

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
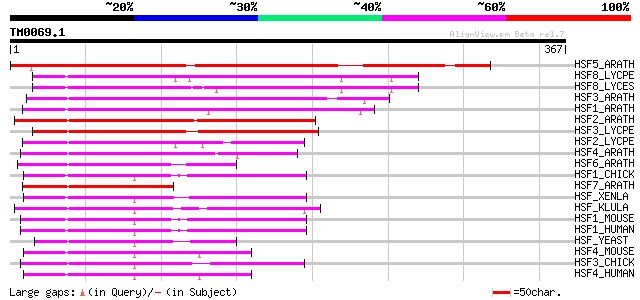
Query= TM0069.1
(367 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
HSF5_ARATH (Q9S7U5) Heat shock factor protein 5 (HSF 5) (Heat sh... 277 3e-74
HSF8_LYCPE (P41153) Heat shock factor protein HSF8 (Heat shock t... 222 1e-57
HSF8_LYCES (Q40152) Heat shock factor protein HSF8 (Heat shock t... 219 7e-57
HSF3_ARATH (O81821) Heat shock factor protein 3 (HSF 3) (Heat sh... 204 3e-52
HSF1_ARATH (P41151) Heat shock factor protein 1 (HSF 1) (Heat sh... 202 1e-51
HSF2_ARATH (Q9SCW5) Heat shock factor protein 2 (HSF 2) (Heat sh... 190 6e-48
HSF3_LYCPE (P41152) Heat shock factor protein HSF30 (Heat shock ... 176 8e-44
HSF2_LYCPE (P22335) Heat shock factor protein HSF24 (Heat shock ... 127 3e-29
HSF4_ARATH (Q96320) Heat shock factor protein 4 (HSF 4) (Heat sh... 126 8e-29
HSF6_ARATH (Q9SCW4) Heat shock factor protein 6 (HSF 6) (Heat sh... 120 5e-27
HSF1_CHICK (P38529) Heat shock factor protein 1 (HSF 1) (Heat sh... 116 1e-25
HSF7_ARATH (Q9T0D3) Heat shock factor protein 7 (HSF 7) (Heat sh... 114 4e-25
HSF_XENLA (P41154) Heat shock factor protein (HSF) (Heat shock t... 108 2e-23
HSF_KLULA (P22121) Heat shock factor protein (HSF) (Heat shock t... 107 5e-23
HSF1_MOUSE (P38532) Heat shock factor protein 1 (HSF 1) (Heat sh... 107 5e-23
HSF1_HUMAN (Q00613) Heat shock factor protein 1 (HSF 1) (Heat sh... 107 5e-23
HSF_YEAST (P10961) Heat shock factor protein (HSF) (Heat shock t... 99 1e-20
HSF4_MOUSE (Q9R0L1) Heat shock factor protein 4 (HSF 4) (Heat sh... 99 1e-20
HSF3_CHICK (P38531) Heat shock factor protein 3 (HSF 3) (Heat sh... 99 2e-20
HSF4_HUMAN (Q9ULV5) Heat shock factor protein 4 (HSF 4) (Heat sh... 97 5e-20
>HSF5_ARATH (Q9S7U5) Heat shock factor protein 5 (HSF 5) (Heat shock
transcription factor 5) (HSTF 5)
Length = 374
Score = 277 bits (709), Expect = 3e-74
Identities = 157/323 (48%), Positives = 204/323 (62%), Gaps = 32/323 (9%)
Query: 1 MVKSKENASGSSS---VAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLL 57
MVKS + GSSS VAPFL KCYDMV+D +TDSIISW+ + ++FVI D T FSV LL
Sbjct: 1 MVKSTDGGGGSSSSSSVAPFLRKCYDMVDDSTTDSIISWSPSADNSFVILDTTVFSVQLL 60
Query: 58 PTYFKHNNFASFIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQK 117
P YFKH+NF+SFIRQLNIYGFRKVD DRWEFAN+ FVRGQK LLKN+ RRK+ ++Q K
Sbjct: 61 PKYFKHSNFSSFIRQLNIYGFRKVDADRWEFANDGFVRGQKDLLKNVIRRKNVQSSEQSK 120
Query: 118 ALPEHNNSDEPSREAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQ 177
+ S + GL KEV+ LK D+ L QEL+ + Q+ E ++KML L DR+Q
Sbjct: 121 -----HESTSTTYAQEKSGLWKEVDILKGDKQVLAQELIKVRQYQEVTDTKMLHLEDRVQ 175
Query: 178 GMEKHQQQMLSFLVMVVQSPGFMVQLLHPKE-NSWRLAEAGNMFDPGKEDDKPVASDGMI 236
