

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0073.15
(616 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
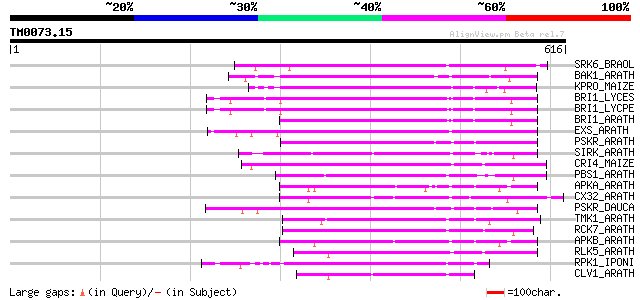
Score E
Sequences producing significant alignments: (bits) Value
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 199 2e-50
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 183 1e-45
KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1 precu... 178 3e-44
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 175 3e-43
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 174 5e-43
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 170 9e-42
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 169 2e-41
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 167 7e-41
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 165 3e-40
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 162 2e-39
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 162 3e-39
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 160 1e-38
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 159 2e-38
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 158 3e-38
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 158 5e-38
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 155 4e-37
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 154 5e-37
RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC... 153 1e-36
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 153 1e-36
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 149 2e-35
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 199 bits (505), Expect = 2e-50
Identities = 132/375 (35%), Positives = 202/375 (53%), Gaps = 32/375 (8%)
Query: 250 SIAGGIGALLICITICIFRRKL----SPIVSKHWKAKKVDQDIEAFIRNNGPQAIKRYSY 305
S+ G+ LL+ I C+++RK + +S + + + + ++ + Y +
Sbjct: 449 SLTVGVSVLLLLIMFCLWKRKQKRAKASAISIANTQRNQNLPMNEMVLSSKREFSGEYKF 508
Query: 306 SEIK----------KVTNSFKS--KLGQGGYGQVYKGNLNNNCPVAVKVLNASKGNG-QE 352
E++ K T +F S KLGQGG+G VYKG L + +AVK L+ + G E
Sbjct: 509 EELELPLIEMETVVKATENFSSCNKLGQGGFGIVYKGRLLDGKEIAVKRLSKTSVQGTDE 568
Query: 353 FLNEVVSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETNLSWERLH 412
F+NEV I R H+N+V +LG C+EG +K LIYE++ N SL+ + K + L+W
Sbjct: 569 FMNEVTLIARLQHINLVQVLGCCIEGDEKMLIYEYLENLSLDSYLFGKTRRSKLNWNERF 628
Query: 413 KIAEGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMH 472
I G+A+GL YLH+ RI+H D+K SNILLDKN PKI+DFG+A++ + +
Sbjct: 629 DITNGVARGLLYLHQDSRFRIIHRDLKVSNILLDKNMIPKISDFGMARIFERDETEANTM 688
Query: 473 DARGTIGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFPHWI 532
GT GY++PE +G S KSDV+S+G+++LEIV K+N R + E ++
Sbjct: 689 KVVGTYGYMSPEY--AMYGIFSEKSDVFSFGVIVLEIVSGKKN-RGFYNLDYENDLLSYV 745
Query: 533 YKHIEVESNLAWHDGM---------SIEENEICKKMVIVGLWCIQTIPSNRPPMSKVVEM 583
+ + L D + SI + + K + +GL C+Q + +RP MS VV M
Sbjct: 746 WSRWKEGRALEIVDPVIVDSLSSQPSIFQPQEVLKCIQIGLLCVQELAEHRPAMSSVVWM 805
Query: 584 LEGSIEQLQIP-PKP 597
GS E +IP PKP
Sbjct: 806 F-GS-EATEIPQPKP 818
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 183 bits (465), Expect = 1e-45
Identities = 127/360 (35%), Positives = 197/360 (54%), Gaps = 31/360 (8%)
Query: 244 SNNCQGSIAGGI--GALLI----CITICIFRRKLSPIVSKHWKAKKVDQDIEAFIRNNGP 297
SN G+IAGG+ GA L+ I + +RRK H+ ++D E +
Sbjct: 220 SNRITGAIAGGVAAGAALLFAVPAIALAWWRRKKP---QDHFFDVPAEEDPEVHLGQ--- 273
Query: 298 QAIKRYSYSEIKKVTNSFKSK--LGQGGYGQVYKGNLNNNCPVAVKVLNASKGNGQE--F 353
+KR+S E++ +++F +K LG+GG+G+VYKG L + VAVK L + G E F
Sbjct: 274 --LKRFSLRELQVASDNFSNKNILGRGGFGKVYKGRLADGTLVAVKRLKEERTQGGELQF 331
