

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
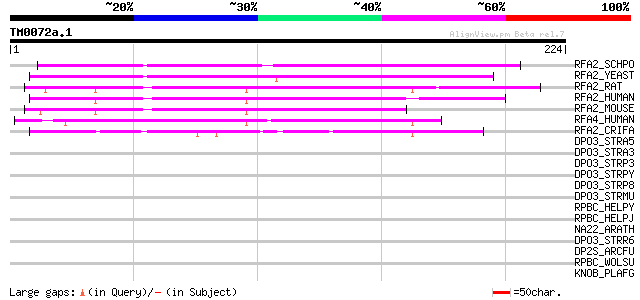
Query= TM0072a.1
(224 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
RFA2_SCHPO (Q92373) Replication factor-A protein 2 (Single-stran... 92 1e-18
RFA2_YEAST (P26754) Replication factor-A protein 2 (RF-A) (DNA b... 83 5e-16
RFA2_RAT (Q63528) Replication protein A 32 kDa subunit (RP-A) (R... 81 2e-15
RFA2_HUMAN (P15927) Replication protein A 32 kDa subunit (RP-A) ... 81 2e-15
RFA2_MOUSE (Q62193) Replication protein A 32 kDa subunit (RP-A) ... 76 7e-14
RFA4_HUMAN (Q13156) Replication protein A 30 kDa subunit (RP-A) ... 57 3e-08
RFA2_CRIFA (Q23697) Replication protein A 28 kDa subunit (RP-A) ... 54 4e-07
DPO3_STRA5 (P63984) DNA polymerase III polC-type (EC 2.7.7.7) (P... 37 0.029
DPO3_STRA3 (P63983) DNA polymerase III polC-type (EC 2.7.7.7) (P... 37 0.029
DPO3_STRP3 (Q8K5S8) DNA polymerase III polC-type (EC 2.7.7.7) (P... 37 0.049
DPO3_STRPY (Q9FDF9) DNA polymerase III polC-type (EC 2.7.7.7) (P... 35 0.11
DPO3_STRP8 (Q8NZB5) DNA polymerase III polC-type (EC 2.7.7.7) (P... 35 0.11
DPO3_STRMU (Q8DWE0) DNA polymerase III polC-type (EC 2.7.7.7) (P... 35 0.11
RPBC_HELPY (O25806) Bifunctional DNA-directed RNA polymerase, be... 35 0.19
RPBC_HELPJ (Q9ZK23) Bifunctional DNA-directed RNA polymerase, be... 33 0.41
NA22_ARATH (Q84TE6) NAC-domain containing protein 21/22 (ANAC021... 32 1.6
DPO3_STRR6 (Q8DRA5) DNA polymerase III polC-type (EC 2.7.7.7) (P... 32 1.6
DP2S_ARCFU (O28484) DNA polymerase II small subunit (EC 2.7.7.7)... 31 2.1
RPBC_WOLSU (Q7MA56) Bifunctional DNA-directed RNA polymerase, be... 31 2.7
KNOB_PLAFG (P09346) Knob-associated histidine-rich protein precu... 31 2.7
>RFA2_SCHPO (Q92373) Replication factor-A protein 2 (Single-stranded
DNA-binding protein P30 subunit)
Length = 279
Score = 91.7 bits (226), Expect = 1e-18
Identities = 58/195 (29%), Positives = 93/195 (46%), Gaps = 5/195 (2%)
Query: 12 GGGGFTSTQLNDSSPAPQKGRESQGLVPVTVKQINEASQSGDEKSSFVINGVELTNVTLV 71
GG GF + ++ L PVT+KQI ASQ + + F I+GVE+ VT V
Sbjct: 25 GGAGFNEYDQSSQPSVDRQQGAGNKLRPVTIKQILNASQVHAD-AEFKIDGVEVGQVTFV 83
Query: 72 GMVFEKAERNTDVNFVLDDGTGRIKCRRWINDAFDTQEVEEIMNGMYVRVNGHLKSFQGV 131
G++ + T+ + ++DGTG I+ R W + + E+ YVRV G++K F G
