIPIFY VDPS VR+Q S+ AF+EH+ +
Sbjct: 72 VFSENYATSRWCLNELVKIMECKTRFKQTVIPIFYDVDPSHVRNQKESFAKAFEEHETKY 131
Query: 138 VPGVV-MQRWKKALLEAADLSGYHSIAARSESMLIDRIAQDIFKKL---NPPFSQGMLGI 193
V +QRW+ AL EAA+L G ++++ I +I I KL + + Q ++GI
Sbjct: 132 KDDVEGIQRWRIALNEAANLKGSCDNRDKTDADCIRQIVDQISSKLCKISLSYLQNIVGI 191
Query: 194 DKHIAQIQSLLQLDSEAVRIIGICGMGGIGKSTLAEALYHKL------GTQFSSRCLIVN 247
D H+ +I+SLL++ VRI+GI GMGG+GK+T+A A++ L QF C + +
Sbjct: 192 DTHLEKIESLLEIGINGVRIMGIWGMGGVGKTTIARAIFDTLLGRMDSSYQFDGACFLKD 251
Query: 248 AQQKIDRYGIYSLRKKYLSKLLGEDIQSN----GLNYAIERVKRAKVLLILDDLKISIPI 303
++ ++ G++SL+ LS+LL E N G + R++ KVL++LDD+
Sbjct: 252 IKE--NKRGMHSLQNALLSELLREKANYNNEEDGKHQMASRLRSKKVLIVLDDIDNKDHY 309
Query: 304 LELIGGHGN-FGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYP 362
LE + G + FG GSRII+T+R +H+++ + D IYEV + +S++LF+ +AF ++ P
Sbjct: 310 LEYLAGDLDWFGNGSRIIITTRDKHLIE--KNDIIYEVTALPDHESIQLFKQHAFGKEVP 367
Query: 363 LEGYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSY 422
E + L +EV+ YAKG+PLAL+V GSLL++ W+S ++ +K I D LK+SY
Sbjct: 368 NENFEKLSLEVVNYAKGLPLALKVWGSLLHNLRLTEWKSAIEHMKNNSYSGIIDKLKISY 427
Query: 423 DGLDDEPKDIFLDITCFYAGDFVDKVVELLDSCGFSADIGMDVLKDRGLISILE-DRIAV 481
DGL+ + +++FLDI CF G+ D ++++L+SC A+ G+ +L D+ L+ I E +++ +
Sbjct: 428 DGLEPKQQEMFLDIACFLRGEEKDYILQILESCHIGAEYGLRILIDKSLVFISEYNQVQM 487
Query: 482 HDLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDMCQIEKVQL 541
HDL+ +MG+ IV Q DPG+ S LW KE+ V+ N GT A++ I++ ++
Sbjct: 488 HDLIQDMGKYIVNFQ--KDPGERSRLWLAKEVEEVMSNNTGTMAMEAIWVSSYS-STLRF 544
Query: 542 HPEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLDFC 601
+ K+M +R+ F G+S+ ++ LPN L+ +P SFP F
Sbjct: 545 SNQAVKNMKRLRV--------FNMGRSSTHYA--IDYLPNNLRCFVCTNYPWESFPSTFE 594
Query: 602 PEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQELPNLERLDLSNSW 661
+ LV L +R + L LW + + LP+L R+DLS S
Sbjct: 595 LKMLVHLQLRHNSLRHLWTETK------------------------HLPSLRRIDLSWSK 630
Query: 662 KLVRIPDLSKSPNIKEI------------------------ILSGCKSLTRLPINLFKLK 697
+L R PD + PN++ + L+ CKSL R P ++
Sbjct: 631 RLTRTPDFTGMPNLEYVNLYQCSNLEEVHHSLGCCSKVIGLYLNDCKSLKRFPC--VNVE 688
Query: 698 FLERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSL-NHLVGLEELSLKFCSK 756
LE L L C ++E +PEI M+ + ++ + I+ELPSS+ + + +L L
Sbjct: 689 SLEYLGLRSCDSLEKLPEIYGRMKPEIQIHMQGSGIRELPSSIFQYKTHVTKLLLWNMKN 748
Query: 757 LESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAES 816
L ++PSSI L L L ++ C LE+ P I DL L+ F A
Sbjct: 749 LVALPSSICRLKSLVSLSVSGCSKLESLPEEIGDL----------DNLRVFDA------- 791
Query: 817 FAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLE--SLPNSICNLKLLSELDCSGCRK 874
++T I + PSS+ L L L + D P L L L+ S C
Sbjct: 792 ------SDTLILRPPSSIIRLNKLIILMFRGFKDGVHFEFPPVAEGLHSLEYLNLSYCNL 845
Query: 875 LTG-IPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIPHLPPFMK 933
+ G +P +IG LSSL+ L L +LP SIA L +L+SLD+ C++L +P LPP +
Sbjct: 846 IDGGLPEEIGSLSSLKKLDLSRNNFEHLPSSIAQLGALQSLDLKDCQRLTQLPELPPELN 905
Query: 934 QL-------LAF-HCPSIRSVMSNSRFLSDSKEGTFELYFTNNEKQDPGAQRDVVRNAEH 985
+L L F H + + L D+ T F Q+ + R + ++
Sbjct: 906 ELHVDCHMALKFIHYLVTKRKKLHRVKLDDAHNDTMYNLFAYTMFQNISSMRHDISASDS 965
Query: 986 RITDVACRFAFYCFPGSEVPNWFHHHCKRNSVTVDKDSLNLSNDNRIIGFAMCFVLQLED 1045
V F +P ++P+WFHH +SV+V+ N ++ +GFA+C+ L D
Sbjct: 966 LSLTV---FTGQPYP-EKIPSWFHHQGWDSSVSVNLPE-NWYIPDKFLGFAVCYSRSLID 1020
>WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY
DNA-binding protein 19)
Length = 1895
Score = 385 bits (988), Expect = e-106
Identities = 275/828 (33%), Positives = 432/828 (51%), Gaps = 55/828 (6%)
Query: 24 YDVFLSFSGED-TGKTFTSHLHAALRRKNVFIDDRSLETGDEICKAIEASTIYVIVFSEN 82
YDV + + D + + F SHL A+L R+ + + ++ E A+ + +IV +
Sbjct: 668 YDVVIRYGRADISNEDFISHLRASLCRRGISVYEKFNEVD-----ALPKCRVLIIVLTST 722
Query: 83 YASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNRLVPGVV 142
Y S L+ I+E + DR V PIFYR+ P + +Y + + + +
Sbjct: 723 YVPSN-----LLNILEHQHTEDRVVYPIFYRLSPYDFVCNSKNYERFYLQDEPK------ 771
Query: 143 MQRWKKALLEAADLSGYHSIAARSESMLIDRIAQDIFKKLNPPFSQGMLGIDKHIAQIQS 202
+W+ AL E + GY ++ +SES LID I +D K L M+G+D + +I S
Sbjct: 772 --KWQAALKEITQMPGY-TLTDKSESELIDEIVRDALKVLCSADKVNMIGMDMQVEEILS 828
Query: 203 LLQLDSEAVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNAQQKIDRYGIYSLRK 262
LL ++S VR IGI G GIGK+T+AE ++ K+ Q+ + ++ + ++++ G ++R+
Sbjct: 829 LLCIESLDVRSIGIWGTVGIGKTTIAEEIFRKISVQYETCVVLKDLHKEVEVKGHDAVRE 888
Query: 263 KYLSKLLGED-----IQSNGLNYAIERVKRAKVLLILDDLKISIPILELIGGHGNFGQGS 317
+LS++L + I ++ R++R ++L+ILDD+ + +G FG GS
Sbjct: 889 NFLSEVLEVEPHVIRISDIKTSFLRSRLQRKRILVILDDVNDYRDVDTFLGTLNYFGPGS 948
Query: 318 RIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPLEGYANLIVEVLKYA 377
RII+TSR+R V + D +YEV+ + SL L + E Y L +E++K++
Sbjct: 949 RIIMTSRNRRVFVLCKIDHVYEVKPLDIPKSLLLLDRGTCQIVLSPEVYKTLSLELVKFS 1008
Query: 378 KGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSYDGLDDEPKDIFLDIT 437
G P LQ L S+ D+E W +++K + I + + S GLDD + IFLDI
Sbjct: 1009 NGNPQVLQFLSSI--DRE---WNKLSQEVKTTSPIYIPGIFEKSCCGLDDNERGIFLDIA 1063
Query: 438 CFYAGDFVDKVVELLDSCGFSADIGMDVLKDRGLISILEDRIA-VHDLVLEMGREIVRQQ 496
CF+ D V LLD CGFSA +G L D+ L++I + + + + GREIVRQ+
Sbjct: 1064 CFFNRIDKDNVAMLLDGCGFSAHVGFRGLVDKSLLTISQHNLVDMLSFIQATGREIVRQE 1123
Query: 497 CVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDMCQIEKVQLHPEIFKSMPNIRMLY 556
PG S LWN I HV + GT AI+ IFLDM + K +P +F+ M N+R+L
Sbjct: 1124 SADRPGDRSRLWNADYIRHVFINDTGTSAIEGIFLDMLNL-KFDANPNVFEKMCNLRLLK 1182
Query: 557 FHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLDFCPEKLVKLHMRESRLE 616
+ S + + V P LE LP+ L++LHW+ +P S P F PE LV+L++ S +
Sbjct: 1183 L--YCSKAEEKHGVSFPQGLEYLPSKLRLLHWEYYPLSSLPKSFNPENLVELNLPSSCAK 1240
Query: 617 QLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQELPNLERLDLSNSWKLVRIPDLSKSPNIK 676
+LW + C + + L L+++ LS S +L +IP LS + N++
Sbjct: 1241 KLWKGKKA--RFC--------------TTNSSLEKLKKMRLSYSDQLTKIPRLSSATNLE 1284
Query: 677 EIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQEL 736
I L GC SL L ++ LK L LNL GCS +ENIP + + +E+L VL L + +L
Sbjct: 1285 HIDLEGCNSLLSLSQSISYLKKLVFLNLKGCSKLENIPSMVD-LESLEVLNL--SGCSKL 1341
Query: 737 PSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLK-LTK 795
+ ++EL + + ++ IPSSI L L KLDL L+ P SI+ LK L
Sbjct: 1342 GNFPEISPNVKELYMG-GTMIQEIPSSIKNLVLLEKLDLENSRHLKNLPTSIYKLKHLET 1400
Query: 796 LDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTL 843
L+L+ C L+ FP + + L+ T IK+LPSS+ L L L
Sbjct: 1401 LNLSGCISLERFPDSSRRMKCLRFLDLSRTDIKELPSSISYLTALDEL 1448
Score = 61.2 bits (147), Expect = 2e-08
Identities = 77/274 (28%), Positives = 119/274 (43%), Gaps = 27/274 (9%)
Query: 698 FLERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQE-----LPSSLNHLVGLEELSLK 752
FL+ LNL +N P + E M NL +L L + +E P L +L L
Sbjct: 1157 FLDMLNLKFDAN----PNVFEKMCNLRLLKLYCSKAEEKHGVSFPQGLEYLPSKLRLLHW 1212
Query: 753 FCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDL--------KLTKLDLNDCSRL 804
L S+P S L +L+L + + + G KL K+ L+ +L
Sbjct: 1213 EYYPLSSLPKSFNP-ENLVELNLPSSCAKKLWKGKKARFCTTNSSLEKLKKMRLSYSDQL 1271
Query: 805 KTFPAILEPAESFAHISLAE-TAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKL 863
P L A + HI L ++ L S+ L L L LK C LE++P S+ +L+
Sbjct: 1272 TKIPR-LSSATNLEHIDLEGCNSLLSLSQSISYLKKLVFLNLKGCSKLENIP-SMVDLES 1329
Query: 864 LSELDCSGCRKLTGIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLE 923
L L+ SGC KL P +++ L + T + +P SI +L LE LD+ R L+
Sbjct: 1330 LEVLNLSGCSKLGNFPE---ISPNVKELYMGGTMIQEIPSSIKNLVLLEKLDLENSRHLK 1386
Query: 924 CIPHLPPFMKQLLAFHCPSIRSVMSNSRFLSDSK 957
+LP + +L ++ +S RF S+
Sbjct: 1387 ---NLPTSIYKLKHLETLNLSGCISLERFPDSSR 1417
Score = 49.3 bits (116), Expect = 6e-05
Identities = 55/224 (24%), Positives = 90/224 (39%), Gaps = 25/224 (11%)
Query: 728 LKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLT--YCESLETFP 785
++ I + +S + L+ L+LKF ++ K+C L L YC E
Sbjct: 1140 IRHVFINDTGTSAIEGIFLDMLNLKF-------DANPNVFEKMCNLRLLKLYCSKAEEKH 1192
Query: 786 GSIFDLKLTKLDLN------DCSRLKTFPAILEPAESFAHISLAETAIKKL--------P 831
G F L L + L + P P E+ ++L + KKL
Sbjct: 1193 GVSFPQGLEYLPSKLRLLHWEYYPLSSLPKSFNP-ENLVELNLPSSCAKKLWKGKKARFC 1251
Query: 832 SSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRTL 891
++ +L L+ + L L +P + + L +D GC L + I L L L
Sbjct: 1252 TTNSSLEKLKKMRLSYSDQLTKIPR-LSSATNLEHIDLEGCNSLLSLSQSISYLKKLVFL 1310
Query: 892 SLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIPHLPPFMKQL 935
+L+ + S+ L SLE L++S C KL P + P +K+L
Sbjct: 1311 NLKGCSKLENIPSMVDLESLEVLNLSGCSKLGNFPEISPNVKEL 1354
>WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY
DNA-binding protein 52) (Disease resistance protein
RRS1) (Resistance to Ralstonia solanacearum 1 protein)
Length = 1378
Score = 360 bits (923), Expect = 2e-98
Identities = 295/938 (31%), Positives = 463/938 (48%), Gaps = 135/938 (14%)
Query: 33 EDTGKTFTSHLHAALRRK---NVFIDDRSLETGDEICKA----IEASTIYVIVFSENYAS 85
E+ +F SHL ALRRK NV +D ++ D + K IE + + V+V N
Sbjct: 17 EEVRYSFVSHLSEALRRKGINNVVVD---VDIDDLLFKESQAKIEKAGVSVMVLPGNCDP 73
Query: 86 SRCCLDELVKIMECKMNY-DRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNRLVPGVVMQ 144
S LD+ K++EC+ N D+AV+ + Y GD+ ++
Sbjct: 74 SEVWLDKFAKVLECQRNNKDQAVVSVLY--------------GDS-----------LLRD 108
