

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0066.6
(137 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
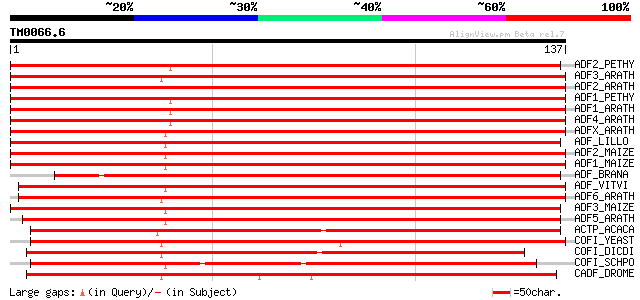
Score E
Sequences producing significant alignments: (bits) Value
ADF2_PETHY (Q9FVI1) Actin-depolymerizing factor 2 (ADF 2) 239 1e-63
ADF3_ARATH (Q9ZSK4) Actin-depolymerizing factor 3 (ADF 3) (AtADF3) 236 8e-63
ADF2_ARATH (Q39251) Actin-depolymerizing factor 2 (ADF-2) (AtADF2) 236 8e-63
ADF1_PETHY (Q9FVI2) Actin-depolymerizing factor 1 (ADF 1) 228 3e-60
ADF1_ARATH (Q39250) Actin-depolymerizing factor 1 (ADF-1) (AtADF1) 224 4e-59
ADF4_ARATH (Q9ZSK3) Actin-depolymerizing factor 4 (ADF-4) (AtADF4) 224 6e-59
ADFX_ARATH (Q9LQ81) Actin-depolymerizing factor like At1g01750 (... 208 2e-54
ADF_LILLO (P30175) Actin-depolymerizing factor (ADF) 206 9e-54
ADF2_MAIZE (Q43694) Actin-depolymerizing factor 2 (ADF 2) (ZmABP... 196 1e-50
ADF1_MAIZE (P46251) Actin-depolymerizing factor 1 (ADF 1) (ZmABP... 194 5e-50
ADF_BRANA (P30174) Actin depolymerizing factor (ADF) (Fragment) 190 7e-49
ADF_VITVI (Q8SAG3) Actin-depolymerizing factor (ADF) 172 1e-43
ADF6_ARATH (Q9ZSK2) Actin-depolymerizing factor 6 (ADF-6) (AtADF6) 166 2e-41
ADF3_MAIZE (Q41764) Actin-depolymerizing factor 3 (ADF 3) (ZmABP... 164 7e-41
ADF5_ARATH (Q9ZNT3) Actin-depolymerizing factor 5 (ADF-5) (AtADF5) 158 4e-39
ACTP_ACACA (P37167) Actophorin 125 4e-29
COFI_YEAST (Q03048) Cofilin 111 5e-25
COFI_DICDI (P54706) Cofilin 107 1e-23
COFI_SCHPO (P78929) Cofilin 102 2e-22
CADF_DROME (P45594) Cofilin/actin depolymerizing factor homolog ... 98 5e-21
>ADF2_PETHY (Q9FVI1) Actin-depolymerizing factor 2 (ADF 2)
Length = 143
Score = 239 bits (611), Expect = 1e-63
Identities = 116/138 (84%), Positives = 129/138 (93%), Gaps = 2/138 (1%)
Query: 1 MANAASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDKQ--VIVEKLGEPGQGYEDFTANL 58
MANAASGMAVHDDCKL+F+ELK KRTYRFI+YKIE+KQ V+VEKLGEP + YEDFTA L
Sbjct: 1 MANAASGMAVHDDCKLKFLELKAKRTYRFIIYKIEEKQKEVVVEKLGEPTESYEDFTAGL 60
Query: 59 PADECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIE 118
PADECRYAVYDF+++T+ N KSRIFFIAWSPDT+RVRSKMIYASSKDRFKRELDGIQ+E
Sbjct: 61 PADECRYAVYDFDFMTKENHQKSRIFFIAWSPDTARVRSKMIYASSKDRFKRELDGIQVE 120
Query: 119 LQATDPTEMGLDVFKSRA 136
LQATDPTEMGLDVF+SRA
Sbjct: 121 LQATDPTEMGLDVFRSRA 138
>ADF3_ARATH (Q9ZSK4) Actin-depolymerizing factor 3 (ADF 3) (AtADF3)
Length = 139
Score = 236 bits (603), Expect = 8e-63
Identities = 113/139 (81%), Positives = 130/139 (93%), Gaps = 2/139 (1%)
Query: 1 MANAASGMAVHDDCKLRFMELKTKRTYRFIVYKIED--KQVIVEKLGEPGQGYEDFTANL 58
MANAASGMAVHDDCKL+FMELKTKRT+RFI+YKIE+ KQVIVEK+GEPGQ +ED A+L