Sbjct: 84 GVLRNIHAQTTNTTYQIEDGTGMIEVRHWEH----IDALSELATDTYVRVYGNIKIFSGK 139
Query: 132 RQVAAFSVRPVVNFDEIPFHFIDCIHHHLRSKLKMEGTISTDPPASDSSLKTPVRNTSNG 191
+A+ +R + + +E+ FHF++ I HL K + P +S N S+
Sbjct: 140 IYIASQYIRTIKDHNEVHFHFLEAIAVHLHFTQKANAVNGANAPGYGTSNALGYNNISSN 199
Query: 192 SQAPSSTPAYAQYGV 206
A S A+Y +
Sbjct: 200 GAANSLEQKLAEYSL 214
>RFA2_YEAST (P26754) Replication factor-A protein 2 (RF-A) (DNA
binding protein BUF1) (Replication protein A 36 kDa
subunit)
Length = 273
Score = 83.2 bits (204), Expect = 5e-16
Identities = 55/201 (27%), Positives = 95/201 (46%), Gaps = 15/201 (7%)
Query: 9 SSSGGGGFTSTQLNDSSPAPQKGRESQGLVPVTVKQINEASQSGDEKSSFVINGVELTNV 68
SS GGGF +++ S + L PVT+KQI E+ Q + FV + EL +V
Sbjct: 11 SSVTGGGFENSESRPGSGESETNTRVNTLTPVTIKQILESKQDIQD-GPFVSHNQELHHV 69
Query: 69 TLVGMVFEKAERNTDVNFVLDDGTGRIKCRRWINDAFD--------------TQEVEEIM 114
VG+V + ++ ++DGTG+I+ R+W DA D +Q ++
Sbjct: 70 CFVGVVRNITDHTANIFLTIEDGTGQIEVRKWSEDANDLAAGNDDSSGKGYGSQVAQQFE 129
Query: 115 NGMYVRVNGHLKSFQGVRQVAAFSVRPVVNFDEIPFHFIDCIHHHLRSKLKMEGTISTDP 174
G YV+V G LK F G + + ++P+ +F+E+ H ++ I H + M+ + +
Sbjct: 130 IGGYVKVFGALKEFGGKKNIQYAVIKPIDSFNEVLTHHLEVIKCHSIASGMMKQPLESAS 189
Query: 175 PASDSSLKTPVRNTSNGSQAP 195
+ SL N ++ +P
Sbjct: 190 NNNGQSLFVKDDNDTSSGSSP 210
>RFA2_RAT (Q63528) Replication protein A 32 kDa subunit (RP-A)
(RF-A) (Replication factor-A protein 2) (Fragment)
Length = 266
Score = 80.9 bits (198), Expect = 2e-15
Identities = 56/217 (25%), Positives = 106/217 (48%), Gaps = 13/217 (5%)
Query: 7 FDSSSGG--GGFTSTQLNDSSPAPQKGRE-----SQGLVPVTVKQINEASQSGDEKSSFV 59
F SSS G GG+T + SP P + + +Q +VP T+ Q+ A+ + + F
Sbjct: 5 FSSSSYGAAGGYTQSPGGFGSPTPSQAEKKSRARAQHIVPCTISQLLSATLTDEV---FK 61
Query: 60 INGVELTNVTLVGMVFEKAERNTDVNFVLDDGTGR-IKCRRWINDAFDTQEVEEIMNGMY 118
I VE++ VT+VG++ + T++ + +DD T + R+W++ + E + Y
Sbjct: 62 IGDVEISQVTIVGIIRHAEKAPTNIVYKIDDMTAAPMDVRQWVDTDDTSGENTVVPPETY 121
Query: 119 VRVNGHLKSFQGVRQVAAFSVRPVVNFDEIPFHFIDCIHHHLR-SKLKMEGTISTDPPAS 177
V+V GHL+SFQ + + AF + P+ + +E H ++ ++ HL SK + ++ P S
Sbjct: 122 VKVAGHLRSFQNKKSLVAFKIIPLEDMNEFTAHILEVVNSHLMLSKANSQASVGR-PSMS 180
Query: 178 DSSLKTPVRNTSNGSQAPSSTPAYAQYGVDGLKDCDK 214
+ + P + N + ++ +K C +
Sbjct: 181 NPGMGEPGNFSGNNFMPANGLTVVQNQVLNLIKACPR 217