Query: 145 RWKKALLEAADLSGYHSIAAR-SESMLIDRIAQDIFKKLNPPFSQGMLGIDKHIAQIQSL 203
+W L + LS H S+S+L++ I +D+++ F G +GI + +I+++
Sbjct: 109 QWLSEL-DFRGLSRIHQSRKECSDSILVEEIVRDVYET---HFYVGRIGIYSKLLEIENM 164
Query: 204 LQLDSEAVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNAQQKIDRYGIYSLRKK 263
+ +R +GI GM GIGK+TLA+A++ ++ + F + C I + + I G+Y L ++
Sbjct: 165 VNKQPIGIRCVGIWGMPGIGKTTLAKAVFDQMSSAFDASCFIEDYDKSIHEKGLYCLLEE 224
Query: 264 YLSKLLGEDIQSNGLNYAIERVKRAKVLLILDDLKISIPILELIGGHGNFGQGSRIIVTS 323
L L G D L+ +R+ +VL++LDD+ ++ + G G GS II+TS
Sbjct: 225 QL--LPGNDATIMKLSSLRDRLNSKRVLVVLDDVCNALVAESFLEGFDWLGPGSLIIITS 282
Query: 324 RHRHVLKNAEADEIYEVQGMSYQDSLELFRLNA-FKEDYPLEGYANLIVEVLKYAKGVPL 382
R + V + ++IYEVQG++ +++ +LF L+A ED + L V V+ YA G PL
Sbjct: 283 RDKQVFRLCGINQIYEVQGLNEKEARQLFLLSASIMEDMGEQNLHELSVRVISYANGNPL 342
Query: 383 ALQVLGSLLYDKER-EVWESELKKLKELPNVDIFDVLKLSYDGLDDEPKDIFLDITCFYA 441
A+ V G L K++ E+ KLK P I D K SYD L D K+IFLDI CF+
Sbjct: 343 AISVYGRELKGKKKLSEMETAFLKLKRRPPFKIVDAFKSSYDTLSDNEKNIFLDIACFFQ 402
Query: 442 GDFVDKVVELLDSCGFSADIGMDVLKDRGLISILEDRIAVHDLVLEMGREIVRQQCVSDP 501
G+ V+ V++LL+ CGF + +DVL D+ L++I E+R+ +H L ++GREI+ + V
Sbjct: 403 GENVNYVIQLLEGCGFFPHVEIDVLVDKCLVTISENRVWLHKLTQDIGREIINGETVQIE 462
Query: 502 GKHSHLWNHKEIYHVVRKN---------------KGTDAIQCIFLDMCQIEKVQLHPEIF 546
+ LW I +++ N +G++ I+ +FLD + + L P F
Sbjct: 463 -RRRRLWEPWSIKYLLEYNEHKANGEPKTTFKRAQGSEEIEGLFLDTSNL-RFDLQPSAF 520
Query: 547 KSMPNIRMLYF-------HKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLD 599
K+M N+R+L H +F G L SLPN L++LHW+ +P +S P +
Sbjct: 521 KNMLNLRLLKIYCSNPEVHPVINFPTGS--------LHSLPNELRLLHWENYPLKSLPQN 572
Query: 600 FCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQELPNLERLDLSN 659
F P LV+++M S+L++LWG + L+ L + L +
Sbjct: 573 FDPRHLVEINMPYSQLQKLWGGTKNLEM------------------------LRTIRLCH 608
Query: 660 SWKLVRIPDLSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIPEIKET 719
S LV I DL K+ N++ I L GC L P +L L +NLSGC ++++ EI
Sbjct: 609 SQHLVDIDDLLKAENLEVIDLQGCTRLQNFPA-AGRLLRLRVVNLSGCIKIKSVLEIPPN 667
Query: 720 MENL-----AVLVLKQTAI----QELPSSLNHLVGLEELS-LKFCSKLESIPSSIGTLTK 769
+E L +L L + + +EL + L + GL E S L+ + L SS L K
Sbjct: 668 IEKLHLQGTGILALPVSTVKPNHRELVNFLTEIPGLSEASKLERLTSLLESNSSCQDLGK 727
Query: 770 LCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKT---FPAILEPAESFAHISLAETA 826
L L+L C L++ P ++ +L L LDL+ CS L + FP L+ + L TA
Sbjct: 728 LICLELKDCSCLQSLP-NMANLDLNVLDLSGCSSLNSIQGFPRFLK------QLYLGGTA 780
Query: 827 IKKLPSSLQNL--VGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGC 884
I+++P Q+L + CL+ SLPN + NL+ L LD SGC +L I
Sbjct: 781 IREVPQLPQSLEILNAHGSCLR------SLPN-MANLEFLKVLDLSGCSELETIQGFPRN 833
Query: 885 LSSLRTLSLQDTGVVNLPESI----AHLSSLESLDVSY 918
L L V LP S+ AH S E L + Y
Sbjct: 834 LKELYFAGTTLREVPQLPLSLEVLNAHGSDSEKLPMHY 871
Score = 50.1 bits (118), Expect = 4e-05
Identities = 29/83 (34%), Positives = 45/83 (53%), Gaps = 1/83 (1%)
Query: 415 FDVLKLSYDGLDDEPKDIFLDITCFYAGDFVDKVVELLDSCGFSADIGMDVLKDRGLISI 474
++VL++SYD L + K +FL I + + VD V L+ G+ VL D LIS+
Sbjct: 1086 YEVLRVSYDDLQEMDKVLFLYIASLFNDEDVDFVAPLIAGIDLDVSSGLKVLADVSLISV 1145
Query: 475 LED-RIAVHDLVLEMGREIVRQQ 496
+ I +H L +MG+EI+ Q
Sbjct: 1146 SSNGEIVMHSLQRQMGKEILHGQ 1168
>WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY
DNA-binding protein 16)
Length = 1372
Score = 358 bits (918), Expect = 6e-98
Identities = 255/842 (30%), Positives = 421/842 (49%), Gaps = 103/842 (12%)
Query: 33 EDTGKTFTSHLHAALRRK---NVFID-DRSLETGDEICKAIEASTIYVIVFSENYASSRC 88
E+ +F SHL AL+RK +VFID D SL +E +E + + V++ N S
Sbjct: 14 EEVRYSFVSHLSKALQRKGVNDVFIDSDDSLS--NESQSMVERARVSVMILPGNRTVS-- 69
Query: 89 CLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNRLVPGVVMQRWKK 148
LD+LVK+++C+ N D+ V+P+ Y V S W
Sbjct: 70 -LDKLVKVLDCQKNKDQVVVPVLYGVRSSETE-------------------------WLS 103
Query: 149 ALLEAADLSGYHSIAARSESMLIDRIAQDIFKKLNPPFSQGMLGIDKHIAQIQSLLQLDS 208
AL S +HS S+S L+ +D+++KL F +GI + +I+ ++
Sbjct: 104 ALDSKGFSSVHHSRKECSDSQLVKETVRDVYEKL---FYMERIGIYSKLLEIEKMINKQP 160
Query: 209 EAVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNAQQKIDRYGIYSL-RKKYLSK 267
+R +GI GM GIGK+TLA+A++ ++ +F + C I + + I G+Y L +++L +
Sbjct: 161 LDIRCVGIWGMPGIGKTTLAKAVFDQMSGEFDAHCFIEDYTKAIQEKGVYCLLEEQFLKE 220
Query: 268 LLGEDIQSNGLNYAIERVKRAKVLLILDDLKISIPILELIGGHGNFGQGSRIIVTSRHRH 327
G L+ +R+ +VL++LDD++ + + +GG FG S II+TS+ +
Sbjct: 221 NAGASGTVTKLSLLRDRLNNKRVLVVLDDVRSPLVVESFLGGFDWFGPKSLIIITSKDKS 280
Query: 328 VLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPLEGYANLIVEVLKYAKGVPLALQVL 387
V + ++IYEVQG++ +++L+LF L A +D + + ++V+KYA G PLAL +
Sbjct: 281 VFRLCRVNQIYEVQGLNEKEALQLFSLCASIDDMAEQNLHEVSMKVIKYANGHPLALNLY 340
Query: 388 GSLLYDKEREV-WESELKKLKELPNVDIFDVLKLSYDGLDDEPKDIFLDITCFYAGDFVD 446
G L K+R E KLKE P D +K SYD L+D K+IFLDI CF+ G+ VD
Sbjct: 341 GRELMGKKRPPEMEIAFLKLKECPPAIFVDAIKSSYDTLNDREKNIFLDIACFFQGENVD 400
Query: 447 KVVELLDSCGFSADIGMDVLKDRGLISILEDRIAVHDLVLEMGREIVRQQCVSDPGKHSH 506
V++LL+ CGF +G+DVL ++ L++I E+R+ +H+L+ ++GR+I+ ++ + S
Sbjct: 401 YVMQLLEGCGFFPHVGIDVLVEKSLVTISENRVRMHNLIQDVGRQIINRE-TRQTKRRSR 459
Query: 507 LWNHKEIYHVVRKNKGT---------------DAIQCIFLDMCQIEKVQLHPEIFKSMPN 551
LW I +++ + + I+ +FLD + + F +M N
Sbjct: 460 LWEPCSIKYLLEDKEQNENEEQKTTFERAQVPEEIEGMFLDTSNL-SFDIKHVAFDNMLN 518
Query: 552 IRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLDFCPEKLVKLHMR 611
+R+ + S+ + N + L SLPN L++LHW+ +P + P +F P LV+++M
Sbjct: 519 LRLFKIYS-SNPEVHHVNNFLKGSLSSLPNVLRLLHWENYPLQFLPQNFDPIHLVEINMP 577
Query: 612 ESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQELPNLERLDLSNSWKLVRIPDLSK 671
S+L++LWG ++L L+ + L +S +LV I DL K
Sbjct: 578 YSQLKKLWGGT------------------------KDLEMLKTIRLCHSQQLVDIDDLLK 613
Query: 672 SPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIPEIKETMENLAVLVLKQT 731
+ N++ + L GC L P +L L +NLSGC+ +++ PEI N+ L L+ T
Sbjct: 614 AQNLEVVDLQGCTRLQSFPAT-GQLLHLRVVNLSGCTEIKSFPEIP---PNIETLNLQGT 669
Query: 732 AIQELPSS------------------LNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKL 773
I ELP S L+ + LE+ LK + L I +S KL L
Sbjct: 670 GIIELPLSIVKPNYRELLNLLAEIPGLSGVSNLEQSDLKPLTSLMKISTSYQNPGKLSCL 729
Query: 774 DLTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSS 833
+L C L + P + L LDL+ CS L+T + + A + +LP S
Sbjct: 730 ELNDCSRLRSLPNMVNLELLKALDLSGCSELETIQGFPRNLKELYLVGTAVRQVPQLPQS 789
Query: 834 LQ 835
L+
Sbjct: 790 LE 791
Score = 53.5 bits (127), Expect = 3e-06
Identities = 49/165 (29%), Positives = 76/165 (45%), Gaps = 13/165 (7%)
Query: 773 LDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPS 832
+DL C L++FP + L L ++L+ C+ +K+FP I E+ ++L T I +LP
Sbjct: 620 VDLQGCTRLQSFPATGQLLHLRVVNLSGCTEIKSFPEIPPNIET---LNLQGTGIIELPL 676
Query: 833 SL--QNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRT 890
S+ N L L L E P L + N L + D L I L
Sbjct: 677 SIVKPNYRELLNL-LAEIPGLSGVSN-------LEQSDLKPLTSLMKISTSYQNPGKLSC 728
Query: 891 LSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIPHLPPFMKQL 935
L L D + ++ +L L++LD+S C +LE I P +K+L
Sbjct: 729 LELNDCSRLRSLPNMVNLELLKALDLSGCSELETIQGFPRNLKEL 773
Score = 45.8 bits (107), Expect = 7e-04
Identities = 33/109 (30%), Positives = 54/109 (49%), Gaps = 7/109 (6%)
Query: 651 NLERLDLSNSWKLVRIPDLSKSPN-IKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSN 709
NLE+ DL L++I ++P + + L+ C L LP N+ L+ L+ L+LSGCS
Sbjct: 701 NLEQSDLKPLTSLMKISTSYQNPGKLSCLELNDCSRLRSLP-NMVNLELLKALDLSGCSE 759
Query: 710 VENIPEIKETMENLAVLVLKQTAIQELPSSLNHL-----VGLEELSLKF 753
+E I ++ L ++ + +LP SL V L+ + L F
Sbjct: 760 LETIQGFPRNLKELYLVGTAVRQVPQLPQSLEFFNAHGCVSLKSIRLDF 808
Score = 35.8 bits (81), Expect = 0.69
Identities = 25/83 (30%), Positives = 43/83 (51%), Gaps = 2/83 (2%)
Query: 416 DVLKLSYDGLDDEPKDIFLDITCFYAGDFVDKVVELL-DSCGFSADIGMDVLKDRGLISI 474
+VL++ Y GL + K +FL I + + V V L+ + G+ VL R LI +
Sbjct: 1049 EVLRVRYAGLQEIYKALFLYIAGLFNDEDVGLVAPLIANIIDMDVSYGLKVLAYRSLIRV 1108
Query: 475 LED-RIAVHDLVLEMGREIVRQQ 496
+ I +H L+ +MG+EI+ +
Sbjct: 1109 SSNGEIVMHYLLRQMGKEILHTE 1131
>DR25_ARATH (O50052) Putative disease resistance protein At4g19050
Length = 1181
Score = 115 bits (288), Expect = 7e-25
Identities = 88/292 (30%), Positives = 147/292 (50%), Gaps = 39/292 (13%)
Query: 648 ELPNLERLDLSNSWKLVRIP---------DLSKSPNIKEIILSGCKSLTRLPINLFKLKF 698
+L LE LD S + K++R+P D S P + ++L C L RLP L L
Sbjct: 598 QLQLLEHLDFSET-KIIRLPIFHLKDSTNDFSTMPILTRLLLRNCTRLKRLP-QLRPLTN 655
Query: 699 LERLNLSGCSNVENIPEI-KETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKL 757
L+ L+ G +++ + E+ E + L +L + +T++ EL ++ +V L +L L+ CS +
Sbjct: 656 LQILDACGATDLVEMLEVCLEEKKELRILDMSKTSLPELADTIADVVNLNKLLLRNCSLI 715
Query: 758 ESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESF 817
E +PS I LT L D++ C L+ GS ++ S+
Sbjct: 716 EELPS-IEKLTHLEVFDVSGCIKLKNINGSFGEM------------------------SY 750
Query: 818 AH-ISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLT 876
H ++L+ET + +LP + L L+ L +++C L++LPN + L L D SGC +L
Sbjct: 751 LHEVNLSETNLSELPDKISELSNLKELIIRKCSKLKTLPN-LEKLTNLEIFDVSGCTELE 809
Query: 877 GIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIPHL 928
I LS L ++L +T + LP I+ LS+L+ L + C KL+ +P+L