Query: 354 LNEVVSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKK-NFETNLSWERLH 412
EV I H N++ L GFC+ ++ L+Y +M NGS+ ++ + L W +
Sbjct: 332 QTEVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPESQPPLDWPKRQ 391
Query: 413 KIAEGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLC--SETHSIIS 470
+IA G A+GL YLH C+ +I+H D+K +NILLD+ F + DFGLAKL +TH +
Sbjct: 392 RIALGSARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHVTTA 451
Query: 471 MHDARGTIGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIV-GAKQNIRNEASDSSETYFP 529
+ RGTIG+IAPE + G S K+DV+ YG+++LE++ G + ++ +
Sbjct: 452 V---RGTIGHIAPEYLST--GKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLL 506
Query: 530 HWIYKHIEVESNLAWHDGMSIEEN---EICKKMVIVGLWCIQTIPSNRPPMSKVVEMLEG 586
W+ K + E L + ++ N E ++++ V L C Q+ P RP MS+VV MLEG
Sbjct: 507 DWV-KGLLKEKKLEALVDVDLQGNYKDEEVEQLIQVALLCTQSSPMERPKMSEVVRMLEG 565
>KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1
precursor (EC 2.7.1.37)
Length = 817
Score = 178 bits (452), Expect = 3e-44
Identities = 117/330 (35%), Positives = 177/330 (53%), Gaps = 25/330 (7%)
Query: 266 IFRRKLSPIVSKHWKAKKVDQDIEAFIRNNGPQAIKRYSYSEIKKVTNSFKSKLGQGGYG 325
+ +R+L P S+ W ++K +A N +RYSY E+ K T FK +LG+G G
Sbjct: 497 VLKRELRP--SELWASEK---GYKAMTSN-----FRRYSYRELVKATRKFKVELGRGESG 546
Query: 326 QVYKGNLNNNCPVAVKVLNASKGNGQEFLNEVVSIGRTSHVNVVNLLGFCLEGQKKALIY 385
VYKG L ++ VAVK L + + F E+ IGR +H+N+V + GFC EG + L+
Sbjct: 547 TVYKGVLEDDRHVAVKKLENVRQGKEVFQAELSVIGRINHMNLVRIWGFCSEGSHRLLVS 606
Query: 386 EFMPNGSLEKFTHKKNFETNLSWERLHKIAEGIAKGLEYLHKGCNTRILHFDIKPSNILL 445
E++ NGSL + L WE IA G+AKGL YLH C ++H D+KP NILL
Sbjct: 607 EYVENGSLANILFSEGGNILLDWEGRFNIALGVAKGLAYLHHECLEWVIHCDVKPENILL 666
Query: 446 DKNFCPKIADFGLAKLCSETHSIISMHDARGTIGYIAPEVWNRNFGGVSHKSDVYSYGML 505
D+ F PKI DFGL KL + S ++ RGT+GYIAPE W + ++ K DVYSYG++
Sbjct: 667 DQAFEPKITDFGLVKLLNRGGSTQNVSHVRGTLGYIAPE-WVSSL-PITAKVDVYSYGVV 724
Query: 506 ILEIVGAKQNIRNEASDSSETY-----FPHWIYKHIEVESNLAWHDG-----MSIEENEI 555
+LE++ + + + E + + +E E +W DG ++ N +
Sbjct: 725 LLELLTGTR-VSELVGGTDEVHSMLRKLVRMLSAKLEGEEQ-SWIDGYLDSKLNRPVNYV 782
Query: 556 -CKKMVIVGLWCIQTIPSNRPPMSKVVEML 584
+ ++ + + C++ S RP M V+ L
Sbjct: 783 QARTLIKLAVSCLEEDRSKRPTMEHAVQTL 812
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 175 bits (444), Expect = 3e-43
Identities = 124/406 (30%), Positives = 205/406 (49%), Gaps = 49/406 (12%)
Query: 219 ASDGVCGYSNTKKELLCFCKDGSTT-------SNNCQGSIAGGIGALLICITICIFRRKL 271
A++ +CGY L C G + S+ Q S+AG + L+ CIF +
Sbjct: 766 ANNSLCGYP-----LPIPCSSGPKSDANQHQKSHRRQASLAGSVAMGLLFSLFCIFGLII 820
Query: 272 SPIVSKHWKAKKVDQDIEAFIRNNGPQA-------------------------IKRYSYS 306
I +K + KK + +EA++ + A +++ +++
Sbjct: 821 VAIETKKRRRKK-EAALEAYMDGHSHSATANSAWKFTSAREALSINLAAFEKPLRKLTFA 879
Query: 307 EIKKVTNSFK--SKLGQGGYGQVYKGNLNNNCPVAVKVLNASKGNG-QEFLNEVVSIGRT 363
++ + TN F S +G GG+G VYK L + VA+K L G G +EF E+ +IG+
Sbjct: 880 DLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAEMETIGKI 939
Query: 364 SHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTH-KKNFETNLSWERLHKIAEGIAKGL 422
H N+V LLG+C G+++ L+YE+M GSLE H +K L+W KIA G A+GL
Sbjct: 940 KHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKIGIKLNWPARRKIAIGAARGL 999
Query: 423 EYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGTIGYIA 482
+LH C I+H D+K SN+LLD+N +++DFG+A+L S + +S+ GT GY+
Sbjct: 1000 AFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTPGYVP 1059
Query: 483 PEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFPHWIYKHIEVESNL 542
PE + ++F S K DVYSYG+++LE++ KQ +++D + W+ H + +
Sbjct: 1060 PEYY-QSF-RCSTKGDVYSYGVVLLELLTGKQ--PTDSADFGDNNLVGWVKLHAKGKITD 1115