>RFA2_HUMAN (P15927) Replication protein A 32 kDa subunit (RP-A)
(RF-A) (Replication factor-A protein 2)
Length = 270
Score = 80.9 bits (198), Expect = 2e-15
Identities = 52/198 (26%), Positives = 97/198 (48%), Gaps = 14/198 (7%)
Query: 9 SSSGGGGFTSTQLNDSSPAPQKGRE-----SQGLVPVTVKQINEASQSGDEKSSFVINGV 63
S G GG+T + SPAP + + +Q +VP T+ Q+ A+ + F I V
Sbjct: 13 SYGGAGGYTQSPGGFGSPAPSQAEKKSRARAQHIVPCTISQLLSATLVDEV---FRIGNV 69
Query: 64 ELTNVTLVGMVFEKAERNTDVNFVLDDGTGR-IKCRRWINDAFDTQEVEEIMNGMYVRVN 122
E++ VT+VG++ + T++ + +DD T + R+W++ + E + YV+V
Sbjct: 70 EISQVTIVGIIRHAEKAPTNIVYKIDDMTAAPMDVRQWVDTDDTSSENTVVPPETYVKVA 129
Query: 123 GHLKSFQGVRQVAAFSVRPVVNFDEIPFHFIDCIHHHLRSKLKMEGTISTDPPASDSSLK 182
GHL+SFQ + + AF + P+ + +E H ++ I+ H+ + ++ P A + +
Sbjct: 130 GHLRSFQNKKSLVAFKIMPLEDMNEFTTHILEVINAHM-----VLSKANSQPSAGRAPIS 184
Query: 183 TPVRNTSNGSQAPSSTPA 200
P + + S PA
Sbjct: 185 NPGMSEAGNFGGNSFMPA 202
>RFA2_MOUSE (Q62193) Replication protein A 32 kDa subunit (RP-A)
(RF-A) (Replication factor-A protein 2)
Length = 270
Score = 75.9 bits (185), Expect = 7e-14
Identities = 46/162 (28%), Positives = 87/162 (53%), Gaps = 11/162 (6%)
Query: 7 FDSSS--GGGGFTSTQLNDSSPAPQKGRE-----SQGLVPVTVKQINEASQSGDEKSSFV 59
F SS+ G GG+T + SP P + + +Q +VP T+ Q+ A+ + + F
Sbjct: 9 FTSSTYGGRGGYTQSPGGFGSPTPSQAEKKSRVRAQHIVPCTISQLLSATLTDEV---FR 65
Query: 60 INGVELTNVTLVGMVFEKAERNTDVNFVLDDGTGR-IKCRRWINDAFDTQEVEEIMNGMY 118
I VE++ VT+VG++ + T++ + +DD T + R+W++ + E + Y
Sbjct: 66 IGDVEISQVTIVGIIRHAEKAPTNIVYKIDDMTAPPMDVRQWVDTDDASGENAVVPPETY 125
Query: 119 VRVNGHLKSFQGVRQVAAFSVRPVVNFDEIPFHFIDCIHHHL 160
V+V GHL+SFQ + + AF + P+ + +E H ++ ++ H+
Sbjct: 126 VKVAGHLRSFQNKKSLVAFKIIPLEDMNEFTAHILEVVNSHM 167
>RFA4_HUMAN (Q13156) Replication protein A 30 kDa subunit (RP-A)
(RF-A) (Replication factor-A protein 4)
Length = 261
Score = 57.4 bits (137), Expect = 3e-08
Identities = 41/176 (23%), Positives = 86/176 (48%), Gaps = 9/176 (5%)
Query: 3 SVTQFDSSSGGGGFTSTQL--NDSSPAPQKGRESQGLVPVTVKQINEASQSGDEKSSFVI 60
S++ D +SGG S QL D++PA + R + V +N+ S F +
Sbjct: 11 SISAADGASGG----SDQLCERDATPAIKTQRPKVRIQDVVPCNVNQLLSSTVFDPVFKV 66
Query: 61 NGVELTNVTLVGMVFEKAERNTDVNFVLDDGTGR-IKCRRWINDAFDTQEVEEIMNGMYV 119
G+ ++ V++VG++ + + + + +DD T + I+ R+W ++V + G+YV