Sbjct: 810 TIEGSFENLSCLHKVNLSETNLGELPNKISELSNLKELILRNCSKLKALPNL 861
Score = 100 bits (248), Expect = 3e-20
Identities = 86/330 (26%), Positives = 143/330 (43%), Gaps = 72/330 (21%)
Query: 669 LSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIPE-IKETMENLAVLV 727
LSK ++ +++ C + + L L+ L L +SG S++ NIP+ + M L L
Sbjct: 464 LSKLKKLRVLVIRDCDLIDNID-KLSGLQGLHVLEVSGASSLVNIPDDFFKNMTQLQSLN 522
Query: 728 LKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGS 787
L AI+ PS++ L L L+ CS+L+ +P+ I KL +D+ LE++
Sbjct: 523 LSGLAIKSSPSTIEKLSMLRCFILRHCSELQDLPNFIVETRKLEVIDIHGARKLESYFDR 582
Query: 788 IFDLK---------------------------------------------LTKLDLNDCS 802
+ D K LT+L L +C+
Sbjct: 583 VKDWKDYKGKNKNFAQLQLLEHLDFSETKIIRLPIFHLKDSTNDFSTMPILTRLLLRNCT 642
Query: 803 RLKTFPAI------------------------LEPAESFAHISLAETAIKKLPSSLQNLV 838
RLK P + LE + + +++T++ +L ++ ++V
Sbjct: 643 RLKRLPQLRPLTNLQILDACGATDLVEMLEVCLEEKKELRILDMSKTSLPELADTIADVV 702
Query: 839 GLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRTLSLQDTGV 898
L L L+ C +E LP SI L L D SGC KL I G +S L ++L +T +
Sbjct: 703 NLNKLLLRNCSLIEELP-SIEKLTHLEVFDVSGCIKLKNINGSFGEMSYLHEVNLSETNL 761
Query: 899 VNLPESIAHLSSLESLDVSYCRKLECIPHL 928
LP+ I+ LS+L+ L + C KL+ +P+L
Sbjct: 762 SELPDKISELSNLKELIIRKCSKLKTLPNL 791
Score = 99.4 bits (246), Expect = 5e-20
Identities = 72/257 (28%), Positives = 122/257 (47%), Gaps = 26/257 (10%)
Query: 647 QELPNLERLDLSNSWKLVRIPDLSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSG 706
+E L LD+S + ++ N+ +++L C + LP ++ KL LE ++SG
Sbjct: 676 EEKKELRILDMSKTSLPELADTIADVVNLNKLLLRNCSLIEELP-SIEKLTHLEVFDVSG 734
Query: 707 CSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGT 766
C ++NI M L + L +T + ELP ++ L L+EL ++ CSKL+++P+
Sbjct: 735 CIKLKNINGSFGEMSYLHEVNLSETNLSELPDKISELSNLKELIIRKCSKLKTLPN---- 790
Query: 767 LTKLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFAHISLAETA 826
L KL L++ D++ C+ L+T E ++L+ET
Sbjct: 791 LEKLTNLEI--------------------FDVSGCTELETIEGSFENLSCLHKVNLSETN 830
Query: 827 IKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLS 886
+ +LP+ + L L+ L L+ C L++LPN + L L D SGC L I +S
Sbjct: 831 LGELPNKISELSNLKELILRNCSKLKALPN-LEKLTHLVIFDVSGCTNLDKIEESFESMS 889
Query: 887 SLRTLSLQDTGVVNLPE 903
L ++L T + PE
Sbjct: 890 YLCEVNLSGTNLKTFPE 906
Score = 45.4 bits (106), Expect = 9e-04
Identities = 80/325 (24%), Positives = 134/325 (40%), Gaps = 55/325 (16%)
Query: 648 ELPNLERLDLSNSWKLVRIPDLSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGC 707
EL NL+ L L N KL +P+L K ++ +SGC +L ++ + + +L +NLSG
Sbjct: 840 ELSNLKELILRNCSKLKALPNLEKLTHLVIFDVSGCTNLDKIEESFESMSYLCEVNLSG- 898
Query: 708 SNVENIPEI--KETMENLAVLVLKQTA---------IQELPSSLNHLVGLEELSLKFCSK 756
+N++ PE+ + + + +VL ++ I+E +S + + K K
Sbjct: 899 TNLKTFPELPKQSILCSSKRIVLADSSCIERDQWSQIKECLTSKSEGSSFSNVGEKTREK 958
Query: 757 L-----------ESIPSSIGTLTKLCKLDL-----------TYCESLETFPGSIFDLKLT 794
L +P +I + DL + E+ S+FD +L
Sbjct: 959 LLYHGNRYRVIDPEVPLNIDIVDIKRSTDLKTEYIAKAEYVSIAENGSKSVSSLFD-ELQ 1017
Query: 795 KLDLNDC--SRLKTFPAILEPAESF-----AHISLAETAIKKLP--SSLQNLVG------ 839
+ C R K + E E + SL I LP +SL + G
Sbjct: 1018 MASVKGCWVERCKNMDVLFESDEQLEKEKSSSPSLQTLWISNLPLLTSLYSSKGGFIFKN 1077
Query: 840 LQTLCLKECPDLESL-PNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRTLSLQDTGV 898
L+ L + CP ++ L P NL++L C +L + G LS LR L L D V
Sbjct: 1078 LKKLSVDCCPSIKWLFPEIPDNLEILRVKFCDKLERLFEV--KAGELSKLRKLHLLDLPV 1135
Query: 899 VNLPESIAHLSSLESLDVSYCRKLE 923
+++ A+ +LE + C KL+
Sbjct: 1136 LSVLG--ANFPNLEKCTIEKCPKLK 1158
>DR39_ARATH (Q9LVT1) Putative disease resistance protein At5g47280
Length = 623
Score = 114 bits (284), Expect = 2e-24
Identities = 146/620 (23%), Positives = 280/620 (44%), Gaps = 87/620 (14%)
Query: 203 LLQLDSEAVRIIGICGMGGIGKSTLAEALYH--KLGTQFSSRCLIVNAQQKIDRYGIYSL 260
L L+ EA RIIGI GM G GK+ LA+ L ++ F++R L + Q + + SL
Sbjct: 2 LFNLNDEA-RIIGISGMIGSGKTILAKELARDEEVRGHFANRVLFLTVSQSPNLEELRSL 60
Query: 261 RKKYLSKLLGEDIQSNGLNYAI-ERVKRAKVLLILDDLKISIPILELIGGHGNFGQGSRI 319
+ +L+ G A+ E V + L+ILDD++ + +L+ G+
Sbjct: 61 IRDFLTG------HEAGFGTALPESVGHTRKLVILDDVRTRESLDQLMFNI----PGTTT 110
Query: 320 IVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPLEGYA-NLIVEVLKYAK 378
+V S+ + V + Y+V+ ++ D+ LF L+AF + G++ +L+ +V+ +K
Sbjct: 111 LVVSQSKLV----DPRTTYDVELLNEHDATSLFCLSAFNQKSVPSGFSKSLVKQVVGESK 166
Query: 379 GVPLALQVLGSLLYDKEREVWESELKKLKELPNVD------IFDVLKLSYDGLDDEPKDI 432
G+PL+L+VLG+ L D+ W +++L VD +F ++ + + LD + K+
Sbjct: 167 GLPLSLKVLGASLNDRPETYWAIAVERLSRGEPVDETHESKVFAQIEATLENLDPKTKEC 226
Query: 433 FLDITCFYAG---------DFVDKVVELLDSCGFSADIGMDVLKDRGLISILEDR----- 478
FLD+ F G + + K+ +L D+ F D+ +D L +R L+++++D
Sbjct: 227 FLDMGAFPEGKKIPVDVLINMLVKIHDLEDAAAF--DVLVD-LANRNLLTLVKDPTFVAM 283
Query: 479 --------IAVHDLVLEMGREIVRQQCVSD------PGKHSHLWNHKEIYHVVRKNKGTD 524
+ HD++ ++ + + VS P + + L + E R N
Sbjct: 284 GTSYYDIFVTQHDVLRDVALHLTNRGKVSRRDRLLMPKRETMLPSEWE-----RSNDEPY 338
Query: 525 AIQCIFLDMCQIEKVQLHPEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLES------ 578
+ + + ++ ++ F P +L + S N ++P +
Sbjct: 339 NARVVSIHTGEMTEMDWFDMDF---PKAEVLIVNFSS------DNYVLPPFIAKMGMLRV 389
Query: 579 ---LPNGLKVLHWDEFPQRSFPLDFCPEKLVKLHMRE--SRLEQLWGDDQVLQSLCHGLS 633
+ NG H +FP + + L ++H+ E S + L ++ +C ++
Sbjct: 390 FVIINNGTSPAHLHDFPIPTSLTNLRSLWLERVHVPELSSSMIPLKNLHKLYLIICK-IN 448
Query: 634 YIFFGHLIVFSLHQELPNLERLDLSNSWKLVRIPD-LSKSPNIKEIILSGCKSLTRLPIN 692
F + Q P L + + L +P + ++ I ++ C ++ LP N
Sbjct: 449 NSF--DQTAIDIAQIFPKLTDITIDYCDDLAELPSTICGITSLNSISITNCPNIKELPKN 506
Query: 693 LFKLKFLERLNLSGCSNVENIP-EIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSL 751
+ KL+ L+ L L C ++++P EI E + V + ++ LP + ++ LE++ +
Sbjct: 507 ISKLQALQLLRLYACPELKSLPVEICELPRLVYVDISHCLSLSSLPEKIGNVRTLEKIDM 566
Query: 752 KFCSKLESIPSSIGTLTKLC 771
+ CS L SIPSS +LT LC
Sbjct: 567 RECS-LSSIPSSAVSLTSLC 585
Score = 82.0 bits (201), Expect = 8e-15
Identities = 60/197 (30%), Positives = 93/197 (46%), Gaps = 20/197 (10%)
Query: 716 IKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDL 775
I ++ NL L L++ + EL SS+ L L +L L C S D
Sbjct: 407 IPTSLTNLRSLWLERVHVPELSSSMIPLKNLHKLYLIICKINNSF-------------DQ 453
Query: 776 TYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFAHISLAETA-IKKLPSSL 834
T + + FP KLT + ++ C L P+ + S IS+ IK+LP ++
Sbjct: 454 TAIDIAQIFP------KLTDITIDYCDDLAELPSTICGITSLNSISITNCPNIKELPKNI 507
Query: 835 QNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRTLSLQ 894
L LQ L L CP+L+SLP IC L L +D S C L+ +P IG + +L + ++
Sbjct: 508 SKLQALQLLRLYACPELKSLPVEICELPRLVYVDISHCLSLSSLPEKIGNVRTLEKIDMR 567
Query: 895 DTGVVNLPESIAHLSSL 911
+ + ++P S L+SL
Sbjct: 568 ECSLSSIPSSAVSLTSL 584
Score = 63.2 bits (152), Expect = 4e-09
Identities = 53/199 (26%), Positives = 100/199 (49%), Gaps = 15/199 (7%)
Query: 639 HLIVFSLHQELPNLERLDLSNSWKLVRIPDLSKS----PNIKEIILSGCK---SLTRLPI 691
HL F + L NL L L V +P+LS S N+ ++ L CK S + I
Sbjct: 401 HLHDFPIPTSLTNLRSLWLER----VHVPELSSSMIPLKNLHKLYLIICKINNSFDQTAI 456
Query: 692 NLFKL-KFLERLNLSGCSNVENIPEIKETMENL-AVLVLKQTAIQELPSSLNHLVGLEEL 749
++ ++ L + + C ++ +P + +L ++ + I+ELP +++ L L+ L
Sbjct: 457 DIAQIFPKLTDITIDYCDDLAELPSTICGITSLNSISITNCPNIKELPKNISKLQALQLL 516
Query: 750 SLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLK-LTKLDLNDCSRLKTFP 808
L C +L+S+P I L +L +D+++C SL + P I +++ L K+D+ +CS L + P
Sbjct: 517 RLYACPELKSLPVEICELPRLVYVDISHCLSLSSLPEKIGNVRTLEKIDMRECS-LSSIP 575
Query: 809 AILEPAESFAHISLAETAI 827
+ S +++ A+
Sbjct: 576 SSAVSLTSLCYVTCYREAL 594
Score = 49.3 bits (116), Expect = 6e-05
Identities = 26/88 (29%), Positives = 45/88 (50%), Gaps = 1/88 (1%)
Query: 840 LQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRTLSLQDTGVV 899
L + + C DL LP++IC + L+ + + C + +P +I L +L+ L L +
Sbjct: 465 LTDITIDYCDDLAELPSTICGITSLNSISITNCPNIKELPKNISKLQALQLLRLYACPEL 524
Query: 900 -NLPESIAHLSSLESLDVSYCRKLECIP 926
+LP I L L +D+S+C L +P
Sbjct: 525 KSLPVEICELPRLVYVDISHCLSLSSLP 552
>POPC_RALSO (Q9RBS2) PopC protein
Length = 1024
Score = 104 bits (260), Expect = 1e-21
Identities = 92/299 (30%), Positives = 144/299 (47%), Gaps = 34/299 (11%)
Query: 652 LERLDLSNSWKLVRIPD-LSKSPNIKEIILS--GCKSLTRLPINLFKLKFLERLNLSGCS 708
LE L L + +PD + + P ++E+ LS G KSL + L+RL + S
Sbjct: 249 LETLSLKGAKNFKALPDAVWRLPALQELKLSETGLKSLPPVGGG----SALQRLTIED-S 303
Query: 709 NVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLT 768
+E +P ++ LA L L T +++L S + L L+ LSL+ KLE +P S+G +
Sbjct: 304 PLEQLPAGFADLDQLASLSLSNTKLEKLSSGIGQLPALKSLSLQDNPKLERLPKSLGQVE 363
Query: 769 KLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIK 828
+L + + P + L KL + D S L PA + AH+SL+ T ++
Sbjct: 364 ELTLIG----GRIHALPSASGMSSLQKLTV-DNSSLAKLPADFGALGNLAHVSLSNTKLR 418
Query: 829 KLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCR--------------- 873
LP+S+ NL L+TL L++ P L SLP S L L EL +G R
Sbjct: 419 DLPASIGNLFTLKTLSLQDNPKLGSLPASFGQLSGLQELTLNGNRIHELPSMGGASSLQT 478
Query: 874 ------KLTGIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIP 926
L G+P D G L +L LSL +T + LP + +L +L++L + ++L +P
Sbjct: 479 LTVDDTALAGLPADFGALRNLAHLSLSNTQLRELPANTGNLHALKTLSLQGNQQLATLP 537
Score = 101 bits (252), Expect = 1e-20
Identities = 118/396 (29%), Positives = 186/396 (46%), Gaps = 37/396 (9%)