Query: 543 AWHDGMSIEENEI---CKKMVIVGLWCIQTIPSNRPPMSKVVEMLE 585
+ + E+ I + + V C+ RP M +V+ M +
Sbjct: 1116 VFDRELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAMFK 1161
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 174 bits (442), Expect = 5e-43
Identities = 124/406 (30%), Positives = 205/406 (49%), Gaps = 49/406 (12%)
Query: 219 ASDGVCGYSNTKKELLCFCKDGSTT-------SNNCQGSIAGGIGALLICITICIFRRKL 271
A++ +CGY L C G + S+ Q S+AG + L+ CIF +
Sbjct: 766 ANNSLCGYP-----LPLPCSSGPKSDANQHQKSHRRQASLAGSVAMGLLFSLFCIFGLII 820
Query: 272 SPIVSKHWKAKKVDQDIEAFIRNNGPQA-------------------------IKRYSYS 306
I +K + KK + +EA++ + A +++ +++
Sbjct: 821 VAIETKKRRRKK-EAALEAYMDGHSHSATANSAWKFTSAREALSINLAAFEKPLRKLTFA 879
Query: 307 EIKKVTNSFK--SKLGQGGYGQVYKGNLNNNCPVAVKVLNASKGNG-QEFLNEVVSIGRT 363
++ + TN F S +G GG+G VYK L + VA+K L G G +EF E+ +IG+
Sbjct: 880 DLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAEMETIGKI 939
Query: 364 SHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTH-KKNFETNLSWERLHKIAEGIAKGL 422
H N+V LLG+C G+++ L+YE+M GSLE H +K L+W KIA G A+GL
Sbjct: 940 KHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKTGIKLNWPARRKIAIGAARGL 999
Query: 423 EYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGTIGYIA 482
+LH C I+H D+K SN+LLD+N +++DFG+A+L S + +S+ GT GY+
Sbjct: 1000 AFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTPGYVP 1059
Query: 483 PEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFPHWIYKHIEVESNL 542
PE + ++F S K DVYSYG+++LE++ KQ +++D + W+ H + +
Sbjct: 1060 PEYY-QSF-RCSTKGDVYSYGVVLLELLTGKQ--PTDSADFGDNNLVGWVKLHAKGKITD 1115
Query: 543 AWHDGMSIEENEI---CKKMVIVGLWCIQTIPSNRPPMSKVVEMLE 585
+ + E+ I + + V C+ RP M +V+ M +
Sbjct: 1116 VFDRELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAMFK 1161
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 170 bits (431), Expect = 9e-42
Identities = 100/293 (34%), Positives = 168/293 (57%), Gaps = 11/293 (3%)
Query: 300 IKRYSYSEIKKVTNSFK--SKLGQGGYGQVYKGNLNNNCPVAVKVLNASKGNG-QEFLNE 356
+++ +++++ + TN F S +G GG+G VYK L + VA+K L G G +EF+ E
Sbjct: 868 LRKLTFADLLQATNGFHNDSLIGSGGFGDVYKAILKDGSAVAIKKLIHVSGQGDREFMAE 927
Query: 357 VVSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHK-KNFETNLSWERLHKIA 415
+ +IG+ H N+V LLG+C G ++ L+YEFM GSLE H K L+W KIA
Sbjct: 928 METIGKIKHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDVLHDPKKAGVKLNWSTRRKIA 987
Query: 416 EGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDAR 475
G A+GL +LH C+ I+H D+K SN+LLD+N +++DFG+A+L S + +S+
Sbjct: 988 IGSARGLAFLHHNCSPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLA 1047
Query: 476 GTIGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFPHWIYKH 535
GT GY+ PE + ++F S K DVYSYG+++LE++ K+ ++ D + W+ +H
Sbjct: 1048 GTPGYVPPEYY-QSF-RCSTKGDVYSYGVVLLELLTGKR--PTDSPDFGDNNLVGWVKQH 1103
Query: 536 IEVESNLAWHDGMSIEENEI---CKKMVIVGLWCIQTIPSNRPPMSKVVEMLE 585
++ + + + E+ + + + V + C+ RP M +V+ M +
Sbjct: 1104 AKLRISDVFDPELMKEDPALEIELLQHLKVAVACLDDRAWRRPTMVQVMAMFK 1156
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 169 bits (429), Expect = 2e-41
Identities = 129/406 (31%), Positives = 205/406 (49%), Gaps = 45/406 (11%)
Query: 220 SDGVCGYSNTKKELLCFCKD--GSTTSNNC--QGS---IAGGIGALLICITICIF----- 267
SDGVC + K LL K+ G ++C +G+ A GI L++ TI +F
Sbjct: 790 SDGVC--QDPSKALLSGNKELCGRVVGSDCKIEGTKLRSAWGIAGLMLGFTIIVFVFVFS 847
Query: 268 --RRKLSPIVSKHWKAKKVDQD-IEAFIRNN-------------------GPQAIKRYSY 305
R ++ V + +++++ ++ F+ N Q + +
Sbjct: 848 LRRWAMTKRVKQRDDPERMEESRLKGFVDQNLYFLSGSRSREPLSINIAMFEQPLLKVRL 907
Query: 306 SEIKKVTNSFKSK--LGQGGYGQVYKGNLNNNCPVAVKVLNASKGNG-QEFLNEVVSIGR 362