Sbjct: 67 RGIIVSQVSIVGVIRGAEKASNHICYKIDDMTAKPIEARQWFGRE-KVKQVTPLSVGVYV 125
Query: 120 RVNGHLKSFQGVRQVAAFSVRPVVNFDEIPFHFIDCIHHHLR-SKLKMEGTISTDP 174
+V G LK G + + + + + +E H ++ ++ H+ K + + T+ + P
Sbjct: 126 KVFGILKCPTGTKSLEVLKIHVLEDMNEFTVHILETVNAHMMLDKARRDTTVESVP 181
>RFA2_CRIFA (Q23697) Replication protein A 28 kDa subunit (RP-A)
(RF-A) (Replication factor-A protein 2)
Length = 258
Score = 53.5 bits (127), Expect = 4e-07
Identities = 52/196 (26%), Positives = 89/196 (44%), Gaps = 20/196 (10%)
Query: 9 SSSGGGGFTSTQLNDSSPAPQKGRESQGLVPVTVKQINEASQSGDEKSSFVINGVELTNV 68
+S G F++ ++ Q+ R+ + P+T+KQ+ EA G V++G E+T
Sbjct: 3 ASQAGSNFSAAASSNGGQQQQQRRQHP-IRPLTIKQMLEAQSVGG--GVMVVDGREVTQA 59
Query: 69 TLVGMV--FEKAERNT--------DVNFVLDDGTGRIKCRRWINDAFDTQEVEEIMNGMY 118
T+VG V +E A + + + D TG I R+WI DA Q E I +
Sbjct: 60 TVVGRVVGYENANMASGGGAITAKHFGYRITDNTGMIVVRQWI-DADRAQ--EPIPLNTH 116
Query: 119 VRVNGHLKSFQGVRQVAAFSVRPVVNFDEIPFHFIDCIHHHLR---SKLKMEGTISTDPP 175
VR +G + +Q + +V + + +E+ +H +D I HLR + G I +
Sbjct: 117 VRASGTVNVWQ-QSPIVTGTVVSMADSNEMNYHMLDAILTHLRLTQGNKRAAGNIGSGAS 175
Query: 176 ASDSSLKTPVRNTSNG 191
+S+ V+N G
Sbjct: 176 VQNSAAAVGVQNMLPG 191
>DPO3_STRA5 (P63984) DNA polymerase III polC-type (EC 2.7.7.7)
(PolIII)
Length = 1468
Score = 37.4 bits (85), Expect = 0.029
Identities = 28/116 (24%), Positives = 55/116 (47%), Gaps = 13/116 (11%)
Query: 21 LNDSSPAPQKGRESQGLVPVTVKQINEASQSGDEKSSFVINGVELTN----VTLVGMVFE 76
L DS+P ++ +Q K+ + Q+G EK+ + +E+T + GMVF
Sbjct: 200 LEDSAPPSEEVTPTQNY---DFKERIKQRQAGFEKAE-ITPMIEVTTEENRIVFEGMVFS 255
Query: 77 KAERNTD-----VNFVLDDGTGRIKCRRWINDAFDTQEVEEIMNGMYVRVNGHLKS 127
+ T +NF + D T ++W D + ++ + I G ++RV G++++
Sbjct: 256 VERKTTRTGRHIINFKMTDYTSSFAMQKWAKDDEELKKYDMISKGSWLRVRGNIEN 311
>DPO3_STRA3 (P63983) DNA polymerase III polC-type (EC 2.7.7.7)
(PolIII)
Length = 1468
Score = 37.4 bits (85), Expect = 0.029
Identities = 28/116 (24%), Positives = 55/116 (47%), Gaps = 13/116 (11%)
Query: 21 LNDSSPAPQKGRESQGLVPVTVKQINEASQSGDEKSSFVINGVELTN----VTLVGMVFE 76
L DS+P ++ +Q K+ + Q+G EK+ + +E+T + GMVF
Sbjct: 200 LEDSAPPSEEVTPTQNY---DFKERIKQRQAGFEKAE-ITPMIEVTTEENRIVFEGMVFS 255
Query: 77 KAERNTD-----VNFVLDDGTGRIKCRRWINDAFDTQEVEEIMNGMYVRVNGHLKS 127