GME+ QQ+MLSFLVMV+++P +VQLL PKE N+WR A G I
Sbjct: 176 GMEESQQEMLSFLVMVMKNPSLLVQLLQPKEKNTWRKAGEG----------------AKI 219
Query: 237 VQYKPPVGEKRKHVIP-IPLSPGFDRQPEPELSADRLKDLCISSEFLKVLLDEKVSPLDN 295
V+ GE + +P + P D + +++ + D +++ LK LDE N
Sbjct: 220 VEEVTDEGESNSYGLPLVTYQPPSDNNGTAKSNSNDVNDFLRNADMLKFCLDE------N 273
Query: 296 HSPFLLPDLPDDGSWEQLFLGSP 318
H P ++PDL DDG+WE+L L SP
Sbjct: 274 HVPLIIPDLYDDGAWEKLLLLSP 296
>HSF8_LYCPE (P41153) Heat shock factor protein HSF8 (Heat shock
transcription factor 8) (HSTF 8) (Heat stress
transcription factor)
Length = 527
Score = 222 bits (565), Expect = 1e-57
Identities = 129/278 (46%), Positives = 165/278 (58%), Gaps = 24/278 (8%)
Query: 16 PFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASFIRQLNI 75
PFL K YDMV+D STD I+SW+ P+ ++FV+ D F+ LLP YFKHNNF+SF+RQLN
Sbjct: 41 PFLVKTYDMVDDPSTDKIVSWS-PTNNSFVVWDPPEFAKDLLPKYFKHNNFSSFVRQLNT 99
Query: 76 YGFRKVDTDRWEFANENFVRGQKHLLKNIRRRK--HPHVTDQQK---------ALPEHNN 124
YGFRKVD DRWEFANE F+RGQKHLLK+I RRK H H QQ+ P H+
Sbjct: 100 YGFRKVDPDRWEFANEGFLRGQKHLLKSISRRKPAHGHAQQQQQPHGHAQQQMQPPGHSA 159
Query: 125 SDEPSREAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQQ 184
S E GL +EVE LK D+N LMQELV L Q +S ++++ + RLQGME QQ
Sbjct: 160 SVGACVEVGKFGLEEEVERLKRDKNVLMQELVRLRQQQQSTDNQLQGMVQRLQGMELRQQ 219
Query: 185 QMLSFLVMVVQSPGFMVQLLHPK-ENSWRLAEAGN----MFDPGKEDDKPVASDGMIVQY 239
QM+SFL V SPGF+ Q + + E++ R+AE D +D +DG IV+Y
Sbjct: 220 QMMSFLAKAVNSPGFLAQFVQQQNESNKRIAEGSKKRRIKQDIESQDPSVTPADGQIVKY 279
Query: 240 KPPVGEKRKHVI-------PIPLSPGFDRQPEPELSAD 270
+P + E K ++ P F PE L D
Sbjct: 280 QPGINEAAKAMLRELSKLDSSPRLENFSNSPESFLIGD 317
>HSF8_LYCES (Q40152) Heat shock factor protein HSF8 (Heat shock
transcription factor 8) (HSTF 8) (Heat stress
transcription factor)
Length = 527
Score = 219 bits (559), Expect = 7e-57
Identities = 127/280 (45%), Positives = 166/280 (58%), Gaps = 28/280 (10%)
Query: 16 PFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASFIRQLNI 75
PFL K YDMV+D STD I+SW+ P+ ++FV+ D F+ LLP YFKHNNF+SF+RQLN
Sbjct: 39 PFLVKTYDMVDDPSTDKIVSWS-PTNNSFVVWDPPEFAKDLLPKYFKHNNFSSFVRQLNT 97
Query: 76 YGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNSDEPSREAPNH 135
YGFRKVD DRWEFANE F+RGQKHLLK+I RRK H QQ+ P H N+ + + P H
Sbjct: 98 YGFRKVDPDRWEFANEGFLRGQKHLLKSISRRKPAHGHAQQQQQP-HGNAQQ-QMQPPGH 155
Query: 136 -------------GLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKH 182
GL +EVE LK D+N LMQELV L Q ++ ++++ + RLQGME
Sbjct: 156 SASVGACVEVGKFGLEEEVERLKRDKNVLMQELVRLRQQQQATDNQLQGMVQRLQGMELR 215
Query: 183 QQQMLSFLVMVVQSPGFMVQLLHPK-ENSWRLAEAGN----MFDPGKEDDKPVASDGMIV 237
QQQM+SFL V PGF+ Q + + E++ R+AE D +D +DG IV
Sbjct: 216 QQQMMSFLAKAVNRPGFLAQFVQQQNESNKRIAEGSKKRRIKQDIESQDPSVTPADGQIV 275