+I + T+ F K +G GG+G VYK L VAVK L+ +K G +EF+ E+ ++G+
Sbjct: 908 GDIVEATDHFSKKNIIGDGGFGTVYKACLPGEKTVAVKKLSEAKTQGNREFMAEMETLGK 967
Query: 363 TSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETN-LSWERLHKIAEGIAKG 421
H N+V+LLG+C ++K L+YE+M NGSL+ + + L W + KIA G A+G
Sbjct: 968 VKHPNLVSLLGYCSFSEEKLLVYEYMVNGSLDHWLRNQTGMLEVLDWSKRLKIAVGAARG 1027
Query: 422 LEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGTIGYI 481
L +LH G I+H DIK SNILLD +F PK+ADFGLA+L S S +S A GT GYI
Sbjct: 1028 LAFLHHGFIPHIIHRDIKASNILLDGDFEPKVADFGLARLISACESHVSTVIA-GTFGYI 1086
Query: 482 APEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFPHWIYKHIEVESN 541
PE + K DVYS+G+++LE+V K+ + +S W + I
Sbjct: 1087 PPEYGQS--ARATTKGDVYSFGVILLELVTGKEPTGPDFKESEGGNLVGWAIQKINQGKA 1144
Query: 542 LAWHDGM--SIEENEICKKMVIVGLWCIQTIPSNRPPMSKVVEMLE 585
+ D + S+ +++ + + C+ P+ RP M V++ L+
Sbjct: 1145 VDVIDPLLVSVALKNSQLRLLQIAMLCLAETPAKRPNMLDVLKALK 1190
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 167 bits (423), Expect = 7e-41
Identities = 108/291 (37%), Positives = 160/291 (54%), Gaps = 10/291 (3%)
Query: 301 KRYSYSEIKKVTNSFKSK--LGQGGYGQVYKGNLNNNCPVAVKVLNASKGN-GQEFLNEV 357
K SY ++ TNSF +G GG+G VYK L + VA+K L+ G +EF EV
Sbjct: 720 KELSYDDLLDSTNSFDQANIIGCGGFGMVYKATLPDGKKVAIKKLSGDCGQIEREFEAEV 779
Query: 358 VSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNF-ETNLSWERLHKIAE 416
++ R H N+V L GFC + LIY +M NGSL+ + H++N L W+ +IA+
Sbjct: 780 ETLSRAQHPNLVLLRGFCFYKNDRLLIYSYMENGSLDYWLHERNDGPALLKWKTRLRIAQ 839
Query: 417 GIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARG 476
G AKGL YLH+GC+ ILH DIK SNILLD+NF +ADFGLA+L S + +S D G
Sbjct: 840 GAAKGLLYLHEGCDPHILHRDIKSSNILLDENFNSHLADFGLARLMSPYETHVST-DLVG 898
Query: 477 TIGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFPHWI--YK 534
T+GYI PE + ++K DVYS+G+++LE++ K+ + + W+ K
Sbjct: 899 TLGYIPPEYGQASV--ATYKGDVYSFGVVLLELLTDKRPV-DMCKPKGCRDLISWVVKMK 955
Query: 535 HIEVESNLAWHDGMSIEENEICKKMVIVGLWCIQTIPSNRPPMSKVVEMLE 585
H S + S E ++ +++ + C+ P RP ++V L+
Sbjct: 956 HESRASEVFDPLIYSKENDKEMFRVLEIACLCLSENPKQRPTTQQLVSWLD 1006
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 165 bits (418), Expect = 3e-40
Identities = 125/343 (36%), Positives = 185/343 (53%), Gaps = 36/343 (10%)
Query: 255 IGALLICIT-ICIFRRKLSPIVSKHWKAKKVDQDIEAFIRNNGPQAIKRY-SYSEIKKVT 312
+G +++ +T + +FRR KK Q RN + KRY YSE+ +T
Sbjct: 525 VGIIVVLLTALALFRR-----------FKKKQQRGTLGERNGPLKTAKRYFKYSEVVNIT 573
Query: 313 NSFKSKLGQGGYGQVYKGNLNNNCPVAVKVLNASKGNG-QEFLNEVVSIGRTSHVNVVNL 371
N+F+ +G+GG+G+VY G +N VAVKVL+ G +EF EV + R H N+ +L
Sbjct: 574 NNFERVIGKGGFGKVYHGVINGE-QVAVKVLSEESAQGYKEFRAEVDLLMRVHHTNLTSL 632
Query: 372 LGFCLEGQKKALIYEFMPNGSL-EKFTHKKNFETNLSWERLHKIAEGIAKGLEYLHKGCN 430
+G+C E LIYE+M N +L + K++F LSWE KI+ A+GLEYLH GC
Sbjct: 633 VGYCNEINHMVLIYEYMANENLGDYLAGKRSF--ILSWEERLKISLDAAQGLEYLHNGCK 690
Query: 431 TRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGTIGYIAPEVWNRNF 490
I+H D+KP+NILL++ K+ADFGL++ S S G+IGY+ PE ++
Sbjct: 691 PPIVHRDVKPTNILLNEKLQAKMADFGLSRSFSVEGSGQISTVVAGSIGYLDPEYYSTR- 749
Query: 491 GGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFPHWIYKHIEVESNLAWHDGMSI 550
++ KSDVYS G+++LE++ + I SS+T H I H V S LA D I
Sbjct: 750 -QMNEKSDVYSLGVVLLEVITGQPAIA-----SSKTEKVH-ISDH--VRSILANGDIRGI 800
Query: 551 EENEICK--------KMVIVGLWCIQTIPSNRPPMSKVVEMLE 585
+ + + KM + L C + + RP MS+VV L+
Sbjct: 801 VDQRLRERYDVGSAWKMSEIALACTEHTSAQRPTMSQVVMELK 843
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 162 bits (411), Expect = 2e-39
Identities = 111/358 (31%), Positives = 188/358 (52%), Gaps = 25/358 (6%)