Query: 555 LYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLDFCPEKLVKLHMRESR 614
L+ + S K ++ +PD + LP L+ L E +S P L +L + +S
Sbjct: 246 LFLLETLSLKGAKNFKALPDAVWRLP-ALQELKLSETGLKSLPPVGGGSALQRLTIEDSP 304
Query: 615 LEQL---WGDDQVLQSLCHGLSYIFFGHLIVFSLHQELPNLERLDLSNSWKLVRIPDLSK 671
LEQL + D L SL LS L S +LP L+ L L ++ KL R+P
Sbjct: 305 LEQLPAGFADLDQLASL--SLSNTKLEKLS--SGIGQLPALKSLSLQDNPKLERLP--KS 358
Query: 672 SPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIPEIKETMENLAVLVLKQT 731
++E+ L G + + LP + + L++L + S++ +P + NLA + L T
Sbjct: 359 LGQVEELTLIGGR-IHALP-SASGMSSLQKLTVDN-SSLAKLPADFGALGNLAHVSLSNT 415
Query: 732 AIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDL 791
+++LP+S+ +L L+ LSL+ KL S+P+S G L+ L +L L + P
Sbjct: 416 KLRDLPASIGNLFTLKTLSLQDNPKLGSLPASFGQLSGLQELTLNG-NRIHELPSMGGAS 474
Query: 792 KLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDL 851
L L ++D + L PA + AH+SL+ T +++LP++ NL L+TL L+ L
Sbjct: 475 SLQTLTVDDTA-LAGLPADFGALRNLAHLSLSNTQLRELPANTGNLHALKTLSLQGNQQL 533
Query: 852 ESLPNSICNLKLLSELDCSGC---------------------RKLTGIPNDIGC-LSSLR 889
+LP+S+ L L EL LT IP DIG L
Sbjct: 534 ATLPSSLGYLSGLEELTLKNSSVSELPPMGPGSALKTLTVENSPLTSIPADIGIQCERLT 593
Query: 890 TLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECI 925
LSL +T + LP SI LS+L+ L + +LE +
Sbjct: 594 QLSLSNTQLRALPSSIGKLSNLKGLTLKNNARLELL 629
Score = 93.6 bits (231), Expect = 3e-18
Identities = 92/286 (32%), Positives = 142/286 (49%), Gaps = 34/286 (11%)
Query: 649 LPNLERLDLSNSWKLVRIP-DLSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGC 707
L NL + LSN+ KL +P + +K + L L LP + +L L+ L L+G
Sbjct: 404 LGNLAHVSLSNT-KLRDLPASIGNLFTLKTLSLQDNPKLGSLPASFGQLSGLQELTLNG- 461
Query: 708 SNVENIPE-----------IKET-----------MENLAVLVLKQTAIQELPSSLNHLVG 745
+ + +P + +T + NLA L L T ++ELP++ +L
Sbjct: 462 NRIHELPSMGGASSLQTLTVDDTALAGLPADFGALRNLAHLSLSNTQLRELPANTGNLHA 521
Query: 746 LEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFP-GSIFDLKLTKLDLNDCSRL 804
L+ LSL+ +L ++PSS+G L+ L +L L E P G LK ++ S L
Sbjct: 522 LKTLSLQGNQQLATLPSSLGYLSGLEELTLKNSSVSELPPMGPGSALKTLTVEN---SPL 578
Query: 805 KTFPAILE-PAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNS-ICNLK 862
+ PA + E +SL+ T ++ LPSS+ L L+ L LK LE L S + L+
Sbjct: 579 TSIPADIGIQCERLTQLSLSNTQLRALPSSIGKLSNLKGLTLKNNARLELLSESGVRKLE 638
Query: 863 LLSELDCSGCRKLTGIPNDIGCLSSLRTLSLQD-TG--VVNLPESI 905
+ ++D SGC +LTG+P+ IG L LRTL L TG + +LP S+
Sbjct: 639 SVRKIDLSGCVRLTGLPSSIGKLPKLRTLDLSGCTGLSMASLPRSL 684
Score = 84.7 bits (208), Expect = 1e-15
Identities = 82/289 (28%), Positives = 140/289 (48%), Gaps = 37/289 (12%)
Query: 657 LSNSWKLVRIPD-LSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIPE 715
L+N ++ VR+ D LS++ + KS+ R+ + +LK L V +P+
Sbjct: 175 LANDFEQVRVYDRLSRA-------VDHLKSVLRMSGDSVQLKSLP---------VPELPD 218
Query: 716 IKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDL 775
+ + +L L + LP++L +L LE LSLK +++P ++ L L +L L
Sbjct: 219 VTFEIAHLKNLETVDCDLHALPATLENLFLLETLSLKGAKNFKALPDAVWRLPALQELKL 278
Query: 776 TYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQ 835
+ L++ P L +L + D S L+ PA + A +SL+ T ++KL S +
Sbjct: 279 SE-TGLKSLPPVGGGSALQRLTIED-SPLEQLPAGFADLDQLASLSLSNTKLEKLSSGIG 336
Query: 836 NLVGLQTLCLKECPDLESLPNSICNLK-----------LLSELDCSGCRKLT-------G 877
L L++L L++ P LE LP S+ ++ L S S +KLT
Sbjct: 337 QLPALKSLSLQDNPKLERLPKSLGQVEELTLIGGRIHALPSASGMSSLQKLTVDNSSLAK 396
Query: 878 IPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIP 926
+P D G L +L +SL +T + +LP SI +L +L++L + KL +P
Sbjct: 397 LPADFGALGNLAHVSLSNTKLRDLPASIGNLFTLKTLSLQDNPKLGSLP 445
>DR33_ARATH (Q9FG90) Probable disease resistance protein At5g43740
Length = 862
Score = 93.6 bits (231), Expect = 3e-18
Identities = 175/745 (23%), Positives = 317/745 (42%), Gaps = 105/745 (14%)
Query: 174 IAQDIFKKLNPPFSQGMLGIDKHIAQIQSLLQLDSEAVRIIGICGMGGIGKSTLAEALYH 233
+AQ+I K+ Q +G+DK + S L D + +G+ GMGG+GK+TL E+L +
Sbjct: 136 VAQEIIHKVEKKLIQTTVGLDKLVEMAWSSLMNDE--IGTLGLYGMGGVGKTTLLESLNN 193
Query: 234 K---LGTQFSSRCLIVNAQQKIDRYGIYSLRKKYLSKLLGE-----DIQSNGLNYAIERV 285
K L ++F +V ++ + ++ + L +L + + +S + +
Sbjct: 194 KFVELESEFDVVIWVVVSKD----FQFEGIQDQILGRLRSDKEWERETESKKASLIYNNL 249
Query: 286 KRAKVLLILDDLKISIPILELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSY 345
+R K +L+LDDL + + ++ GS+I+ T+R V K+ +AD+ +V +S
Sbjct: 250 ERKKFVLLLDDLWSEVDMTKIGVPPPTRENGSKIVFTTRSTEVCKHMKADKQIKVACLSP 309
Query: 346 QDSLELFRLNAFKEDYPLEGYAN---LIVEVLKYAKGVPLALQVLGSLLYDKER-EVWES 401
++ ELFRL D L + + L V G+PLAL V+G + KE + W
Sbjct: 310 DEAWELFRLTV--GDIILRSHQDIPALARIVAAKCHGLPLALNVIGKAMSCKETIQEWSH 367
Query: 402 ELKKLK----ELPNVD--IFDVLKLSYDGL-DDEPKDIFLDITCFYAGDFV--DKVVELL 452
+ L E P ++ I +LK SYD L + E K FL + F + +K +E
Sbjct: 368 AINVLNSAGHEFPGMEERILPILKFSYDSLKNGEIKLCFLYCSLFPEDSEIPKEKWIEYW 427
Query: 453 DSCGF---------SADIGMDV--LKDRGLISI---LEDRIAVHDLVLEM----GREIVR 494
GF + G D+ L R + I L D + +HD++ EM + +
Sbjct: 428 ICEGFINPNRYEDGGTNHGYDIIGLLVRAHLLIECELTDNVKMHDVIREMALWINSDFGK 487
Query: 495 QQ---CVSDPGKHSHLWNHKEIYHVVR--------------KNKGTDAIQCIFLDMCQIE 537
QQ CV G H + + + +VR ++K + + LD +
Sbjct: 488 QQETICVKS-GAHVRMIPNDINWEIVRTMSFTCTQIKKISCRSKCPNLSTLLILDNRLL- 545
Query: 538 KVQLHPEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFP 597
V++ F+ MP + +L + +P+ + +L L+ L+ +S P
Sbjct: 546 -VKISNRFFRFMPKLVVL------DLSANLDLIKLPEEISNL-GSLQYLNISLTGIKSLP 597
Query: 598 LDFCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVF---SLHQELPNLER 654
+ +KL KL + G + + L + F + V+ L +EL +LE
Sbjct: 598 VGL--KKLRKLIYLNLEFTGVHGSLVGIAATLPNLQVLKFFYSCVYVDDILMKELQDLEH 655
Query: 655 LDL--SNSWKLVRIPDLSKSPNIKEIILSGC---KSLTRLPINLFKLKFLERLNLSGCSN 709
L + +N + + + + I S C S R+ ++ L L++L + C
Sbjct: 656 LKILTANVKDVTILERIQGDDRLASSIRSLCLEDMSTPRVILSTIALGGLQQLAILMC-- 713
Query: 710 VENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTL-- 767
NI EI+ E+ L T I LPS+ + G ++LS + ++LE L
Sbjct: 714 --NISEIRIDWESKERRELSPTEI--LPSTGS--PGFKQLSTVYINQLEGQRDLSWLLYA 767
Query: 768 TKLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFAHISLAETA- 826
L KL++ + +E + +TKL + I+ P + ++L + A
Sbjct: 768 QNLKKLEVCWSPQIEEIINKEKGMNITKLHRD----------IVVPFGNLEDLALRQMAD 817
Query: 827 IKKLPSSLQNLVGLQTLCLKECPDL 851
+ ++ + + L L+ + +CP L
Sbjct: 818 LTEICWNYRTLPNLRKSYINDCPKL 842
Score = 33.5 bits (75), Expect = 3.4
Identities = 49/218 (22%), Positives = 85/218 (38%), Gaps = 27/218 (12%)
Query: 715 EIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLD 774
EI TM + K + + P+ L+ L +K ++ + KL LD
Sbjct: 510 EIVRTMSFTCTQIKKISCRSKCPNLSTLLILDNRLLVKISNRFFRF------MPKLVVLD 563
Query: 775 LTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKK----L 830
L+ L P I +L + + +K+ P L+ +++L T + +
Sbjct: 564 LSANLDLIKLPEEISNLGSLQYLNISLTGIKSLPVGLKKLRKLIYLNLEFTGVHGSLVGI 623
Query: 831 PSSLQNL---------VGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPND 881
++L NL V + + +KE DLE L N+K ++ L+ I D
Sbjct: 624 AATLPNLQVLKFFYSCVYVDDILMKELQDLEHLKILTANVKDVTILE--------RIQGD 675
Query: 882 IGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYC 919
SS+R+L L+D + S L L+ L + C
Sbjct: 676 DRLASSIRSLCLEDMSTPRVILSTIALGGLQQLAILMC 713
>DR27_ARATH (Q9T048) Putative disease resistance protein At4g27190
Length = 985
Score = 93.6 bits (231), Expect = 3e-18
Identities = 189/783 (24%), Positives = 328/783 (41%), Gaps = 152/783 (19%)
Query: 206 LDSEAVRIIGICGMGGIGKSTLAEALYHKL----GTQFSSRCLIVNAQQKIDRYGIYSLR 261
L SE + IG+ GMGG+GK+TL L +KL TQ + V ++ D +
Sbjct: 159 LTSEKAQKIGVWGMGGVGKTTLVRTLNNKLREEGATQPFGLVIFVIVSKEFDPREV---- 214
Query: 262 KKYLSKLLGEDIQSNGLNYAIER------VKRAKVLLILDDLKISIPILELIG-GHGNFG 314
+K +++ L D Q + R +K K LLILDD+ I L+L+G
Sbjct: 215 QKQIAERLDIDTQMEESEEKLARRIYVGLMKERKFLLILDDVWKPID-LDLLGIPRTEEN 273
Query: 315 QGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPLEGYANLIVE-V 373
+GS++I+TSR V ++ + D V + +D+ ELF NA D + I + V
Sbjct: 274 KGSKVILTSRFLEVCRSMKTDLDVRVDCLLEEDAWELFCKNA--GDVVRSDHVRKIAKAV 331
Query: 374 LKYAKGVPLALQVLGSLLYDKER-EVWESELKKL-KELPNV-----DIFDVLKLSYDGLD 426
+ G+PLA+ +G+ + K+ ++W L KL K +P + IF LKLSYD L+
Sbjct: 332 SQECGGLPLAIITVGTAMRGKKNVKLWNHVLSKLSKSVPWIKSIEEKIFQPLKLSYDFLE 391
Query: 427 DEPKDIFLDITCFYAGDF---VDKVVELLDSCGFSADIG------------MDVLKDRGL 471
D+ K FL + + D+ V +VV + GF ++G ++ LKD L
Sbjct: 392 DKAKFCFL-LCALFPEDYSIEVTEVVRYWMAEGFMEELGSQEDSMNEGITTVESLKDYCL 450
Query: 472 I--SILEDRIAVHDLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCI 529
+ D + +HD+V + I+ S HS + + + +R++K +++ +
Sbjct: 451 LEDGDRRDTVKMHDVVRDFAIWIMSS---SQDDSHSLVMSGTGLQD-IRQDKLAPSLRRV 506
Query: 530 FLDMCQIEKVQLHPEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWD 589
L ++E + E F ++ +L Q N LL+ +P G
Sbjct: 507 SLMNNKLESLPDLVEEFCVKTSVLLL-----------QGNF----LLKEVPIGF------ 545
Query: 590 EFPQRSFPLDFCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQEL 649
++FP L L++ +R++ S+ L +FSLH
Sbjct: 546 ---LQAFP------TLRILNLSGTRIK----------------SFPSCSLLRLFSLHS-- 578
Query: 650 PNLERLDLSNSWKLVRIPDLSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSN 709
L L + +KLV++P L ++ + L G L P L +LK L+LS +
Sbjct: 579 -----LFLRDCFKLVKLPSLETLAKLELLDLCGTHIL-EFPRGLEELKRFRHLDLSRTLH 632