+ T +NF + D T ++W D + ++ + I G ++RV G++++
Sbjct: 256 VERKTTRTGRHIINFKMTDYTSSFAMQKWAKDDEELKKYDMISKGSWLRVRGNIEN 311
>DPO3_STRP3 (Q8K5S8) DNA polymerase III polC-type (EC 2.7.7.7)
(PolIII)
Length = 1465
Score = 36.6 bits (83), Expect = 0.049
Identities = 30/127 (23%), Positives = 55/127 (42%), Gaps = 20/127 (15%)
Query: 50 QSGDEKSSF---VINGVELTNVTLVGMVFEKAERNTD-----VNFVLDDGTGRIKCRRWI 101
Q+G EK++ + E + GMVF+ + T +NF + D T ++W
Sbjct: 223 QAGFEKATITPMIEIETEENRIVFEGMVFDVERKTTRTGRHIINFKMTDYTSSFALQKWA 282
Query: 102 NDAFDTQEVEEIMNGMYVRVNGHLKSFQGVRQVAAFSVRPVVNFDEIPFHFIDCIHHHLR 161
D + ++ + I G+++RV G++ + F+ +N ++ I HH R
Sbjct: 283 KDDEELRKFDMIAKGVWLRVQGNI-------ETNPFTKSLTMNVQQVKE-----IVHHER 330
Query: 162 SKLKMEG 168
L EG
Sbjct: 331 KDLMPEG 337
>DPO3_STRPY (Q9FDF9) DNA polymerase III polC-type (EC 2.7.7.7)
(PolIII)
Length = 1465
Score = 35.4 bits (80), Expect = 0.11
Identities = 21/86 (24%), Positives = 42/86 (48%), Gaps = 8/86 (9%)
Query: 50 QSGDEKSSF---VINGVELTNVTLVGMVFEKAERNTD-----VNFVLDDGTGRIKCRRWI 101
Q+G EK++ + E + GMVF+ + T +NF + D T ++W
Sbjct: 223 QAGFEKATITPMIEIETEENRIVFEGMVFDVERKTTRTGRHIINFKMTDYTSSFALQKWA 282
Query: 102 NDAFDTQEVEEIMNGMYVRVNGHLKS 127
D + ++ + I G ++RV G++++
Sbjct: 283 KDDEELRKFDMIAKGAWLRVQGNIET 308
>DPO3_STRP8 (Q8NZB5) DNA polymerase III polC-type (EC 2.7.7.7)
(PolIII)
Length = 1465
Score = 35.4 bits (80), Expect = 0.11
Identities = 21/86 (24%), Positives = 42/86 (48%), Gaps = 8/86 (9%)
Query: 50 QSGDEKSSF---VINGVELTNVTLVGMVFEKAERNTD-----VNFVLDDGTGRIKCRRWI 101
Q+G EK++ + E + GMVF+ + T +NF + D T ++W
Sbjct: 223 QAGFEKATITPMIEIETEENRIVFEGMVFDVERKTTRTGRHIINFKMTDYTSSFALQKWA 282
Query: 102 NDAFDTQEVEEIMNGMYVRVNGHLKS 127
D + ++ + I G ++RV G++++
Sbjct: 283 KDDEELRKFDMIAKGAWLRVQGNIET 308
>DPO3_STRMU (Q8DWE0) DNA polymerase III polC-type (EC 2.7.7.7)
(PolIII)
Length = 1465
Score = 35.4 bits (80), Expect = 0.11
Identities = 17/69 (24%), Positives = 35/69 (50%), Gaps = 5/69 (7%)
Query: 64 ELTNVTLVGMVFEKAERNTD-----VNFVLDDGTGRIKCRRWINDAFDTQEVEEIMNGMY 118
E +T G+VF+ + T +NF + D T ++W D + ++ + I G +
Sbjct: 238 EENRITFEGLVFDVERKTTRTGRHIINFKMTDYTSSFPMQKWAKDDEELKKYDMISKGAW 297
Query: 119 VRVNGHLKS 127
+RV G++++
Sbjct: 298 LRVRGNIEN 306
>RPBC_HELPY (O25806) Bifunctional DNA-directed RNA polymerase, beta