Query: 258 LLICITICIF-----------RRKLSPIVSKHWKAKKVDQDIEAFIRNNGPQAIKRYSYS 306
L + +T C++ R+L S + +K + I+ + + + + +SY
Sbjct: 437 LSVSVTTCLYVRHKLRHCQCSNRELRLAKSTAYSFRKDNMKIQPDMEDLKIRRAQEFSYE 496
Query: 307 EIKKVTNSFK--SKLGQGGYGQVYKGNLNNNCPVAVK-VLNAS--KGNGQEFLNEVVSIG 361
E+++ T F S++G+G + V+KG L + VAVK + AS K + +EF NE+ +
Sbjct: 497 ELEQATGGFSEDSQVGKGSFSCVFKGILRDGTVVAVKRAIKASDVKKSSKEFHNELDLLS 556
Query: 362 RTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKK--NFETNLSWERLHKIAEGIA 419
R +H +++NLLG+C +G ++ L+YEFM +GSL + H K N + L+W R IA A
Sbjct: 557 RLNHAHLLNLLGYCEDGSERLLVYEFMAHGSLYQHLHGKDPNLKKRLNWARRVTIAVQAA 616
Query: 420 KGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGTIG 479
+G+EYLH ++H DIK SNIL+D++ ++ADFGL+ L GT+G
Sbjct: 617 RGIEYLHGYACPPVIHRDIKSSNILIDEDHNARVADFGLSILGPADSGTPLSELPAGTLG 676
Query: 480 YIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFPHWIYKHIEVE 539
Y+ PE + ++ ++ KSDVYS+G+++LEI+ ++ I + + + W I+
Sbjct: 677 YLDPEYYRLHY--LTTKSDVYSFGVVLLEILSGRKAIDMQFEEGN---IVEWAVPLIKAG 731
Query: 540 SNLAWHDGM--SIEENEICKKMVIVGLWCIQTIPSNRPPMSKVVEMLEGSIEQLQIPP 595
A D + + E KK+ V C++ +RP M KV LE ++ L P
Sbjct: 732 DIFAILDPVLSPPSDLEALKKIASVACKCVRMRGKDRPSMDKVTTALEHALALLMGSP 789
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 162 bits (409), Expect = 3e-39
Identities = 118/324 (36%), Positives = 165/324 (50%), Gaps = 41/324 (12%)
Query: 296 GPQAIKRYSYSEIKKVTNSFKSK--LGQGGYGQVYKGNLNNNCPV-AVKVL--NASKGNG 350
G A +++ E+ T +F LG+GG+G+VYKG L++ V AVK L N +GN
Sbjct: 67 GQIAAHTFAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGN- 125
Query: 351 QEFLNEVVSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETN-LSWE 409
+EFL EV+ + H N+VNL+G+C +G ++ L+YEFMP GSLE H + L W
Sbjct: 126 REFLVEVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEALDWN 185
Query: 410 RLHKIAEGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSII 469
KIA G AKGLE+LH N +++ D K SNILLD+ F PK++DFGLAKL
Sbjct: 186 MRMKIAAGAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSH 245
Query: 470 SMHDARGTIGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFP 529
GT GY APE G ++ KSDVYS+G++ LE++ ++ I +E P
Sbjct: 246 VSTRVMGTYGYCAPEY--AMTGQLTVKSDVYSFGVVFLELITGRKAIDSE--------MP 295
Query: 530 HWIYKHIEVESNL-AWHDGMSIEENEICK----------------KMVIVGLWCIQTIPS 572
H E NL AW + + + K + + V CIQ +
Sbjct: 296 HG-------EQNLVAWARPLFNDRRKFIKLADPRLKGRFPTRALYQALAVASMCIQEQAA 348
Query: 573 NRPPMSKVVEMLEGSIEQLQIPPK 596
RP ++ VV L Q P K
Sbjct: 349 TRPLIADVVTALSYLANQAYDPSK 372
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 160 bits (404), Expect = 1e-38
Identities = 114/309 (36%), Positives = 175/309 (55%), Gaps = 33/309 (10%)
Query: 300 IKRYSYSEIKKVTNSFK--SKLGQGGYGQVYKG-----NLNNNCP-----VAVKVLNASK 347
+K +S++E+K T +F+ S LG+GG+G V+KG +L + P +AVK LN
Sbjct: 53 LKSFSFAELKSATRNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIAVKKLNQDG 112
Query: 348 GNG-QEFLNEVVSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKN-FETN 405
G QE+L EV +G+ SH ++V L+G+CLE + + L+YEFMP GSLE ++ +
Sbjct: 113 WQGHQEWLAEVNYLGQFSHRHLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGLYFQP 172
Query: 406 LSWERLHKIAEGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAK---LC 462
LSW+ K+A G AKGL +LH TR+++ D K SNILLD + K++DFGLAK +
Sbjct: 173 LSWKLRLKVALGAAKGLAFLHSS-ETRVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPIG 231
Query: 463 SETHSIISMHDARGTIGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASD 522
++H +S GT GY APE G ++ KSDVYS+G+++LE++ ++ + ++
Sbjct: 232 DKSH--VSTR-VMGTHGYAAPEYLAT--GHLTTKSDVYSFGVVLLELLSGRRAV-DKNRP 285