Query: 710 VENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTK 769
+E+IP A +V + ++++ L + +H + + K ++ IG L +
Sbjct: 633 LESIP---------ARVVSRLSSLETLDMTSSHYRWSVQGETQ---KGQATVEEIGCLQR 680
Query: 770 LCKLDL-------------TYCESLETFP---GSIFDL-------KLTKLDLNDCSRLKT 806
L L + T+ + L+ F GS + L +LT LN
Sbjct: 681 LQVLSIRLHSSPFLLNKRNTWIKRLKKFQLVVGSRYILRTRHDKRRLTISHLNVSQVSIG 740
Query: 807 FPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSE 866
+ + + H E +KKL S + L++L ++ N I N E
Sbjct: 741 WLLAYTTSLALNHCQGIEAMMKKLVSDNKGFKNLKSL---------TIENVIINTNSWVE 791
Query: 867 LDCSGCRKLTGIPNDIGCLSSLRTLSLQDTGVVNLPESIAH----LSSLESLDVSYCRKL 922
+ + K + D+ L +L L L+ + E H L +L+ ++++ CRKL
Sbjct: 792 MVSTNTSKQSSDILDL--LPNLEELHLRRVDLETFSELQTHLGLKLETLKIIEITMCRKL 849
Query: 923 ECI 925
+
Sbjct: 850 RTL 852
>DR32_ARATH (Q9FG91) Probable disease resistance protein At5g43730
Length = 848
Score = 91.3 bits (225), Expect = 1e-17
Identities = 116/425 (27%), Positives = 196/425 (45%), Gaps = 66/425 (15%)
Query: 174 IAQDIFKKLNPPFSQGMLGIDKHIAQIQSLLQLDSEAVRIIGICGMGGIGKSTLAEALYH 233
+AQ I K Q +G+D + L +D E +R +G+ GMGGIGK+TL E+L +
Sbjct: 137 VAQKIIPKAEKKHIQTTVGLDTMVGIAWESL-IDDE-IRTLGLYGMGGIGKTTLLESLNN 194
Query: 234 K---LGTQFSSRCLIVNAQQKIDRYGIYSLRKKYLSKLLGE-----DIQSNGLNYAIERV 285
K L ++F +V ++ + + ++ + L +L + + +S + +
Sbjct: 195 KFVELESEFDVVIWVVVSKD----FQLEGIQDQILGRLRPDKEWERETESKKASLINNNL 250
Query: 286 KRAKVLLILDDLKISIPILELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSY 345
KR K +L+LDDL + ++++ + GS+I+ T+R + V K+ +AD+ +V +S
Sbjct: 251 KRKKFVLLLDDLWSEVDLIKIGVPPPSRENGSKIVFTTRSKEVCKHMKADKQIKVDCLSP 310
Query: 346 QDSLELFRLNAFKEDYPLEGYAN---LIVEVLKYAKGVPLALQVLGSLLYDKER-EVWES 401
++ ELFRL D L + + L V G+PLAL V+G + KE + W
Sbjct: 311 DEAWELFRLTV--GDIILRSHQDIPALARIVAAKCHGLPLALNVIGKAMVCKETVQEWRH 368
Query: 402 ELKKLK----ELPNVD--IFDVLKLSYDGL-DDEPKDIFLDITCFYAGDF---VDKVVEL 451
+ L + P ++ I +LK SYD L + E K FL + F DF DK++E
Sbjct: 369 AINVLNSPGHKFPGMEERILPILKFSYDSLKNGEIKLCFLYCSLF-PEDFEIEKDKLIEY 427
Query: 452 LDSCGF---------SADIGMDV--LKDRGLISI---LEDRIAVHDLVLEM----GREIV 493
G+ + G D+ L R + I L D++ +HD++ EM +
Sbjct: 428 WICEGYINPNRYEDGGTNQGYDIIGLLVRAHLLIECELTDKVKMHDVIREMALWINSDFG 487
Query: 494 RQQ---CVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDMCQIEKVQLHPEIFKSMP 550
QQ CV G H L + + +VR+ + L Q+EK+ P + P
Sbjct: 488 NQQETICVKS-GAHVRLIPNDISWEIVRQ---------MSLISTQVEKIACSP----NCP 533
Query: 551 NIRML 555
N+ L
Sbjct: 534 NLSTL 538
Score = 44.7 bits (104), Expect = 0.001
Identities = 31/83 (37%), Positives = 49/83 (58%), Gaps = 6/83 (7%)
Query: 649 LPNLERLDLSNSWKLVRIP-DLSKSPNIK--EIILSGCKSLTRLPINLFKLKFLERLNLS 705
+P L LDLS +W L+ +P ++S +++ + L+G KS LP+ L KL+ L LNL
Sbjct: 556 MPKLVVLDLSTNWSLIELPEEISNLGSLQYLNLSLTGIKS---LPVGLKKLRKLIYLNLE 612
Query: 706 GCSNVENIPEIKETMENLAVLVL 728
+ +E++ I T+ NL VL L
Sbjct: 613 FTNVLESLVGIATTLPNLQVLKL 635
Score = 41.6 bits (96), Expect = 0.013
Identities = 55/207 (26%), Positives = 84/207 (40%), Gaps = 20/207 (9%)
Query: 660 SWKLVRIPDLSKSPNIKEIILSGCKSLTRLPINLFKLK--------FLERLNLSGCSNVE 711
SW++VR L + K C +L+ L + KL F+ +L + S
Sbjct: 509 SWEIVRQMSLISTQVEKIACSPNCPNLSTLLLPYNKLVDISVGFFLFMPKLVVLDLSTNW 568
Query: 712 NIPEIKETMENLAVLV---LKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLT 768
++ E+ E + NL L L T I+ LP L L L L+L+F + LES+ TL
Sbjct: 569 SLIELPEEISNLGSLQYLNLSLTGIKSLPVGLKKLRKLIYLNLEFTNVLESLVGIATTLP 628
Query: 769 KLCKLDLTYCESLETFPGSIFD-------LKLTKLDLNDCSRLKTFPAILEPAESFAHIS 821
L L L Y SL I + LK+ + D L+ + A S +
Sbjct: 629 NLQVLKLFY--SLFCVDDIIMEELQRLKHLKILTATIEDAMILERVQGVDRLASSIRGLC 686
Query: 822 LAETAIKKLPSSLQNLVGLQTLCLKEC 848
L + ++ + L GLQ L + C
Sbjct: 687 LRNMSAPRVILNSVALGGLQQLGIVSC 713
Score = 37.4 bits (85), Expect = 0.24
Identities = 32/111 (28%), Positives = 50/111 (44%), Gaps = 1/111 (0%)
Query: 815 ESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRK 874
E +SL T ++K+ S N L TL L ++ + L LD S
Sbjct: 511 EIVRQMSLISTQVEKIACS-PNCPNLSTLLLPYNKLVDISVGFFLFMPKLVVLDLSTNWS 569
Query: 875 LTGIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECI 925
L +P +I L SL+ L+L TG+ +LP + L L L++ + LE +
Sbjct: 570 LIELPEEISNLGSLQYLNLSLTGIKSLPVGLKKLRKLIYLNLEFTNVLESL 620
>SHO2_HUMAN (Q9UQ13) Leucine-rich repeat protein SHOC-2 (Ras-binding
protein Sur-8)
Length = 582
Score = 89.0 bits (219), Expect = 7e-17
Identities = 74/221 (33%), Positives = 118/221 (52%), Gaps = 8/221 (3%)
Query: 708 SNVENIPEIKETME-NLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGT 766
SN E I E+ + E N L L + +I LPSS+ L L EL L + +KL+S+P+ +G
Sbjct: 86 SNAEVIKELNKCREENSMRLDLSKRSIHILPSSIKELTQLTELYL-YSNKLQSLPAEVGC 144
Query: 767 LTKLCKLDLTYCESLETFPGSIFDLK-LTKLDLNDCSRLKTFPAILEPAESFAHISLAET 825
L L L L+ SL + P S+ +LK L LDL ++L+ P+++ +S + L
Sbjct: 145 LVNLMTLALSE-NSLTSLPDSLDNLKKLRMLDLRH-NKLREIPSVVYRLDSLTTLYLRFN 202
Query: 826 AIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCL 885
I + ++NL L L ++E ++ LP I L L LD + +L +P +IG
Sbjct: 203 RITTVEKDIKNLSKLSMLSIRE-NKIKQLPAEIGELCNLITLDVAH-NQLEHLPKEIGNC 260
Query: 886 SSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIP 926
+ + L LQ +++LP++I +LSSL L + Y +L IP
Sbjct: 261 TQITNLDLQHNELLDLPDTIGNLSSLSRLGLRY-NRLSAIP 300
Score = 80.1 bits (196), Expect = 3e-14
Identities = 94/336 (27%), Positives = 156/336 (45%), Gaps = 51/336 (15%)
Query: 604 KLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQELPN---LERLDLSNS 660
KL L +RE++++QL + + LC+ L + H + L +E+ N + LDL ++
Sbjct: 216 KLSMLSIRENKIKQLPAE---IGELCN-LITLDVAHNQLEHLPKEIGNCTQITNLDLQHN 271
Query: 661 WKLVRIPDLSKSPNIKEIILSGCK--SLTRLPINLFKLKFLERLNLSGCSNVENIPEIKE 718
+L+ +PD N+ + G + L+ +P +L K LE LNL +N+ +PE
Sbjct: 272 -ELLDLPDTIG--NLSSLSRLGLRYNRLSAIPRSLAKCSALEELNLEN-NNISTLPE--- 324
Query: 719 TMENLAVLVLKQTAIQELPSSLNHLVGLEELSL-KFCSKLESI--PSSIGTLTKLCKLDL 775
S L+ LV L L+L + C +L + PS T+ L ++
Sbjct: 325 -------------------SLLSSLVKLNSLTLARNCFQLYPVGGPSQFSTIYSL---NM 362
Query: 776 TYCESLETFPGSIFDLK--LTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSS 833
+ + P IF L+KL++ D ++L + P S ++LA + K+P
Sbjct: 363 EH-NRINKIPFGIFSRAKVLSKLNMKD-NQLTSLPLDFGTWTSMVELNLATNQLTKIPED 420
Query: 834 LQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRTLSL 893
+ LV L+ L L L+ LP+ + NL+ L ELD KL +PN+I L L+ L L
Sbjct: 421 VSGLVSLEVLILSNNL-LKKLPHGLGNLRKLRELDLEE-NKLESLPNEIAYLKDLQKLVL 478
Query: 894 QDTGVVNLPESIAHLSSLESLDVSYCRKLECIPHLP 929
+ + LP I HL++L L + + HLP
Sbjct: 479 TNNQLTTLPRGIGHLTNLTHLGLGE----NLLTHLP 510
Score = 71.2 bits (173), Expect = 1e-11
Identities = 69/242 (28%), Positives = 116/242 (47%), Gaps = 9/242 (3%)
Query: 644 SLHQELPNLERLDLS-NSWKLVRIPDLSKSPNIKEIILSGCKSLTRLPINLF-KLKFLER 701
SL L L L L+ N ++L + S+ I + + + + ++P +F + K L +
Sbjct: 325 SLLSSLVKLNSLTLARNCFQLYPVGGPSQFSTIYSLNMEHNR-INKIPFGIFSRAKVLSK 383
Query: 702 LNLSGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIP 761
LN+ + + ++P T ++ L L + ++P ++ LV LE L L + L+ +P
Sbjct: 384 LNMKD-NQLTSLPLDFGTWTSMVELNLATNQLTKIPEDVSGLVSLEVLILSN-NLLKKLP 441
Query: 762 SSIGTLTKLCKLDLTYCESLETFPGSIFDLK-LTKLDLNDCSRLKTFPAILEPAESFAHI 820
+G L KL +LDL LE+ P I LK L KL L + ++L T P + + H+
Sbjct: 442 HGLGNLRKLRELDLEE-NKLESLPNEIAYLKDLQKLVLTN-NQLTTLPRGIGHLTNLTHL 499
Query: 821 SLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPN 880
L E + LP + L L+ L L + P+L SLP + LS + C L+ +P
Sbjct: 500 GLGENLLTHLPEEIGTLENLEELYLNDNPNLHSLPFELALCSKLSIMSIENC-PLSHLPP 558
Query: 881 DI 882
I
Sbjct: 559 QI 560
>SHO2_MOUSE (O88520) Leucine-rich repeat protein SHOC-2 (Ras-binding
protein Sur-8)
Length = 582
Score = 87.0 bits (214), Expect = 3e-16
Identities = 73/221 (33%), Positives = 117/221 (52%), Gaps = 8/221 (3%)
Query: 708 SNVENIPEIKETME-NLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGT 766
SN E I E+ + E N L L + +I LP S+ L L EL L + +KL+S+P+ +G
Sbjct: 86 SNAEVIKELNKCREENSMRLDLSKRSIHILPPSVKELTQLTELYL-YSNKLQSLPAEVGC 144
Query: 767 LTKLCKLDLTYCESLETFPGSIFDLK-LTKLDLNDCSRLKTFPAILEPAESFAHISLAET 825
L L L L+ SL + P S+ +LK L LDL ++L+ P+++ +S + L
Sbjct: 145 LVNLMTLALSE-NSLTSLPDSLDNLKKLRMLDLRH-NKLREIPSVVYRLDSLTTLYLRFN 202
Query: 826 AIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCL 885
I + ++NL L L ++E ++ LP I L L LD + +L +P +IG
Sbjct: 203 RITTVEKDIKNLPKLSMLSIRE-NKIKQLPAEIGELCNLITLDVAH-NQLEHLPKEIGNC 260
Query: 886 SSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIP 926
+ + L LQ +++LP++I +LSSL L + Y +L IP
Sbjct: 261 TQITNLDLQHNDLLDLPDTIGNLSSLNRLGLRY-NRLSAIP 300
Score = 79.7 bits (195), Expect = 4e-14
Identities = 104/395 (26%), Positives = 177/395 (44%), Gaps = 54/395 (13%)
Query: 543 PEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLDFCP 602
P+ ++ +RML +H+ ++ S V D L +L + E ++ P
Sbjct: 162 PDSLDNLKKLRMLDL-RHNKLREIPSVVYRLDSLTTLYLRFNRITTVEKDIKNLP----- 215
Query: 603 EKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQELPN---LERLDLSN 659
KL L +RE++++QL + + LC+ L + H + L +E+ N + LDL +
Sbjct: 216 -KLSMLSIRENKIKQLPAE---IGELCN-LITLDVAHNQLEHLPKEIGNCTQITNLDLQH 270
Query: 660 SWKLVRIPDLSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIPEIKET 719
+ L+ +PD + + + L+ +P +L K LE LNL +N+ +PE
Sbjct: 271 N-DLLDLPDTIGNLSSLNRLGLRYNRLSAIPRSLAKCSALEELNLEN-NNISTLPE---- 324