Query: 238 QYKPPVGEKRKHVI-------PIPLSPGFDRQPEPELSAD 270
+Y+P + E K ++ P F PE L D
Sbjct: 276 KYQPGINEAAKAMLRELSKLDSSPRLDNFSNSPESFLIGD 315
>HSF3_ARATH (O81821) Heat shock factor protein 3 (HSF 3) (Heat shock
transcription factor 3) (HSTF 3)
Length = 481
Score = 204 bits (519), Expect = 3e-52
Identities = 111/254 (43%), Positives = 153/254 (59%), Gaps = 21/254 (8%)
Query: 12 SSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASFIR 71
+SV PFL+K YDMV+D T+ ++SW+ ++FV+ FS LLP YFKHNNF+SF+R
Sbjct: 23 NSVPPFLSKTYDMVDDPLTNEVVSWSS-GNNSFVVWSAPEFSKVLLPKYFKHNNFSSFVR 81
Query: 72 QLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNSDEPSRE 131
QLN YGFRKVD DRWEFANE F+RG+K LLK+I RRK HV Q+ ++S E
Sbjct: 82 QLNTYGFRKVDPDRWEFANEGFLRGRKQLLKSIVRRKPSHVQQNQQQTQVQSSSVGACVE 141
Query: 132 APNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQQQMLSFLV 191
G+ +EVE LK D+N LMQELV L Q ++ E+++ + ++Q ME+ QQQM+SFL
Sbjct: 142 VGKFGIEEEVERLKRDKNVLMQELVRLRQQQQATENQLQNVGQKVQVMEQRQQQMMSFLA 201
Query: 192 MVVQSPGFMVQLLHPKENSWRLAEAGNMFDPGKEDDKPVASD--------------GMIV 237
VQSPGF+ QL+ N GN PG + + D IV
Sbjct: 202 KAVQSPGFLNQLVQQNNND------GNRQIPGSNKKRRLPVDEQENRGDNVANGLNRQIV 255
Query: 238 QYKPPVGEKRKHVI 251
+Y+P + E ++++
Sbjct: 256 RYQPSINEAAQNML 269
>HSF1_ARATH (P41151) Heat shock factor protein 1 (HSF 1) (Heat shock
transcription factor 1) (HSTF 1)
Length = 495
Score = 202 bits (514), Expect = 1e-51
Identities = 114/253 (45%), Positives = 150/253 (59%), Gaps = 21/253 (8%)
Query: 9 SGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFAS 68
+ +S PFL+K YDMVED +TD+I+SW+ P+ ++F++ D FS LLP YFKHNNF+S
Sbjct: 45 NANSLPPPFLSKTYDMVEDPATDAIVSWS-PTNNSFIVWDPPEFSRDLLPKYFKHNNFSS 103
Query: 69 FIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNSDEP 128
F+RQLN YGFRKVD DRWEFANE F+RGQKHLLK I RRK + P+ +
Sbjct: 104 FVRQLNTYGFRKVDPDRWEFANEGFLRGQKHLLKKISRRKSVQGHGSSSSNPQSQQLSQG 163
Query: 129 SR---------EAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGM 179
E GL +EVE LK D+N LMQELV L Q ++ ++K+ VL LQ M
Sbjct: 164 QGSMAALSSCVEVGKFGLEEEVEQLKRDKNVLMQELVKLRQQQQTTDNKLQVLVKHLQVM 223
Query: 180 EKHQQQMLSFLVMVVQSPGFMVQLLHPKENSWRLAEAGNMFDPGKEDDKPV--------- 230
E+ QQQ++SFL VQ+P F+ Q + + +S N +ED
Sbjct: 224 EQRQQQIMSFLAKAVQNPTFLSQFIQKQTDSNMHVTEANKKRRLREDSTAATESNSHSHS 283
Query: 231 --ASDGMIVQYKP 241
ASDG IV+Y+P
Sbjct: 284 LEASDGQIVKYQP 296
>HSF2_ARATH (Q9SCW5) Heat shock factor protein 2 (HSF 2) (Heat shock
transcription factor 2) (HSTF 2)
Length = 468
Score = 190 bits (482), Expect = 6e-48
Identities = 98/199 (49%), Positives = 132/199 (66%), Gaps = 2/199 (1%)
Query: 4 SKENASGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKH 63
+K + + SS+ PFL+K YDMV+D TD ++SW+ ++FV+ ++ F+ LP YFKH