Query: 523 SSETYFPHWIYKHIEVESNL------AWHDGMSIEENEICKKMVIVGLWCIQTIPSNRPP 576
S E W ++ + + D S+EE K+ + L C+ T RP
Sbjct: 286 SGERNLVEWAKPYLVNKRKIFRVIDNRLQDQYSMEE---ACKVATLSLRCLTTEIKLRPN 342
Query: 577 MSKVVEMLE 585
MS+VV LE
Sbjct: 343 MSEVVSHLE 351
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 159 bits (403), Expect = 2e-38
Identities = 113/331 (34%), Positives = 171/331 (51%), Gaps = 26/331 (7%)
Query: 300 IKRYSYSEIKKVTNSFK--SKLGQGGYGQVYKG----------NLNNNCPVAVKVLNASK 347
+K Y++ ++K T +FK S LGQGG+G+VY+G + + VA+K LN+
Sbjct: 71 LKVYNFLDLKTATKNFKPDSMLGQGGFGKVYRGWVDATTLAPSRVGSGMIVAIKRLNSES 130
Query: 348 GNG-QEFLNEVVSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETNL 406
G E+ +EV +G SH N+V LLG+C E ++ L+YEFMP GSLE ++N
Sbjct: 131 VQGFAEWRSEVNFLGMLSHRNLVKLLGYCREDKELLLVYEFMPKGSLESHLFRRN--DPF 188
Query: 407 SWERLHKIAEGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETH 466
W+ KI G A+GL +LH +++ D K SNILLD N+ K++DFGLAKL
Sbjct: 189 PWDLRIKIVIGAARGLAFLH-SLQREVIYRDFKASNILLDSNYDAKLSDFGLAKLGPADE 247
Query: 467 SIISMHDARGTIGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSET 526
GT GY APE G + KSDV+++G+++LEI+ N +
Sbjct: 248 KSHVTTRIMGTYGYAAPEYMAT--GHLYVKSDVFAFGVVLLEIM-TGLTAHNTKRPRGQE 304
Query: 527 YFPHWIYKHIEVESNLAWHDGMSIE---ENEICKKMVIVGLWCIQTIPSNRPPMSKVVEM 583
W+ + + + I+ ++ +M + L CI+ P NRP M +VVE+
Sbjct: 305 SLVDWLRPELSNKHRVKQIMDKGIKGQYTTKVATEMARITLSCIEPDPKNRPHMKEVVEV 364
Query: 584 LEGSIEQLQIPPKPFMFSPTKTEVESGTTSN 614
LE I+ L + P S TK V + + S+
Sbjct: 365 LE-HIQGLNVVPNR---SSTKQAVANSSRSS 391
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 158 bits (400), Expect = 3e-38
Identities = 117/385 (30%), Positives = 191/385 (49%), Gaps = 22/385 (5%)
Query: 218 EASDGVCGYSNTKKELLCFCKDGSTTSN--NCQGSIAGGIGA-----LLICITICIFRRK 270
E + G+CG + + GS + N + +A +G L+ +T+ I R
Sbjct: 636 EGNQGLCGEHASPCHITDQSPHGSAVKSKKNIRKIVAVAVGTGLGTVFLLTVTLLIILRT 695
Query: 271 LSP---IVSKHWKAKKVDQDIEAFIRNNGPQAIKRYSYSEIKKVTNSFKSK--LGQGGYG 325
S K A +++ + + + + S +I K T+SF +G GG+G
Sbjct: 696 TSRGEVDPEKKADADEIELGSRSVVLFHNKDSNNELSLDDILKSTSSFNQANIIGCGGFG 755
Query: 326 QVYKGNLNNNCPVAVKVLNASKGN-GQEFLNEVVSIGRTSHVNVVNLLGFCLEGQKKALI 384
VYK L + VA+K L+ G +EF EV ++ R H N+V+LLG+C K LI
Sbjct: 756 LVYKATLPDGTKVAIKRLSGDTGQMDREFQAEVETLSRAQHPNLVHLLGYCNYKNDKLLI 815
Query: 385 YEFMPNGSLEKFTHKK-NFETNLSWERLHKIAEGIAKGLEYLHKGCNTRILHFDIKPSNI 443
Y +M NGSL+ + H+K + +L W+ +IA G A+GL YLH+ C ILH DIK SNI
Sbjct: 816 YSYMDNGSLDYWLHEKVDGPPSLDWKTRLRIARGAAEGLAYLHQSCEPHILHRDIKSSNI 875
Query: 444 LLDKNFCPKIADFGLAKLCSETHSIISMHDARGTIGYIAPEVWNRNFGGVSHKSDVYSYG 503
LL F +ADFGLA+L + ++ D GT+GYI PE + ++K DVYS+G
Sbjct: 876 LLSDTFVAHLADFGLARLILPYDTHVTT-DLVGTLGYIPPEYGQASV--ATYKGDVYSFG 932
Query: 504 MLILEIVGAKQNIRNEASDSSETYFPHWIYKHIEVESNLAWHDGMSIEENEICKKMVIV- 562
+++LE++ ++ + + W+ + ++ E + I + + ++M++V
Sbjct: 933 VVLLELLTGRRPM-DVCKPRGSRDLISWVLQ-MKTEKRESEIFDPFIYDKDHAEEMLLVL 990
Query: 563 --GLWCIQTIPSNRPPMSKVVEMLE 585
C+ P RP ++V LE
Sbjct: 991 EIACRCLGENPKTRPTTQQLVSWLE 1015
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 158 bits (399), Expect = 5e-38
Identities = 101/299 (33%), Positives = 161/299 (53%), Gaps = 18/299 (6%)
Query: 304 SYSEIKKVTNSFKSK--LGQGGYGQVYKGNLNNNCPVAVKVLN----ASKGNGQEFLNEV 357
S ++ VTN+F S LG GG+G VYKG L++ +AVK + A KG EF +E+
Sbjct: 577 SIQVLRSVTNNFSSDNILGSGGFGVVYKGELHDGTKIAVKRMENGVIAGKGFA-EFKSEI 635
Query: 358 VSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFE--TNLSWERLHKIA 415
+ + H ++V LLG+CL+G +K L+YE+MP G+L + + + E L W++ +A
Sbjct: 636 AVLTKVRHRHLVTLLGYCLDGNEKLLVYEYMPQGTLSRHLFEWSEEGLKPLLWKQRLTLA 695