Query: 720 MENLAVLVLKQTAIQELPSSLNHLVGLEELSL-KFCSKLESI--PSSIGTLTKLCKLDLT 776
S L+ LV L L+L + C +L + PS T+ L ++
Sbjct: 325 ------------------SLLSSLVKLNSLTLARNCFQLYPVGGPSQFSTIYSL---NME 363
Query: 777 YCESLETFPGSIFDLK--LTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSL 834
+ + P IF L+KL++ D ++L + P S ++LA + K+P +
Sbjct: 364 H-NRINKIPFGIFSRAKVLSKLNMKD-NQLTSLPLDFGTWTSMVELNLATNQLTKIPEDV 421
Query: 835 QNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRTLSLQ 894
LV L+ L L L+ LP+ + NL+ L ELD KL +PN+I L L+ L L
Sbjct: 422 SGLVSLEVLILSNNL-LKKLPHGLGNLRKLRELDLEE-NKLESLPNEIAYLKDLQKLVLT 479
Query: 895 DTGVVNLPESIAHLSSLESLDVSYCRKLECIPHLP 929
+ + LP I HL++L L + + HLP
Sbjct: 480 NNQLSTLPRGIGHLTNLTHLGLGE----NLLTHLP 510
Score = 71.6 bits (174), Expect = 1e-11
Identities = 69/242 (28%), Positives = 116/242 (47%), Gaps = 9/242 (3%)
Query: 644 SLHQELPNLERLDLS-NSWKLVRIPDLSKSPNIKEIILSGCKSLTRLPINLF-KLKFLER 701
SL L L L L+ N ++L + S+ I + + + + ++P +F + K L +
Sbjct: 325 SLLSSLVKLNSLTLARNCFQLYPVGGPSQFSTIYSLNMEHNR-INKIPFGIFSRAKVLSK 383
Query: 702 LNLSGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIP 761
LN+ + + ++P T ++ L L + ++P ++ LV LE L L + L+ +P
Sbjct: 384 LNMKD-NQLTSLPLDFGTWTSMVELNLATNQLTKIPEDVSGLVSLEVLILSN-NLLKKLP 441
Query: 762 SSIGTLTKLCKLDLTYCESLETFPGSIFDLK-LTKLDLNDCSRLKTFPAILEPAESFAHI 820
+G L KL +LDL LE+ P I LK L KL L + ++L T P + + H+
Sbjct: 442 HGLGNLRKLRELDLEE-NKLESLPNEIAYLKDLQKLVLTN-NQLSTLPRGIGHLTNLTHL 499
Query: 821 SLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPN 880
L E + LP + L L+ L L + P+L SLP + LS + C L+ +P
Sbjct: 500 GLGENLLTHLPEEIGTLENLEELYLNDNPNLHSLPFELALCSKLSIMSIENC-PLSHLPP 558
Query: 881 DI 882
I
Sbjct: 559 QI 560
>LRR1_HUMAN (Q9BTT6) Leucine-rich repeat-containing protein 1 (LAP
and no PDZ protein) (LANO adapter protein)
Length = 524
Score = 86.7 bits (213), Expect = 3e-16
Identities = 108/380 (28%), Positives = 176/380 (45%), Gaps = 55/380 (14%)
Query: 587 HWDEFPQRSFPLDFCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLH 646
H + +R L + PE++ + LE+L D L+ L FF + + L
Sbjct: 13 HVESIDKRHCSLVYVPEEIYRY---ARSLEELLLDANQLRELPEQ----FFQLVKLRKLG 65
Query: 647 QELPNLERL--DLSNSWKLVRIPDLSKS--PNIKEIILSGCKSL----------TRLPIN 692
++RL +++N +LV + D+S++ P I E I S CK+L TRLP +
Sbjct: 66 LSDNEIQRLPPEIANFMQLVEL-DVSRNEIPEIPESI-SFCKALQVADFSGNPLTRLPES 123
Query: 693 LFKLKFLERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLK 752
+L+ L L+++ S ++++PE + NLA L L++ + LP SL L LEEL L
Sbjct: 124 FPELQNLTCLSVNDIS-LQSLPENIGNLYNLASLELRENLLTYLPDSLTQLRRLEELDLG 182
Query: 753 FCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLK-LTKLDLNDCSRLKTFPAIL 811
+++ ++P SIG L L L L L P I +LK L LD+++ +RL+ P +
Sbjct: 183 -NNEIYNLPESIGALLHLKDLWLD-GNQLSELPQEIGNLKNLLCLDVSE-NRLERLPEEI 239
Query: 812 EPAESFAHISLAETAIKKLPSSLQNLVGLQTL------------CLKECPDLE------- 852
S + +++ ++ +P + L L L + EC L
Sbjct: 240 SGLTSLTDLVISQNLLETIPDGIGKLKKLSILKVDQNRLTQLPEAVGECESLTELVLTEN 299
Query: 853 ---SLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLS 909
+LP SI LK LS L+ KL +P +IG SL ++D + +P ++ +
Sbjct: 300 QLLTLPKSIGKLKKLSNLNADR-NKLVSLPKEIGGCCSLTVFCVRDNRLTRIPAEVSQAT 358
Query: 910 SLESLDVSYCRKLECIPHLP 929
L LDV+ R L HLP
Sbjct: 359 ELHVLDVAGNRLL----HLP 374
Score = 79.3 bits (194), Expect = 5e-14
Identities = 67/230 (29%), Positives = 114/230 (49%), Gaps = 5/230 (2%)
Query: 697 KFLERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSK 756
+ +E ++ CS V EI +L L+L ++ELP LV L +L L ++
Sbjct: 12 RHVESIDKRHCSLVYVPEEIYRYARSLEELLLDANQLRELPEQFFQLVKLRKLGLS-DNE 70
Query: 757 LESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAES 816
++ +P I +L +LD++ E + P SI K ++ + L P ++
Sbjct: 71 IQRLPPEIANFMQLVELDVSRNE-IPEIPESISFCKALQVADFSGNPLTRLPESFPELQN 129
Query: 817 FAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLT 876
+S+ + +++ LP ++ NL L +L L+E L LP+S+ L+ L ELD G ++
Sbjct: 130 LTCLSVNDISLQSLPENIGNLYNLASLELRE-NLLTYLPDSLTQLRRLEELDL-GNNEIY 187
Query: 877 GIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIP 926
+P IG L L+ L L + LP+ I +L +L LDVS +LE +P
Sbjct: 188 NLPESIGALLHLKDLWLDGNQLSELPQEIGNLKNLLCLDVSE-NRLERLP 236
Score = 77.8 bits (190), Expect = 2e-13
Identities = 102/377 (27%), Positives = 159/377 (42%), Gaps = 35/377 (9%)
Query: 558 HKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLDFCP-EKLVKLHMRESRLE 616
H S K+ S V VP+ + L+ L D R P F KL KL + ++ ++
Sbjct: 13 HVESIDKRHCSLVYVPEEIYRYARSLEELLLDANQLRELPEQFFQLVKLRKLGLSDNEIQ 72
Query: 617 QLWGDD----QVLQ---------------SLCHGLSYIFFGHLIVFSLHQELPNLERLDL 657
+L + Q+++ S C L F + L + P L+ L
Sbjct: 73 RLPPEIANFMQLVELDVSRNEIPEIPESISFCKALQVADFSGNPLTRLPESFPELQNLTC 132
Query: 658 S--NSWKLVRIPD-LSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIP 714
N L +P+ + N+ + L LT LP +L +L+ LE L+L G + + N+P
Sbjct: 133 LSVNDISLQSLPENIGNLYNLASLELRE-NLLTYLPDSLTQLRRLEELDL-GNNEIYNLP 190
Query: 715 EIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLD 774
E + +L L L + ELP + +L L L + ++LE +P I LT L L
Sbjct: 191 ESIGALLHLKDLWLDGNQLSELPQEIGNLKNLLCLDVSE-NRLERLPEEISGLTSLTDLV 249
Query: 775 LTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSL 834
++ LET P I LK + D +RL P + ES + L E + LP S+
Sbjct: 250 ISQ-NLLETIPDGIGKLKKLSILKVDQNRLTQLPEAVGECESLTELVLTENQLLTLPKSI 308
Query: 835 QNLVGLQTLCLKECPDLESLPNSI---CNLKLLSELDCSGCRKLTGIPNDIGCLSSLRTL 891
L L L L SLP I C+L + C +LT IP ++ + L L
Sbjct: 309 GKLKKLSNLNADR-NKLVSLPKEIGGCCSLTVF----CVRDNRLTRIPAEVSQATELHVL 363
Query: 892 SLQDTGVVNLPESIAHL 908
+ +++LP S+ L
Sbjct: 364 DVAGNRLLHLPLSLTAL 380
>LAP4_DROME (Q7KRY7) LAP4 protein (Scribble protein) (Smell-impaired
protein)
Length = 1851
Score = 86.3 bits (212), Expect = 4e-16
Identities = 88/294 (29%), Positives = 137/294 (45%), Gaps = 29/294 (9%)
Query: 647 QELPNLERLDLSNS-----------WKLVRIPDLSKSP---------NIKEIILSGCK-- 684
Q NL LD+S + + +++ D S +P +K + + G
Sbjct: 80 QNFENLVELDVSRNDIPDIPDDIKHLQSLQVADFSSNPIPKLPSGFSQLKNLTVLGLNDM 139
Query: 685 SLTRLPINLFKLKFLERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLV 744
SLT LP + L LE L L + ++++PE + L L L I++LP L +L
Sbjct: 140 SLTTLPADFGSLTQLESLELRE-NLLKHLPETISQLTKLKRLDLGDNEIEDLPPYLGYLP 198
Query: 745 GLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDL-KLTKLDLNDCSR 803
GL EL L ++L+ +P +G LTKL LD++ LE P I L LT LDL +
Sbjct: 199 GLHELWLDH-NQLQRLPPELGLLTKLTYLDVSE-NRLEELPNEISGLVSLTDLDLAQ-NL 255
Query: 804 LKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKL 863
L+ P + + L + +++L +L N +Q L L E L LP SI +
Sbjct: 256 LEALPDGIAKLSRLTILKLDQNRLQRLNDTLGNCENMQELILTE-NFLSELPASIGQMTK 314
Query: 864 LSELDCSGCRKLTGIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVS 917
L+ L+ L +P +IG ++L LSL+D + LP + + + L LDVS
Sbjct: 315 LNNLNVDR-NALEYLPLEIGQCANLGVLSLRDNKLKKLPPELGNCTVLHVLDVS 367
Score = 80.5 bits (197), Expect = 2e-14
Identities = 88/316 (27%), Positives = 135/316 (41%), Gaps = 19/316 (6%)
Query: 663 LVRIPD--LSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIPEIKETM 720
L ++P+ L S ++E+ L + LP N F+L L +L LS + + +P +
Sbjct: 25 LPQVPEEILRYSRTLEELFLDA-NHIRDLPKNFFRLHRLRKLGLSD-NEIGRLPPDIQNF 82
Query: 721 ENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCES 780
ENL L + + I ++P + HL L+ + + +PS L L L L S
Sbjct: 83 ENLVELDVSRNDIPDIPDDIKHLQSLQVADFS-SNPIPKLPSGFSQLKNLTVLGLNDM-S 140
Query: 781 LETFPGSIFDL-KLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVG 839
L T P L +L L+L + + LK P + + L + I+ LP L L G
Sbjct: 141 LTTLPADFGSLTQLESLELRE-NLLKHLPETISQLTKLKRLDLGDNEIEDLPPYLGYLPG 199
Query: 840 LQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRTLSLQDTGVV 899
L L L L+ LP + L L+ LD S R L +PN+I L SL L L +
Sbjct: 200 LHELWLDH-NQLQRLPPELGLLTKLTYLDVSENR-LEELPNEISGLVSLTDLDLAQNLLE 257
Query: 900 NLPESIAHLSSLESLDVSYCRKLECIPHLPPFMKQLLAFHCPSIRSVMSNSRFLSDSKEG 959
LP+ IA LS L L + R L +C +++ ++ FLS+
Sbjct: 258 ALPDGIAKLSRLTILKLDQNRLQRLNDTLG---------NCENMQELILTENFLSELPAS 308
Query: 960 TFELYFTNNEKQDPGA 975
++ NN D A
Sbjct: 309 IGQMTKLNNLNVDRNA 324
Score = 62.0 bits (149), Expect = 9e-09
Identities = 51/195 (26%), Positives = 92/195 (47%), Gaps = 9/195 (4%)
Query: 746 LEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDL-KLTKLDLNDCSRL 804
+E + + CS L +P I ++ + + P + F L +L KL L+D + +
Sbjct: 15 VEFVDKRHCS-LPQVPEEILRYSRTLEELFLDANHIRDLPKNFFRLHRLRKLGLSD-NEI 72
Query: 805 KTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLL 864
P ++ E+ + ++ I +P +++L LQ P + LP+ LK L
Sbjct: 73 GRLPPDIQNFENLVELDVSRNDIPDIPDDIKHLQSLQVADFSSNP-IPKLPSGFSQLKNL 131
Query: 865 SELDCSGCRKLTGIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLEC 924
+ L + LT +P D G L+ L +L L++ + +LPE+I+ L+ L+ LD+
Sbjct: 132 TVLGLNDM-SLTTLPADFGSLTQLESLELRENLLKHLPETISQLTKLKRLDLGDNE---- 186
Query: 925 IPHLPPFMKQLLAFH 939
I LPP++ L H
Sbjct: 187 IEDLPPYLGYLPGLH 201
>LRR1_MOUSE (Q80VQ1) Leucine-rich repeat-containing protein 1
Length = 524
Score = 84.7 bits (208), Expect = 1e-15
Identities = 107/380 (28%), Positives = 177/380 (46%), Gaps = 55/380 (14%)
Query: 587 HWDEFPQRSFPLDFCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLH 646
H + +R L + PE++ + LE+L D L+ L FF + + L
Sbjct: 13 HVEAIDKRHCSLVYVPEEIYRY---ARSLEELLLDANQLRELPEQ----FFQLVKLRKLG 65