Sbjct: 11 AKSSTAVMSSIPPFLSKTYDMVDDPLTDDVVSWSS-GNNSFVVWNVPEFAKQFLPKYFKH 69
Query: 64 NNFASFIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHN 123
NNF+SF+RQLN YGFRKVD DRWEFANE F+RGQK +LK+I RRK V Q+ +H
Sbjct: 70 NNFSSFVRQLNTYGFRKVDPDRWEFANEGFLRGQKQILKSIVRRKPAQVQPPQQPQVQH- 128
Query: 124 NSDEPSREAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQ 183
+S E GL +EVE L+ D+N LMQELV L Q + E + + ++ ME+ Q
Sbjct: 129 SSVGACVEVGKFGLEEEVERLQRDKNVLMQELVRLRQQQQVTEHHLQNVGQKVHVMEQRQ 188
Query: 184 QQMLSFLVMVVQSPGFMVQ 202
QQM+SFL VQSPGF+ Q
Sbjct: 189 QQMMSFLAKAVQSPGFLNQ 207
>HSF3_LYCPE (P41152) Heat shock factor protein HSF30 (Heat shock
transcription factor 30) (HSTF 30) (Heat stress
transcription factor)
Length = 351
Score = 176 bits (446), Expect = 8e-44
Identities = 88/189 (46%), Positives = 127/189 (66%), Gaps = 8/189 (4%)
Query: 16 PFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASFIRQLNI 75
PFL+K Y+MVED STD +ISW+ + ++F++ D FS TLLP +FKH+NF+SFIRQLN
Sbjct: 31 PFLSKTYEMVEDSSTDQVISWST-TRNSFIVWDSHKFSTTLLPRFFKHSNFSSFIRQLNT 89
Query: 76 YGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNSDEPSREAPNH 135
YGFRKVD DRWEFANE F+ GQKHLLK I+RR++ + Q+ E +
Sbjct: 90 YGFRKVDPDRWEFANEGFLGGQKHLLKTIKRRRNVGQSMNQQ-------GSGACIEIGYY 142
Query: 136 GLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQQQMLSFLVMVVQ 195
G+ +E+E LK D+N LM E+V L Q +S ++++ + ++++ E+ Q QM+SFL +
Sbjct: 143 GMEEELERLKRDKNVLMTEIVKLRQQQQSTRNQIIAMGEKIETQERKQVQMMSFLAKIFS 202
Query: 196 SPGFMVQLL 204
+P F+ Q L
Sbjct: 203 NPTFLQQYL 211
>HSF2_LYCPE (P22335) Heat shock factor protein HSF24 (Heat shock
transcription factor 24) (HSTF 24) (Heat stress
transcription factor)
Length = 301
Score = 127 bits (320), Expect = 3e-29
Identities = 79/197 (40%), Positives = 106/197 (53%), Gaps = 16/197 (8%)
Query: 9 SGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFAS 68
S ++ APFL K Y +V+D +TD +ISW E G TFV+ F+ LLP YFKHNNF+S
Sbjct: 2 SQRTAPAPFLLKTYQLVDDAATDDVISWNE-IGTTFVVWKTAEFAKDLLPKYFKHNNFSS 60
Query: 69 FIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRK--------HPHVTDQQKALP 120
F+RQLN YGFRK+ D+WEFANENF RGQK LL IRRRK V A P
Sbjct: 61 FVRQLNTYGFRKIVPDKWEFANENFKRGQKELLTAIRRRKTVTSTPAGGKSVAAGASASP 120
Query: 121 EHNNSD--EPSREAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQG 178
+++ D S +P+ V D + + LS E + +LS L
Sbjct: 121 DNSGDDIGSSSTSSPDSKNPGSV-----DTPGKLSQFTDLSDENEKLKKDNQMLSSELVQ 175
Query: 179 MEKHQQQMLSFLVMVVQ 195
+K ++++FL V+
Sbjct: 176 AKKQCNELVAFLSQYVK 192
>HSF4_ARATH (Q96320) Heat shock factor protein 4 (HSF 4) (Heat shock
transcription factor 4) (HSTF 4)
Length = 284
Score = 126 bits (317), Expect = 8e-29
Identities = 75/185 (40%), Positives = 105/185 (56%), Gaps = 4/185 (2%)