Query: 416 EGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDAR 475
+A+G+EYLH + +H D+KPSNILL + K+ADFGL +L E I A
Sbjct: 696 LDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRLAPEGKGSIETRIA- 754
Query: 476 GTIGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFPHW---I 532
GT GY+APE G V+ K DVYS+G++++E++ ++++ +E+ + W +
Sbjct: 755 GTFGYLAPEY--AVTGRVTTKVDVYSFGVILMELITGRKSL-DESQPEESIHLVSWFKRM 811
Query: 533 YKHIEVESNLAWHDGMSIEENEICKKMVIVGL--WCIQTIPSNRPPMSKVVEMLEGSIE 589
Y + E A + ++E + + L C P RP M V +L +E
Sbjct: 812 YINKEASFKKAIDTTIDLDEETLASVHTVAELAGHCCAREPYQRPDMGHAVNILSSLVE 870
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 155 bits (391), Expect = 4e-37
Identities = 95/287 (33%), Positives = 159/287 (55%), Gaps = 11/287 (3%)
Query: 303 YSYSEIKKVTNSFKSK--LGQGGYGQVYKGNLNN-NCPVAVKVLNASKGNG-QEFLNEVV 358
+++ E+ + T +F+S LG+GG+G+V+KG + + VA+K L+ + G +EF+ EV+
Sbjct: 91 FTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGIREFVVEVL 150
Query: 359 SIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHK-KNFETNLSWERLHKIAEG 417
++ H N+V L+GFC EG ++ L+YE+MP GSLE H + + L W KIA G
Sbjct: 151 TLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLHVLPSGKKPLDWNTRMKIAAG 210
Query: 418 IAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGT 477
A+GLEYLH +++ D+K SNILL +++ PK++DFGLAK+ GT
Sbjct: 211 AARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTHVSTRVMGT 270
Query: 478 IGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFPHWIYKHIE 537
GY AP+ G ++ KSD+YS+G+++LE++ ++ I N + + W +
Sbjct: 271 YGYCAPDY--AMTGQLTFKSDIYSFGVVLLELITGRKAIDNTKTRKDQN-LVGWARPLFK 327
Query: 538 VESNLAWHDGMSIEENEICK---KMVIVGLWCIQTIPSNRPPMSKVV 581
N ++ + + + + C+Q P+ RP +S VV
Sbjct: 328 DRRNFPKMVDPLLQGQYPVRGLYQALAISAMCVQEQPTMRPVVSDVV 374
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 154 bits (390), Expect = 5e-37
Identities = 108/306 (35%), Positives = 163/306 (52%), Gaps = 27/306 (8%)
Query: 300 IKRYSYSEIKKVTNSFK--SKLGQGGYGQVYKGNLNNNC----------PVAVKVLNASK 347
+K ++++E+K T +F+ S LG+GG+G V+KG ++ +AVK LN
Sbjct: 54 LKSFTFAELKAATRNFRPDSVLGEGGFGSVFKGWIDEQTLTASKPGTGVVIAVKKLNQDG 113
Query: 348 GNG-QEFLNEVVSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKK-NFETN 405
G QE+L EV +G+ SH N+V L+G+CLE + + L+YEFMP GSLE ++ ++
Sbjct: 114 WQGHQEWLAEVNYLGQFSHPNLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGSYFQP 173
Query: 406 LSWERLHKIAEGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSET 465
LSW K+A G AKGL +LH T +++ D K SNILLD + K++DFGLAK
Sbjct: 174 LSWTLRLKVALGAAKGLAFLH-NAETSVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPTG 232
Query: 466 HSIISMHDARGTIGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSE 525
GT GY APE G ++ KSDVYSYG+++LE++ ++ + ++ E
Sbjct: 233 DKSHVSTRIMGTYGYAAPEYLAT--GHLTTKSDVYSYGVVLLEVLSGRRAV-DKNRPPGE 289
Query: 526 TYFPHWIYKHIEVESNL------AWHDGMSIEENEICKKMVIVGLWCIQTIPSNRPPMSK 579
W + + L D S+EE K+ + L C+ RP M++
Sbjct: 290 QKLVEWARPLLANKRKLFRVIDNRLQDQYSMEE---ACKVATLALRCLTFEIKLRPNMNE 346
Query: 580 VVEMLE 585
VV LE
Sbjct: 347 VVSHLE 352
>RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC
2.7.1.37)
Length = 999
Score = 153 bits (387), Expect = 1e-36
Identities = 104/286 (36%), Positives = 148/286 (51%), Gaps = 22/286 (7%)
Query: 316 KSKLGQGGYGQVYKGNLNNNCPVAVKVLNASKGNGQE-----------FLNEVVSIGRTS 364
K+ +G G G+VYK L VAVK LN S G + F EV ++G
Sbjct: 686 KNVIGFGSSGKVYKVELRGGEVVAVKKLNKSVKGGDDEYSSDSLNRDVFAAEVETLGTIR 745
Query: 365 HVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTH-KKNFETNLSWERLHKIAEGIAKGLE 423
H ++V L C G K L+YE+MPNGSL H + L W +IA A+GL