Query: 647 QELPNLERL--DLSNSWKLVRIPDLSKS--PNIKEIILSGCKSL----------TRLPIN 692
++RL +++N +LV + D+S++ P I E I + CK+L TRLP +
Sbjct: 66 LSDNEIQRLPPEIANFMQLVEL-DVSRNDIPEIPESI-AFCKALQVADFSGNPLTRLPES 123
Query: 693 LFKLKFLERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLK 752
+L+ L L+++ S ++++PE + NLA L L++ + LP SL L LEEL L
Sbjct: 124 FPELQNLTCLSVNDIS-LQSLPENIGNLYNLASLELRENLLTYLPDSLTQLRRLEELDLG 182
Query: 753 FCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLK-LTKLDLNDCSRLKTFPAIL 811
+++ ++P SIG L L L L L P I +LK L LD+++ +RL+ P +
Sbjct: 183 N-NEIYNLPESIGALLHLKDLWLDG-NQLSELPQEIGNLKNLLCLDVSE-NRLERLPEEI 239
Query: 812 EPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKE------------CPDLE------- 852
S ++ +++ ++ +P + L L L L + C +L
Sbjct: 240 SGLTSLTYLVISQNLLETIPEGIGKLKKLSILKLDQNRLTQLPEAIGDCENLTELVLTEN 299
Query: 853 ---SLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLS 909
+LP SI LK LS L+ KL +P +IG SL ++D + LP ++
Sbjct: 300 RLLTLPKSIGKLKKLSNLNADR-NKLVSLPKEIGGCCSLTMFCIRDNRLTRLPAEVSQAV 358
Query: 910 SLESLDVSYCRKLECIPHLP 929
L LDV+ R + HLP
Sbjct: 359 ELHVLDVAGNR----LHHLP 374
Score = 78.6 bits (192), Expect = 9e-14
Identities = 66/230 (28%), Positives = 113/230 (48%), Gaps = 5/230 (2%)
Query: 697 KFLERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSK 756
+ +E ++ CS V EI +L L+L ++ELP LV L +L L ++
Sbjct: 12 RHVEAIDKRHCSLVYVPEEIYRYARSLEELLLDANQLRELPEQFFQLVKLRKLGLSD-NE 70
Query: 757 LESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAES 816
++ +P I +L +LD++ + P SI K ++ + L P ++
Sbjct: 71 IQRLPPEIANFMQLVELDVSR-NDIPEIPESIAFCKALQVADFSGNPLTRLPESFPELQN 129
Query: 817 FAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLT 876
+S+ + +++ LP ++ NL L +L L+E L LP+S+ L+ L ELD G ++
Sbjct: 130 LTCLSVNDISLQSLPENIGNLYNLASLELRENL-LTYLPDSLTQLRRLEELDL-GNNEIY 187
Query: 877 GIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIP 926
+P IG L L+ L L + LP+ I +L +L LDVS +LE +P
Sbjct: 188 NLPESIGALLHLKDLWLDGNQLSELPQEIGNLKNLLCLDVSE-NRLERLP 236
>LAP4_HUMAN (Q14160) LAP4 protein (Scribble homolog protein)
(hScrib)
Length = 1630
Score = 84.3 bits (207), Expect = 2e-15
Identities = 85/298 (28%), Positives = 142/298 (47%), Gaps = 30/298 (10%)
Query: 652 LERLDLSNSWKLVRIPDLSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVE 711
L LD+S + + IP+ K EI L+RLP +L+ L L L+ S ++
Sbjct: 84 LVELDVSRN-DIPEIPESIKFCKALEIADFSGNPLSRLPDGFTQLRSLAHLALNDVS-LQ 141
Query: 712 NIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLC 771
+P + NL L L++ ++ LP+SL+ LV LE+L L + LE +P ++G L L
Sbjct: 142 ALPGDVGNLANLVTLELRENLLKSLPASLSFLVKLEQLDLGG-NDLEVLPDTLGALPNLR 200
Query: 772 KLDLTYCESLETFPGSIFDLK-LTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKL 830
+L L L P + +L+ L LD+++ +RL+ PA L + L++ +++L
Sbjct: 201 ELWLDR-NQLSALPPELGNLRRLVCLDVSE-NRLEELPAELGGLVLLTDLLLSQNLLRRL 258
Query: 831 PSSLQNLVGLQTL------------CLKECPDLE----------SLPNSICNLKLLSELD 868
P + L L L + +C +L +LP S+ L L+ L+
Sbjct: 259 PDGIGQLKQLSILKVDQNRLCEVTEAIGDCENLSELILTENLLMALPRSLGKLTKLTNLN 318
Query: 869 CSGCRKLTGIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIP 926
L +P +IG +L LSL+D + LP +AH + L LDV+ +L+ +P
Sbjct: 319 VDR-NHLEALPPEIGGCVALSVLSLRDNRLAVLPPELAHTTELHVLDVA-GNRLQSLP 374
Score = 77.0 bits (188), Expect = 3e-13
Identities = 67/230 (29%), Positives = 111/230 (48%), Gaps = 5/230 (2%)
Query: 697 KFLERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSK 756
+ +E ++ CS EI +L L+L ++ELP L+ L +L L ++
Sbjct: 12 RHVESVDKRHCSLQAVPEEIYRYSRSLEELLLDANQLRELPKPFFRLLNLRKLGLS-DNE 70
Query: 757 LESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAES 816
++ +P + +L +LD++ + P SI K ++ + L P S
Sbjct: 71 IQRLPPEVANFMQLVELDVSR-NDIPEIPESIKFCKALEIADFSGNPLSRLPDGFTQLRS 129
Query: 817 FAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLT 876
AH++L + +++ LP + NL L TL L+E L+SLP S+ L L +LD G L
Sbjct: 130 LAHLALNDVSLQALPGDVGNLANLVTLELRE-NLLKSLPASLSFLVKLEQLDLGG-NDLE 187
Query: 877 GIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIP 926
+P+ +G L +LR L L + LP + +L L LDVS +LE +P
Sbjct: 188 VLPDTLGALPNLRELWLDRNQLSALPPELGNLRRLVCLDVSE-NRLEELP 236
Score = 69.3 bits (168), Expect = 6e-11
Identities = 73/253 (28%), Positives = 112/253 (43%), Gaps = 8/253 (3%)
Query: 672 SPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIPEIKETMENLAVLVLKQT 731
S +++E++L L LP F+L L +L LS + ++ +P L L + +
Sbjct: 35 SRSLEELLLDA-NQLRELPKPFFRLLNLRKLGLSD-NEIQRLPPEVANFMQLVELDVSRN 92
Query: 732 AIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDL 791
I E+P S+ LE + L +P L L L L SL+ PG + +L
Sbjct: 93 DIPEIPESIKFCKALEIADFS-GNPLSRLPDGFTQLRSLAHLALNDV-SLQALPGDVGNL 150
Query: 792 -KLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPD 850
L L+L + + LK+ PA L + L ++ LP +L L L+ L L +
Sbjct: 151 ANLVTLELRE-NLLKSLPASLSFLVKLEQLDLGGNDLEVLPDTLGALPNLRELWL-DRNQ 208
Query: 851 LESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSS 910
L +LP + NL+ L LD S +L +P ++G L L L L + LP+ I L
Sbjct: 209 LSALPPELGNLRRLVCLDVSE-NRLEELPAELGGLVLLTDLLLSQNLLRRLPDGIGQLKQ 267
Query: 911 LESLDVSYCRKLE 923
L L V R E
Sbjct: 268 LSILKVDQNRLCE 280
Score = 53.9 bits (128), Expect = 2e-06
Identities = 82/310 (26%), Positives = 137/310 (43%), Gaps = 39/310 (12%)
Query: 572 VPDLLESLP--NGLKVLHWDEFPQRSFPLDFCP-EKLVKLHMRESRLEQLWGD------- 621
+P++ ES+ L++ + P P F L L + + L+ L GD
Sbjct: 94 IPEIPESIKFCKALEIADFSGNPLSRLPDGFTQLRSLAHLALNDVSLQALPGDVGNLANL 153
Query: 622 ------DQVLQSLCHGLSYIFFGHLIVFSLHQELPNLERLDLSNSWKLVRIPD-LSKSPN 674
+ +L+SL LS+ L LE+LDL + L +PD L PN
Sbjct: 154 VTLELRENLLKSLPASLSF--------------LVKLEQLDLGGN-DLEVLPDTLGALPN 198
Query: 675 IKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQ 734
++E+ L L+ LP L L+ L L++S + +E +P + L L+L Q ++
Sbjct: 199 LRELWLDR-NQLSALPPELGNLRRLVCLDVSE-NRLEELPAELGGLVLLTDLLLSQNLLR 256
Query: 735 ELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDL-KL 793
LP + L L L + ++L + +IG L +L LT L P S+ L KL
Sbjct: 257 RLPDGIGQLKQLSILKVD-QNRLCEVTEAIGDCENLSELILTE-NLLMALPRSLGKLTKL 314
Query: 794 TKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLES 853
T L++ D + L+ P + + + +SL + + LP L + L L + L+S
Sbjct: 315 TNLNV-DRNHLEALPPEIGGCVALSVLSLRDNRLAVLPPELAHTTELHVLDVAG-NRLQS 372
Query: 854 LPNSICNLKL 863
LP ++ +L L
Sbjct: 373 LPFALTHLNL 382
>DR16_ARATH (O22727) Putative disease resistance protein At1g61190
Length = 967
Score = 84.3 bits (207), Expect = 2e-15
Identities = 178/753 (23%), Positives = 292/753 (38%), Gaps = 158/753 (20%)
Query: 205 QLDSEAVRIIGICGMGGIGKSTLAEALYHKL----GTQFSSRCLIVNAQQKIDRYGIYSL 260
+L + V I+G+ GMGG+GK+TL + +++K GT ++V+ K+ +
Sbjct: 167 RLMEDGVGIMGLHGMGGVGKTTLFKKIHNKFAETGGTFDIVIWIVVSQGAKLSKLQEDIA 226
Query: 261 RKKYLSKLLGEDIQSNGLNYAIERVKRAK-VLLILDDLKISIPILELIG-GHGNFGQGSR 318
K +L L ++ + I RV + K +L+LDD+ + LE IG + + +
Sbjct: 227 EKLHLCDDLWKNKNESDKATDIHRVLKGKRFVLMLDDIWEKVD-LEAIGIPYPSEVNKCK 285
Query: 319 IIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRL----NAFKEDYPLEGYANLIVEVL 374
+ T+R + V + +V+ + +D+ ELF+ N + D + G A EV
Sbjct: 286 VAFTTRDQKVCGQMGDHKPMQVKCLEPEDAWELFKNKVGDNTLRSDPVIVGLAR---EVA 342
Query: 375 KYAKGVPLALQVLGSLLYDKER-EVWESELKKL-------KELPNVDIFDVLKLSYDGLD 426
+ +G+PLAL +G + K + WE + L ++ N I +LK SYD L+
Sbjct: 343 QKCRGLPLALSCIGETMASKTMVQEWEHAIDVLTRSAAEFSDMQN-KILPILKYSYDSLE 401
Query: 427 DEP-KDIFLDITCFYAGDFVDKVVELLDSCGFSADIGMD--------------------- 464
DE K FL F D +D L++ IG D
Sbjct: 402 DEHIKSCFLYCALFPEDDKID-TKTLINKWICEGFIGEDQVIKRARNKGYEMLGTLIRAN 460
Query: 465 -VLKDRGLISILEDRIAVHDLVLEMGREIVRQQCVSDPG--KHSHLWNHKEIYHVVRKNK 521
+ DRG + + + +HD+V EM I SD G K +++ + H + K K
Sbjct: 461 LLTNDRGFV---KWHVVMHDVVREMALWI-----ASDFGKQKENYVVRARVGLHEIPKVK 512
Query: 522 GTDAIQCIFLDMCQIEKV--------------------QLHPEIFKSMPNIRMLYFHKHS 561
A++ + L M +IE++ L E + M + +L +
Sbjct: 513 DWGAVRRMSLMMNEIEEITCESKCSELTTLFLQSNQLKNLSGEFIRYMQKLVVLDLSHNP 572
Query: 562 SFKQ------GQSNVIVPDL----LESLPNGLKVLHWDEFPQRSFPLDFCP----EKLVK 607
F + G ++ DL +E LP GLK L F F C +L+
Sbjct: 573 DFNELPEQISGLVSLQYLDLSWTRIEQLPVGLKELKKLIFLNLCFTERLCSISGISRLLS 632
Query: 608 LHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQELPNLERLDLSNSWKLVRIP 667
L R + GD VL+ L Q+L NL+ L ++ S +L+ +
Sbjct: 633 LRWLSLRESNVHGDASVLKEL------------------QQLENLQDLRITESAELISLD 674
Query: 668 D-LSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIPEIKETMENLAVL 726
L+K ++ I + + P F L FL +MENL L
Sbjct: 675 QRLAKLISVLRI-----EGFLQKP---FDLSFL------------------ASMENLYGL 708
Query: 727 VLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPG 786
+++ + E + ++ + S I I T L L + C S++
Sbjct: 709 LVENSYFSE--------INIKCRESETESSYLHINPKIPCFTNLTGLIIMKCHSMKDLTW 760
Query: 787 SIFDLKLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLK 846
+F L LD+ D + I E AI L S + L+ L L
Sbjct: 761 ILFAPNLVNLDIRD-------------SREVGEIINKEKAI-NLTSIITPFQKLERLFLY 806
Query: 847 ECPDLESLPNSICNLKLLSELDCSGCRKLTGIP 879
P LES+ S LLS + C KL +P
Sbjct: 807 GLPKLESIYWSPLPFPLLSNIVVKYCPKLRKLP 839
Score = 33.9 bits (76), Expect = 2.6
Identities = 30/86 (34%), Positives = 42/86 (47%), Gaps = 2/86 (2%)
Query: 829 KLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSL 888
+LP + LV LQ L L +E LP + LK L L+ +L I + I L SL