and beta' chain (EC 2.7.7.6) [Includes: DNA-directed RNA
polymerase beta chain (Transcriptase beta chain) (RNA
polymerase beta subunit); DNA-directed RNA polymerase
beta' chain (Transcrip
Length = 2890
Score = 34.7 bits (78), Expect = 0.19
Identities = 22/77 (28%), Positives = 41/77 (52%), Gaps = 10/77 (12%)
Query: 54 EKSSFVINGVELTNVTLV----GMVFEKAERNTDVNFVLDDGTGRIKCRRWINDAFDTQE 109
E++SF+INGVE V + G++F++ E +T +N ++ G W+ +D+++
Sbjct: 135 ERTSFIINGVERVVVNQLHRSPGVIFKEEESSTSLNKLIYTGQIIPDRGSWLYFEYDSKD 194
Query: 110 VEEIMNGMYVRVNGHLK 126
V +Y R+N K
Sbjct: 195 V------LYARINKRRK 205
>RPBC_HELPJ (Q9ZK23) Bifunctional DNA-directed RNA polymerase, beta
and beta' chain (EC 2.7.7.6) [Includes: DNA-directed RNA
polymerase beta chain (Transcriptase beta chain) (RNA
polymerase beta subunit); DNA-directed RNA polymerase
beta' chain (Transcrip
Length = 2890
Score = 33.5 bits (75), Expect = 0.41
Identities = 22/77 (28%), Positives = 40/77 (51%), Gaps = 10/77 (12%)
Query: 54 EKSSFVINGVELTNVTLV----GMVFEKAERNTDVNFVLDDGTGRIKCRRWINDAFDTQE 109
E++SF+INGVE V + G++F++ E +T N ++ G W+ +D+++
Sbjct: 135 ERTSFIINGVERVVVNQLYRSPGVIFKEEESSTSSNKLIYTGQIIPDRGSWLYFEYDSKD 194
Query: 110 VEEIMNGMYVRVNGHLK 126
V +Y R+N K
Sbjct: 195 V------LYARINKRRK 205
>NA22_ARATH (Q84TE6) NAC-domain containing protein 21/22 (ANAC021)
(ANAC022)
Length = 324
Score = 31.6 bits (70), Expect = 1.6
Identities = 23/88 (26%), Positives = 41/88 (46%), Gaps = 17/88 (19%)
Query: 141 PVVNFDEIPFHFIDCIHHHL-------RSKLKMEGTISTDPPASDSSLKTPVRNT----S 189
P +NFD+ P ++ HH++ S L T++++ S S LK P +N +
Sbjct: 200 PYINFDQEPSSYLSDDHHYIINEHVPCFSNLSQNQTLNSNLTNSVSELKIPCKNPNPLFT 259
Query: 190 NGSQAPSSTPAYAQYGVDGLKDCDKLVI 217
GS + + T G+D D++V+
Sbjct: 260 GGSASATLT------GLDSFCSSDQMVL 281
>DPO3_STRR6 (Q8DRA5) DNA polymerase III polC-type (EC 2.7.7.7)
(PolIII)
Length = 1463
Score = 31.6 bits (70), Expect = 1.6
Identities = 24/116 (20%), Positives = 52/116 (44%), Gaps = 15/116 (12%)
Query: 20 QLNDSSPAPQKGRESQGLVPVTVKQINEASQSGDEKSSFVINGVELTN----VTLVGMVF 75
QL +P P + + PV Q +A+ + + +E+T + G+VF
Sbjct: 196 QLEQMAPPPAEEK------PVFDFQAKKAAAKPKLDKAEITPMIEVTTEENRLVFEGVVF 249
Query: 76 EKAERNTD-----VNFVLDDGTGRIKCRRWINDAFDTQEVEEIMNGMYVRVNGHLK 126
+ ++ T +NF + D T ++W+ + + Q+ + I ++RV G+++