Query: 8 ASGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFA 67
A+ S APFL+K Y +V+D STD ++SW E G FV+ F+ LLP YFKHNNF+
Sbjct: 6 AAQRSVPAPFLSKTYQLVDDHSTDDVVSWNE-EGTAFVVWKTAEFAKDLLPQYFKHNNFS 64
Query: 68 SFIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNSDE 127
SFIRQLN YGFRK D+WEFAN+ F RG + LL +IRRRK + K + + S+
Sbjct: 65 SFIRQLNTYGFRKTVPDKWEFANDYFRRGGEDLLTDIRRRKSVIASTAGKCVVVGSPSES 124
Query: 128 PSREAPNHGLRKEVENLKSDRN--SLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQQQ 185
S +HG + S +N S+ + LS E + + LS L +K + +
Sbjct: 125 NSGGGDDHG-SSSTSSPGSSKNPGSVENMVADLSGENEKLKRENNNLSSELAAAKKQRDE 183
Query: 186 MLSFL 190
+++FL
Sbjct: 184 LVTFL 188
>HSF6_ARATH (Q9SCW4) Heat shock factor protein 6 (HSF 6) (Heat shock
transcription factor 6) (HSTF 6)
Length = 299
Score = 120 bits (301), Expect = 5e-27
Identities = 64/145 (44%), Positives = 86/145 (59%), Gaps = 10/145 (6%)
Query: 6 ENASGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNN 65
E++S S PFL K +++VED S D +ISW E G +F++ + T F+ LLP +FKHNN
Sbjct: 13 ESSSQRSIPTPFLTKTFNLVEDSSIDDVISWNE-DGSSFIVWNPTDFAKDLLPKHFKHNN 71
Query: 66 FASFIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNS 125
F+SF+RQLN YGF+KV DRWEF+N+ F RG+K LL+ I+RR K H
Sbjct: 72 FSSFVRQLNTYGFKKVVPDRWEFSNDFFKRGEKRLLREIQRR---------KITTTHQTV 122
Query: 126 DEPSREAPNHGLRKEVENLKSDRNS 150
PS E N + N D N+
Sbjct: 123 VAPSSEQRNQTMVVSPSNSGEDNNN 147
>HSF1_CHICK (P38529) Heat shock factor protein 1 (HSF 1) (Heat shock
transcription factor 1) (HSTF 1) (HSF 3A) (HSTF 3A)
Length = 491
Score = 116 bits (290), Expect = 1e-25
Identities = 70/198 (35%), Positives = 112/198 (56%), Gaps = 20/198 (10%)
Query: 10 GSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASF 69
G S+V+ FL K + +VED TD +I W+ PSG++F + D F+ +LP YFKHNN ASF
Sbjct: 16 GGSNVSAFLTKLWTLVEDPETDPLICWS-PSGNSFHVFDQGQFAKEVLPKYFKHNNMASF 74
Query: 70 IRQLNIYGFRKV-----------DTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKA 118
+RQLN+YGFRKV + D EF + F+RGQ+HLL+NI+R+ VT +
Sbjct: 75 VRQLNMYGFRKVVHIEQGGLVKPEKDDTEFQHPYFIRGQEHLLENIKRK----VT----S 126
Query: 119 LPEHNNSDEPSREAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQG 178
+ N D R+ L +++ +K + S+ +L+ + E+ ++ L +
Sbjct: 127 VSSIKNEDIKVRQDNVTKLLTDIQVMKGKQESMDSKLIAMKHENEALWREVASLRQKHAQ 186
Query: 179 MEKHQQQMLSFLVMVVQS 196
+K +++ FL+ +VQS
Sbjct: 187 QQKVVNKLIQFLISLVQS 204
>HSF7_ARATH (Q9T0D3) Heat shock factor protein 7 (HSF 7) (Heat shock
transcription factor 7) (HSTF 7)
Length = 377
Score = 114 bits (285), Expect = 4e-25
Identities = 57/100 (57%), Positives = 70/100 (70%), Gaps = 1/100 (1%)
Query: 9 SGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFAS 68
S S PFL K Y +VED D +ISW E G TF++ F+ LLP YFKHNNF+S