Sbjct: 576 ELPEQISGLVSLQYLDLS-WTRIEQLPVGLKELKKLIFLNLCFTERLCSI-SGISRLLSL 633
Query: 889 RTLSLQDTGVVNLPESIAHLSSLESL 914
R LSL+++ V + L LE+L
Sbjct: 634 RWLSLRESNVHGDASVLKELQQLENL 659
>LAP1_DROME (Q9V780) Lap1 protein
Length = 849
Score = 82.8 bits (203), Expect = 5e-15
Identities = 95/340 (27%), Positives = 151/340 (43%), Gaps = 23/340 (6%)
Query: 589 DEFPQRSFPLDFCPEKLVKLHMRESRLEQLWGDDQVLQSL------CHGLSYIFFGHLIV 642
D+ + PL PE + E LE+L+ LQ+L C GL + +
Sbjct: 20 DKLDYSNTPLTDFPE----VWQHERTLEELYLSTTRLQALPPQLFYCQGLRVLHVNSNNL 75
Query: 643 FSLHQ---ELPNLERLDLSNSWKLVRIPDLSKSPNIKEIILSGCKSLTRLPINLFKLKFL 699
S+ Q L L+ LDL+ + +V +P+ KS + C SL RLP + L L
Sbjct: 76 ESIPQAIGSLRQLQHLDLNRNL-IVNVPEEIKSCKHLTHLDLSCNSLQRLPDAITSLISL 134
Query: 700 ERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLES 759
+ L L+ + +E +P + NL +L L+ + LP S+ L+ L+ L + ++
Sbjct: 135 QELLLNE-TYLEFLPANFGRLVNLRILELRLNNLMTLPKSMVRLINLQRLDIGG-NEFTE 192
Query: 760 IPSSIGTLTKLCKL--DLTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESF 817
+P +G L L +L D + G + DL+ + + N L T P+ L +
Sbjct: 193 LPEVVGELKSLRELWIDFNQIRRVSANIGKLRDLQHFEANGN---LLDTLPSELSNWRNV 249
Query: 818 AHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTG 877
+S+ +++ P S+ L L T E L LP+SI L+ L EL S KL
Sbjct: 250 EVLSICSNSLEAFPFSVGMLKSLVTFKC-ESNGLTELPDSISYLEQLEELVLSH-NKLIR 307
Query: 878 IPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVS 917
+P+ IG L SLR L D + LP+ + L L V+
Sbjct: 308 LPSTIGMLRSLRFLFADDNQLRQLPDELCSCQQLSVLSVA 347
Score = 50.8 bits (120), Expect = 2e-05
Identities = 58/257 (22%), Positives = 108/257 (41%), Gaps = 27/257 (10%)
Query: 649 LPNLERLDLSNSWKLVRIPDL-SKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSG- 706
L NL+RLD+ + + +P++ + +++E+ + + R+ N+ KL+ L+ +G
Sbjct: 177 LINLQRLDIGGN-EFTELPEVVGELKSLRELWID-FNQIRRVSANIGKLRDLQHFEANGN 234
Query: 707 --------------------CSN-VENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVG 745
CSN +E P +++L + + ELP S+++L
Sbjct: 235 LLDTLPSELSNWRNVEVLSICSNSLEAFPFSVGMLKSLVTFKCESNGLTELPDSISYLEQ 294
Query: 746 LEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLK 805
LEEL L +KL +PS+IG L L L L P + + + ++L
Sbjct: 295 LEELVLSH-NKLIRLPSTIGMLRSLRFL-FADDNQLRQLPDELCSCQQLSVLSVANNQLS 352
Query: 806 TFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLS 865
P + +++ I LP S+ NLV L ++ L + +P + +
Sbjct: 353 ALPQNIGNLSKMKVLNVVNNYINALPVSMLNLVNLTSMWLSDNQSQPLVPLQYLDASTKT 412
Query: 866 ELDCSGCRKLTGIPNDI 882
+L C ++T N I
Sbjct: 413 QLTCFMLPQVTFKMNSI 429
Score = 40.0 bits (92), Expect = 0.037
Identities = 35/134 (26%), Positives = 54/134 (40%), Gaps = 27/134 (20%)
Query: 793 LTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLE 852
+ KLD ++ + L FP + + + + L+ T ++ LP L GL+ L +
Sbjct: 19 IDKLDYSN-TPLTDFPEVWQHERTLEELYLSTTRLQALPPQLFYCQGLRVLHVNS----- 72
Query: 853 SLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLE 912
L IP IG L L+ L L +VN+PE I L
Sbjct: 73 --------------------NNLESIPQAIGSLRQLQHLDLNRNLIVNVPEEIKSCKHLT 112
Query: 913 SLDVSYCRKLECIP 926
LD+S C L+ +P
Sbjct: 113 HLDLS-CNSLQRLP 125
>LAP4_MOUSE (Q80U72) LAP4 protein (Scribble homolog protein)
Length = 1612
Score = 82.0 bits (201), Expect = 8e-15
Identities = 101/368 (27%), Positives = 171/368 (46%), Gaps = 51/368 (13%)
Query: 587 HWDEFPQRSFPLDFCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLH 646
H + +R L PE++ + LE+L D L+ L FF L + L
Sbjct: 13 HVESVDKRHCSLQVVPEEIYRY---SRSLEELLLDANQLRELPKP----FFRLLNLRKLG 65
Query: 647 QELPNLERL--DLSNSWKLVRIPDLSKS--PNIKEIILSGCKSL----------TRLPIN 692
++RL +++N +LV + D+S++ P I E I CK+L +RLP
Sbjct: 66 LSDNEIQRLPPEVANFMQLVEL-DVSRNDIPEIPESI-KFCKALEIADFSGNPLSRLPDG 123
Query: 693 LFKLKFLERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLK 752
+L+ L L L+ S ++ +P + NL L L++ ++ LP+SL+ LV LE+L L
Sbjct: 124 FTQLRSLAHLALNDVS-LQALPGDVGNLANLVTLELRENLLKSLPASLSFLVKLEQLDLG 182
Query: 753 FCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLK-LTKLDLNDCSRLKTFPAIL 811
+ LE +P ++G L L +L L L P + +L+ L LD+++ +RL+ P L
Sbjct: 183 G-NDLEVLPDTLGALPNLRELWLDR-NQLSALPPELGNLRRLVCLDVSE-NRLEELPVEL 239
Query: 812 EPAESFAHISLAETAIKKLPSSLQNLVGLQTL------------CLKECPDLE------- 852
+ L++ +++LP + L L L + +C +L
Sbjct: 240 GGLALLTDLLLSQNLLQRLPEGIGQLKQLSILKVDQNRLCEVTEAIGDCENLSELILTEN 299
Query: 853 ---SLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLS 909
+LP+S+ L L+ L+ L +P +IG +L LSL+D + LP +AH +
Sbjct: 300 LLTALPHSLGKLTKLTNLNVDR-NHLEVLPPEIGGCVALSVLSLRDNRLAVLPPELAHTA 358
Query: 910 SLESLDVS 917
L LDV+
Sbjct: 359 ELHVLDVA 366
Score = 76.6 bits (187), Expect = 4e-13
Identities = 67/230 (29%), Positives = 111/230 (48%), Gaps = 5/230 (2%)
Query: 697 KFLERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSK 756
+ +E ++ CS EI +L L+L ++ELP L+ L +L L ++
Sbjct: 12 RHVESVDKRHCSLQVVPEEIYRYSRSLEELLLDANQLRELPKPFFRLLNLRKLGLS-DNE 70
Query: 757 LESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAES 816
++ +P + +L +LD++ + P SI K ++ + L P S
Sbjct: 71 IQRLPPEVANFMQLVELDVSR-NDIPEIPESIKFCKALEIADFSGNPLSRLPDGFTQLRS 129
Query: 817 FAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLT 876
AH++L + +++ LP + NL L TL L+E L+SLP S+ L L +LD G L
Sbjct: 130 LAHLALNDVSLQALPGDVGNLANLVTLELRE-NLLKSLPASLSFLVKLEQLDLGG-NDLE 187
Query: 877 GIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIP 926
+P+ +G L +LR L L + LP + +L L LDVS +LE +P
Sbjct: 188 VLPDTLGALPNLRELWLDRNQLSALPPELGNLRRLVCLDVSE-NRLEELP 236
>LAP1_CAEEL (O61967) Lap1 protein (Lethal protein 413)
Length = 679
Score = 81.6 bits (200), Expect = 1e-14
Identities = 98/357 (27%), Positives = 171/357 (47%), Gaps = 38/357 (10%)
Query: 604 KLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQELPNLERL-DLS-NSW 661
KL L++ + +++L D L SL H L + + L E+ NL +L +L+ N
Sbjct: 37 KLEDLNLTMNNIKEL---DHRLFSLRH-LRILDVSDNELAVLPAEIGNLTQLIELNLNRN 92
Query: 662 KLVRIPDLSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIPEIKETME 721
+ ++PD ++ + + TRLP + + + L+L+ S + +P ++
Sbjct: 93 SIAKLPDTMQNCKLLTTLNLSSNPFTRLPETICECSSITILSLNETS-LTLLPSNIGSLT 151
Query: 722 NLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESL 781
NL VL + ++ +P S+ L LEEL L ++LE++P+ IG LT L + + SL
Sbjct: 152 NLRVLEARDNLLRTIPLSIVELRKLEELDLGQ-NELEALPAEIGKLTSLREFYVDI-NSL 209
Query: 782 ETFPGSIFDLK-LTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGL 840
+ P SI + L +LD+++ +++ P L + ++++ I +LPSS L L
Sbjct: 210 TSLPDSISGCRMLDQLDVSE-NQIIRLPENLGRMPNLTDLNISINEIIELPSSFGELKRL 268
Query: 841 QTLC------------LKECPDLESL----------PNSICNLKLLSELDCSGCRKLTGI 878
Q L + +C L L P++I +L+ L+ L+ C L+ I
Sbjct: 269 QMLKADRNSLHNLTSEIGKCQSLTELYLGQNFLTDLPDTIGDLRQLTTLNVD-CNNLSDI 327
Query: 879 PNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIPHLPPFMKQL 935
P+ IG SL LSL+ + LP +I +L LDV+ + +PHLP +K L
Sbjct: 328 PDTIGNCKSLTVLSLRQNILTELPMTIGKCENLTVLDVASNK----LPHLPFTVKVL 380
Score = 78.6 bits (192), Expect = 9e-14
Identities = 71/213 (33%), Positives = 110/213 (51%), Gaps = 10/213 (4%)
Query: 708 SNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTL 767
SN++ IP L L L I+EL L L L L + ++L +P+ IG L
Sbjct: 23 SNLQAIPSDIFRFRKLEDLNLTMNNIKELDHRLFSLRHLRILDVSD-NELAVLPAEIGNL 81
Query: 768 TKLCKLDLTYCESLETFPGSIFDLKL-TKLDLNDCSRLKTFPAILEPAESFAHISLAETA 826
T+L +L+L S+ P ++ + KL T L+L+ + P + S +SL ET+
Sbjct: 82 TQLIELNLNR-NSIAKLPDTMQNCKLLTTLNLSS-NPFTRLPETICECSSITILSLNETS 139
Query: 827 IKKLPSSLQNLVGLQTLCLKECPD--LESLPNSICNLKLLSELDCSGCRKLTGIPNDIGC 884
+ LPS++ +L L+ L E D L ++P SI L+ L ELD G +L +P +IG
Sbjct: 140 LTLLPSNIGSLTNLRVL---EARDNLLRTIPLSIVELRKLEELDL-GQNELEALPAEIGK 195
Query: 885 LSSLRTLSLQDTGVVNLPESIAHLSSLESLDVS 917
L+SLR + + +LP+SI+ L+ LDVS
Sbjct: 196 LTSLREFYVDINSLTSLPDSISGCRMLDQLDVS 228
Score = 72.8 bits (177), Expect = 5e-12
Identities = 58/173 (33%), Positives = 90/173 (51%), Gaps = 6/173 (3%)
Query: 755 SKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLK-LTKLDLNDCSRLKTFPAILEP 813
S L++IPS I KL L+LT +++ +F L+ L LD++D + L PA +
Sbjct: 23 SNLQAIPSDIFRFRKLEDLNLTM-NNIKELDHRLFSLRHLRILDVSD-NELAVLPAEIGN 80
Query: 814 AESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCR 873
++L +I KLP ++QN L TL L P LP +IC ++ L +
Sbjct: 81 LTQLIELNLNRNSIAKLPDTMQNCKLLTTLNLSSNP-FTRLPETICECSSITILSLNET- 138
Query: 874 KLTGIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIP 926
LT +P++IG L++LR L +D + +P SI L LE LD+ +LE +P
Sbjct: 139 SLTLLPSNIGSLTNLRVLEARDNLLRTIPLSIVELRKLEELDLGQ-NELEALP 190
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.139 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 136,063,982
Number of Sequences: 164201
Number of extensions: 5892627
Number of successful extensions: 17499
Number of sequences better than 10.0: 211
Number of HSP's better than 10.0 without gapping: 91
Number of HSP's successfully gapped in prelim test: 126
Number of HSP's that attempted gapping in prelim test: 15847
Number of HSP's gapped (non-prelim): 793
length of query: 1156
length of database: 59,974,054
effective HSP length: 121
effective length of query: 1035
effective length of database: 40,105,733
effective search space: 41509433655
effective search space used: 41509433655
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0073.8