Sbjct: 250 DVEQKVTRTGRVLINFKMTDYTSSFSMQKWVKNEEEAQKFDLIKKNSWLRVRGNVE 305
>DP2S_ARCFU (O28484) DNA polymerase II small subunit (EC 2.7.7.7)
(Pol II)
Length = 488
Score = 31.2 bits (69), Expect = 2.1
Identities = 20/60 (33%), Positives = 29/60 (48%), Gaps = 3/60 (5%)
Query: 67 NVTLVGMVFEKAERNTDVNFVLDDGTGRIKCRRWINDAFDTQEVEEIMNGMYVRVNGHLK 126
NV++VGMV E ER L+D TG I C + + + E++ + V G LK
Sbjct: 145 NVSVVGMVNEVYERGDKCYIRLEDTTGTITC---VATGKNAEVARELLGDEVIGVTGLLK 201
>RPBC_WOLSU (Q7MA56) Bifunctional DNA-directed RNA polymerase, beta
and beta' chain (EC 2.7.7.6) [Includes: DNA-directed RNA
polymerase beta chain (Transcriptase beta chain) (RNA
polymerase beta subunit); DNA-directed RNA polymerase
beta' chain (Transcrip
Length = 2883
Score = 30.8 bits (68), Expect = 2.7
Identities = 21/77 (27%), Positives = 38/77 (49%), Gaps = 10/77 (12%)
Query: 54 EKSSFVINGVELTNVTLV----GMVFEKAERNTDVNFVLDDGTGRIKCRRWINDAFDTQE 109
+++SF+INGVE V + G++F++ E T N ++ G W+ +D ++
Sbjct: 135 DRTSFIINGVERVVVNQLHRSPGVIFKEEESATSSNKLIYTGQIIPDRGSWLYFEYDAKD 194
Query: 110 VEEIMNGMYVRVNGHLK 126
+YVR+N K
Sbjct: 195 T------LYVRINKRRK 205
>KNOB_PLAFG (P09346) Knob-associated histidine-rich protein
precursor (KAHRP) (KP)
Length = 634
Score = 30.8 bits (68), Expect = 2.7
Identities = 20/66 (30%), Positives = 33/66 (49%), Gaps = 9/66 (13%)
Query: 157 HHHLRSKLKMEGTISTDPPASDSSLKTPV-RNTSNGSQAPSSTPAY-------AQYGVDG 208
HHH +++GT++ +PP+++ +KT V R G + Y + VDG
Sbjct: 114 HHHQLQPQQLQGTVA-NPPSNEPVVKTQVFREARPGGGFKAYEEKYESKHYKLKENVVDG 172
Query: 209 LKDCDK 214
KDCD+
Sbjct: 173 KKDCDE 178
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.132 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,668,145
Number of Sequences: 164201
Number of extensions: 1093784
Number of successful extensions: 3085
Number of sequences better than 10.0: 33
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 3056
Number of HSP's gapped (non-prelim): 35
length of query: 224
length of database: 59,974,054
effective HSP length: 106
effective length of query: 118
effective length of database: 42,568,748
effective search space: 5023112264
effective search space used: 5023112264
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0072a.1