Sbjct: 52 SQRSIPTPFLTKTYQLVEDPVYDELISWNE-DGTTFIVWRPAEFARDLLPKYFKHNNFSS 110
Query: 69 FIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRK 108
F+RQLN YGFRKV DRWEF+N+ F RG+K LL++I+RRK
Sbjct: 111 FVRQLNTYGFRKVVPDRWEFSNDCFKRGEKILLRDIQRRK 150
>HSF_XENLA (P41154) Heat shock factor protein (HSF) (Heat shock
transcription factor) (HSTF)
Length = 451
Score = 108 bits (271), Expect = 2e-23
Identities = 69/199 (34%), Positives = 107/199 (53%), Gaps = 22/199 (11%)
Query: 10 GSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASF 69
G S+V FL K + +VED TD +I W+ P G++F + D F+ +LP YFKHNN ASF
Sbjct: 8 GGSNVPAFLAKLWTLVEDPDTDPLICWS-PEGNSFHVFDQGQFAKEVLPKYFKHNNMASF 66
Query: 70 IRQLNIYGFRKV-----------DTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKA 118
+RQLN+YGFRKV + D EF + F+RGQ+ LL+NI+R+ +
Sbjct: 67 VRQLNMYGFRKVVHIEQGGLVKPERDDTEFQHPYFIRGQEQLLENIKRKVN--------- 117
Query: 119 LPEHNNSDEPSREAPNHG-LRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQ 177
SDE + G L +V+++K + S+ L+ + E+ ++ L +
Sbjct: 118 TMSATKSDEVKVRQDSVGKLISDVQSMKGKQESIDGRLLSMKHENEALWREVASLRQKHT 177
Query: 178 GMEKHQQQMLSFLVMVVQS 196
+K +++ FLV +VQS
Sbjct: 178 QQQKVVNKLIQFLVSLVQS 196
>HSF_KLULA (P22121) Heat shock factor protein (HSF) (Heat shock
transcription factor) (HSTF)
Length = 677
Score = 107 bits (267), Expect = 5e-23
Identities = 71/220 (32%), Positives = 114/220 (51%), Gaps = 29/220 (13%)
Query: 4 SKENASGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKH 63
SK+ S + + F+NK + MV D S + I W+ SG + V+ + F +LP YFKH
Sbjct: 184 SKKKLSTTRARPAFVNKLWSMVNDKSNEKFIHWST-SGESIVVPNRERFVQEVLPKYFKH 242
Query: 64 NNFASFIRQLNIYGFRKV-----------DTDRWEFANENFVRGQKHLLKNIRRRKHPHV 112
+NFASF+RQLN+YG+ KV + RWEF NENF RG+++LL+NI R+K
Sbjct: 243 SNFASFVRQLNMYGWHKVQDVKSGSMLSNNDSRWEFENENFKRGKEYLLENIVRQK---- 298
Query: 113 TDQQKALPEHNNSDEPSREAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVL 172
L N+ E H L E+E +K ++ ++ ++L +++ E + ++
Sbjct: 299 -SNTNILGGTTNA-----EVDIHILLNELETVKYNQLAIAEDLKRITKDNEMLWKENMMA 352
Query: 173 SDRLQGMEKHQQQMLSFLVMV-------VQSPGFMVQLLH 205
+R Q ++ +++L FL V GF L+H
Sbjct: 353 RERHQSQQQVLEKLLRFLSSVFGPNSAKTIGNGFQPDLIH 392
>HSF1_MOUSE (P38532) Heat shock factor protein 1 (HSF 1) (Heat shock
transcription factor 1) (HSTF 1)
Length = 525
Score = 107 bits (267), Expect = 5e-23
Identities = 68/200 (34%), Positives = 111/200 (55%), Gaps = 20/200 (10%)
Query: 8 ASGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFA 67
A+G S+V FL K + +V D TD++I W+ PSG++F + D F+ +LP YFKHNN A
Sbjct: 9 AAGPSNVPAFLTKLWTLVSDPDTDALICWS-PSGNSFHVFDQGQFAKEVLPKYFKHNNMA 67
Query: 68 SFIRQLNIYGFRKV-----------DTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQ 116