Sbjct: 746 HKSIVRLWCCCSSGDCKLLVYEYMPNGSLADVLHGDRKGGVVLGWPERLRIALDAAEGLS 805
Query: 424 YLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKL--CSETHSIISMHDARGTIGYI 481
YLH C I+H D+K SNILLD ++ K+ADFG+AK+ S + + +M G+ GYI
Sbjct: 806 YLHHDCVPPIVHRDVKSSNILLDSDYGAKVADFGIAKVGQMSGSKTPEAMSGIAGSCGYI 865
Query: 482 APE-VWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFPHWIYKHIE-VE 539
APE V+ V+ KSD+YS+G+++LE+V KQ +E D W+ ++
Sbjct: 866 APEYVYTLR---VNEKSDIYSFGVVLLELVTGKQPTDSELGDKD---MAKWVCTALDKCG 919
Query: 540 SNLAWHDGMSIEENEICKKMVIVGLWCIQTIPSNRPPMSKVVEMLE 585
+ ++ E K++ +GL C +P NRP M KVV ML+
Sbjct: 920 LEPVIDPKLDLKFKEEISKVIHIGLLCTSPLPLNRPSMRKVVIMLQ 965
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 153 bits (386), Expect = 1e-36
Identities = 109/326 (33%), Positives = 164/326 (49%), Gaps = 26/326 (7%)
Query: 214 CAECEASDGVCGYSNTKKELLCFCKDGSTTSNNCQGSIAGG---IGALLICITICIFRRK 270
C C A C S+ +L C S T ++ +GALL I + +F
Sbjct: 727 CINCPADGLACPESS----ILRPCNMQSNTGKGGLSTLGIAMIVLGALLFIICLFLF--- 779
Query: 271 LSPIVSKHWKAKKVDQDIEAFIRNNGPQAIKRYSYSEIKKVTNSFKSK--LGQGGYGQVY 328
S + H KK Q+I A G ++ +++ + T + K +G+G +G +Y
Sbjct: 780 -SAFLFLH--CKKSVQEI-AISAQEGDGSL----LNKVLEATENLNDKYVIGKGAHGTIY 831
Query: 329 KGNLNNNCPVAVK--VLNASKGNGQEFLNEVVSIGRTSHVNVVNLLGFCLEGQKKALIYE 386
K L+ + AVK V K + E+ +IG+ H N++ L F L + ++Y
Sbjct: 832 KATLSPDKVYAVKKLVFTGIKNGSVSMVREIETIGKVRHRNLIKLEEFWLRKEYGLILYT 891
Query: 387 FMPNGSLEKFTHKKNFETNLSWERLHKIAEGIAKGLEYLHKGCNTRILHFDIKPSNILLD 446
+M NGSL H+ N L W H IA G A GL YLH C+ I+H DIKP NILLD
Sbjct: 892 YMENGSLHDILHETNPPKPLDWSTRHNIAVGTAHGLAYLHFDCDPAIVHRDIKPMNILLD 951
Query: 447 KNFCPKIADFGLAKLCSETHSIISMHDARGTIGYIAPEVWNRNFGGVSHKSDVYSYGMLI 506
+ P I+DFG+AKL ++ + I + +GTIGY+APE N S +SDVYSYG+++
Sbjct: 952 SDLEPHISDFGIAKLLDQSATSIPSNTVQGTIGYMAPE--NAFTTVKSRESDVYSYGVVL 1009
Query: 507 LEIVGAKQNIRNEASDSSETYFPHWI 532
LE++ K+ + + S + ET W+
Sbjct: 1010 LELITRKKAL--DPSFNGETDIVGWV 1033
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 149 bits (377), Expect = 2e-35
Identities = 84/201 (41%), Positives = 120/201 (58%), Gaps = 7/201 (3%)
Query: 319 LGQGGYGQVYKGNLNNNCPVAVKVLNASKGNGQE---FLNEVVSIGRTSHVNVVNLLGFC 375
+G+GG G VY+G++ NN VA+K L +G G+ F E+ ++GR H ++V LLG+
Sbjct: 698 IGKGGAGIVYRGSMPNNVDVAIKRL-VGRGTGRSDHGFTAEIQTLGRIRHRHIVRLLGYV 756
Query: 376 LEGQKKALIYEFMPNGSLEKFTHKKNFETNLSWERLHKIAEGIAKGLEYLHKGCNTRILH 435
L+YE+MPNGSL + H +L WE H++A AKGL YLH C+ ILH
Sbjct: 757 ANKDTNLLLYEYMPNGSLGELLHGSK-GGHLQWETRHRVAVEAAKGLCYLHHDCSPLILH 815
Query: 436 FDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGTIGYIAPEVWNRNFGGVSH 495
D+K +NILLD +F +ADFGLAK + + M G+ GYIAPE V
Sbjct: 816 RDVKSNNILLDSDFEAHVADFGLAKFLVDGAASECMSSIAGSYGYIAPEY--AYTLKVDE 873
Query: 496 KSDVYSYGMLILEIVGAKQNI 516
KSDVYS+G+++LE++ K+ +
Sbjct: 874 KSDVYSFGVVLLELIAGKKPV 894
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.136 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 78,725,492
Number of Sequences: 164201
Number of extensions: 3638734
Number of successful extensions: 11079
Number of sequences better than 10.0: 1627
Number of HSP's better than 10.0 without gapping: 814
Number of HSP's successfully gapped in prelim test: 813
Number of HSP's that attempted gapping in prelim test: 8353
Number of HSP's gapped (non-prelim): 1881
length of query: 616
length of database: 59,974,054
effective HSP length: 116
effective length of query: 500
effective length of database: 40,926,738
effective search space: 20463369000
effective search space used: 20463369000
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0073.15