SF+RQLN+YGFRKV + D EF + F+RGQ+ LL+NI+R+ VT
Sbjct: 68 SFVRQLNMYGFRKVVHIEQGGLVKPERDDTEFQHPCFLRGQEQLLENIKRK----VT--- 120
Query: 117 KALPEHNNSDEPSREAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRL 176
++ + D R+ L +V+ +K + + +L+ + E+ ++ L +
Sbjct: 121 -SVSTLKSEDIKIRQDSVTRLLTDVQLMKGKQECMDSKLLAMKHENEALWREVASLRQKH 179
Query: 177 QGMEKHQQQMLSFLVMVVQS 196
+K +++ FL+ +VQS
Sbjct: 180 AQQQKVVNKLIQFLISLVQS 199
>HSF1_HUMAN (Q00613) Heat shock factor protein 1 (HSF 1) (Heat shock
transcription factor 1) (HSTF 1)
Length = 529
Score = 107 bits (267), Expect = 5e-23
Identities = 68/200 (34%), Positives = 111/200 (55%), Gaps = 20/200 (10%)
Query: 8 ASGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFA 67
A+G S+V FL K + +V D TD++I W+ PSG++F + D F+ +LP YFKHNN A
Sbjct: 9 AAGPSNVPAFLTKLWTLVSDPDTDALICWS-PSGNSFHVFDQGQFAKEVLPKYFKHNNMA 67
Query: 68 SFIRQLNIYGFRKV-----------DTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQ 116
SF+RQLN+YGFRKV + D EF + F+RGQ+ LL+NI+R+ VT
Sbjct: 68 SFVRQLNMYGFRKVVHIEQGGLVKPERDDTEFQHPCFLRGQEQLLENIKRK----VT--- 120
Query: 117 KALPEHNNSDEPSREAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRL 176
++ + D R+ L +V+ +K + + +L+ + E+ ++ L +
Sbjct: 121 -SVSTLKSEDIKIRQDSVTKLLTDVQLMKGKQECMDSKLLAMKHENEALWREVASLRQKH 179
Query: 177 QGMEKHQQQMLSFLVMVVQS 196
+K +++ FL+ +VQS
Sbjct: 180 AQQQKVVNKLIQFLISLVQS 199
>HSF_YEAST (P10961) Heat shock factor protein (HSF) (Heat shock
transcription factor) (HSTF)
Length = 833
Score = 99.4 bits (246), Expect = 1e-20
Identities = 55/146 (37%), Positives = 80/146 (54%), Gaps = 24/146 (16%)
Query: 17 FLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASFIRQLNIY 76
F+NK + M+ DDS +I W E G +F++++ F +LP YFKH+NFASF+RQLN+Y
Sbjct: 175 FVNKLWSMLNDDSNTKLIQWAE-DGKSFIVTNREEFVHQILPKYFKHSNFASFVRQLNMY 233
Query: 77 GFRKV-----------DTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNS 125
G+ KV D+W+F NENF+RG++ LL+ I R+K NN
Sbjct: 234 GWHKVQDVKSGSIQSSSDDKWQFENENFIRGREDLLEKIIRQK-----------GSSNNH 282
Query: 126 DEPSREA-PNHGLRKEVENLKSDRNS 150
+ PS P +G ++N NS
Sbjct: 283 NSPSGNGNPANGSNIPLDNAAGSNNS 308
>HSF4_MOUSE (Q9R0L1) Heat shock factor protein 4 (HSF 4) (Heat shock
transcription factor 4) (HSTF 4) (mHSF4)
Length = 492
Score = 99.4 bits (246), Expect = 1e-20
Identities = 59/165 (35%), Positives = 89/165 (53%), Gaps = 15/165 (9%)
Query: 10 GSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASF 69
G S V FL K + +V D TD +I W+ PSG +F++SD + F+ +LP YFKH+N ASF
Sbjct: 13 GPSPVPAFLGKLWALVGDPGTDHLIRWS-PSGTSFLVSDQSRFAKEVLPQYFKHSNMASF 71
Query: 70 IRQLNIYGFRKV-----------DTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKA 118
+RQLN+YGFRKV + D EF + +FVRG++ LL+ +RR+ D +
Sbjct: 72 VRQLNMYGFRKVVSIEQGGLLRPERDHVEFQHPSFVRGREQLLERVRRKVPALRGDDSRW 131
Query: 119 LPEHNN---SDEPSREAPNHGLRKEVENLKSDRNSLMQELVHLSQ 160
PE + + + ++ L+ L +E+V L Q
Sbjct: 132 RPEDLSRLLGEVQALRGVQESTEARLQELRQQNEILWREVVTLRQ 176
>HSF3_CHICK (P38531) Heat shock factor protein 3 (HSF 3) (Heat shock
transcription factor 3) (HSTF 3) (HSF 3C) (HSTF 3C)
Length = 467
Score = 98.6 bits (244), Expect = 2e-20
Identities = 62/199 (31%), Positives = 102/199 (51%), Gaps = 23/199 (11%)
Query: 8 ASGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFA 67
A G++ V FL K + +VED +D +I W+ +G F I D F+ LLP YFKHNN +
Sbjct: 10 APGAAPVPGFLAKLWALVEDPQSDDVICWSR-NGENFCILDEQRFAKELLPKYFKHNNIS 68
Query: 68 SFIRQLNIYGFRKV-----------DTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQ 116
SFIRQLN+YGFRKV EF + F +G HLL+NI+R+ T+
Sbjct: 69 SFIRQLNMYGFRKVVALENGMITAEKNSVIEFQHPFFKQGNAHLLENIKRKVSAVRTEDL 128
Query: 117 KALPEHNNSDEPSREAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRL 176
K E H + EV+ ++ +N++ L ++ + ++ ++ VL +
Sbjct: 129 KVCAE-----------DLHKVLSEVQEMREQQNNMDIRLANMKRENKALWKEVAVLRQKH 177
Query: 177 QGMEKHQQQMLSFLVMVVQ 195
+K ++L F++ +++
Sbjct: 178 SQQQKLLSKILQFILSLMR 196
>HSF4_HUMAN (Q9ULV5) Heat shock factor protein 4 (HSF 4) (Heat shock
transcription factor 4) (HSTF 4) (hHSF4)
Length = 493
Score = 97.4 bits (241), Expect = 5e-20
Identities = 59/165 (35%), Positives = 87/165 (51%), Gaps = 15/165 (9%)
Query: 10 GSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASF 69
G S V FL K + +V D TD +I W+ PSG +F++SD + F+ +LP YFKH+N ASF
Sbjct: 14 GPSPVPAFLGKLWALVGDPGTDHLIRWS-PSGTSFLVSDQSRFAKEVLPQYFKHSNMASF 72
Query: 70 IRQLNIYGFRKV-----------DTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKA 118
+RQLN+YGFRKV + D EF + +FVRG++ LL+ +RR+ D +
Sbjct: 73 VRQLNMYGFRKVVSIEQGGLLRPERDHVEFQHPSFVRGREQLLERVRRKVPALRGDDGRW 132
Query: 119 LPEHNN---SDEPSREAPNHGLRKEVENLKSDRNSLMQELVHLSQ 160
PE + + + L+ L +E+V L Q
Sbjct: 133 RPEDLGRLLGEVQALRGVQESTEARLRELRQQNEILWREVVTLRQ 177
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.133 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 46,183,531
Number of Sequences: 164201
Number of extensions: 2086345
Number of successful extensions: 4453
Number of sequences better than 10.0: 89
Number of HSP's better than 10.0 without gapping: 46
Number of HSP's successfully gapped in prelim test: 43
Number of HSP's that attempted gapping in prelim test: 4318
Number of HSP's gapped (non-prelim): 127
length of query: 367
length of database: 59,974,054
effective HSP length: 112
effective length of query: 255
effective length of database: 41,583,542
effective search space: 10603803210
effective search space used: 10603803210
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0069.1