Sbjct: 1 MANAASGMAVHDDCKLKFMELKTKRTHRFIIYKIEELQKQVIVEKIGEPGQTHEDLAASL 60
Query: 59 PADECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIE 118
PADECRYA++DF++++ VP+SRIFF+AWSPDT+RVRSKMIYASSKDRFKRELDGIQ+E
Sbjct: 61 PADECRYAIFDFDFVSSEGVPRSRIFFVAWSPDTARVRSKMIYASSKDRFKRELDGIQVE 120
Query: 119 LQATDPTEMGLDVFKSRAN 137
LQATDPTEM LDVFKSRAN
Sbjct: 121 LQATDPTEMDLDVFKSRAN 139
>ADF2_ARATH (Q39251) Actin-depolymerizing factor 2 (ADF-2) (AtADF2)
Length = 137
Score = 236 bits (603), Expect = 8e-63
Identities = 112/137 (81%), Positives = 125/137 (90%)
Query: 1 MANAASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDKQVIVEKLGEPGQGYEDFTANLPA 60
MANAASGMAVHDDCKL+FMELK KRT+R IVYKIEDKQVIVEKLGEP Q Y+DF A+LPA
Sbjct: 1 MANAASGMAVHDDCKLKFMELKAKRTFRTIVYKIEDKQVIVEKLGEPEQSYDDFAASLPA 60
Query: 61 DECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIELQ 120
D+CRY +YDF+++T N KS+IFFIAWSPDT++VR KMIYASSKDRFKRELDGIQ+ELQ
Sbjct: 61 DDCRYCIYDFDFVTAENCQKSKIFFIAWSPDTAKVRDKMIYASSKDRFKRELDGIQVELQ 120
Query: 121 ATDPTEMGLDVFKSRAN 137
ATDPTEMGLDVFKSR N
Sbjct: 121 ATDPTEMGLDVFKSRTN 137
>ADF1_PETHY (Q9FVI2) Actin-depolymerizing factor 1 (ADF 1)
Length = 139
Score = 228 bits (581), Expect = 3e-60
Identities = 111/139 (79%), Positives = 125/139 (89%), Gaps = 2/139 (1%)
Query: 1 MANAASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDKQ--VIVEKLGEPGQGYEDFTANL 58
MANAASGMAVHDDCKLRF+ELK KRT+RFIVYKIE+KQ V+VEK+GEP + YEDF A+L
Sbjct: 1 MANAASGMAVHDDCKLRFLELKAKRTHRFIVYKIEEKQKQVVVEKIGEPTESYEDFAASL 60
Query: 59 PADECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIE 118
P +ECRYAVYDF+++T N KSRIFFIAW PDT+RVRSKMIYASSKDRFKRELDGIQ+E
Sbjct: 61 PENECRYAVYDFDFVTAENCQKSRIFFIAWCPDTARVRSKMIYASSKDRFKRELDGIQVE 120
Query: 119 LQATDPTEMGLDVFKSRAN 137
LQA DPTEMGLDV +SRAN
Sbjct: 121 LQACDPTEMGLDVIQSRAN 139
>ADF1_ARATH (Q39250) Actin-depolymerizing factor 1 (ADF-1) (AtADF1)
Length = 139
Score = 224 bits (571), Expect = 4e-59
Identities = 109/139 (78%), Positives = 125/139 (89%), Gaps = 2/139 (1%)
Query: 1 MANAASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDKQ--VIVEKLGEPGQGYEDFTANL 58
MANAASGMAVHDDCKLRF+ELK KRT+RFIVYKIE+KQ V+VEK+G+P Q YE+F A L
Sbjct: 1 MANAASGMAVHDDCKLRFLELKAKRTHRFIVYKIEEKQKQVVVEKVGQPIQTYEEFAACL 60
Query: 59 PADECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIE 118
PADECRYA+YDF+++T N KS+IFFIAW PD ++VRSKMIYASSKDRFKRELDGIQ+E
Sbjct: 61 PADECRYAIYDFDFVTAENCQKSKIFFIAWCPDIAKVRSKMIYASSKDRFKRELDGIQVE 120
Query: 119 LQATDPTEMGLDVFKSRAN 137
LQATDPTEM LDVF+SRAN
Sbjct: 121 LQATDPTEMDLDVFRSRAN 139
>ADF4_ARATH (Q9ZSK3) Actin-depolymerizing factor 4 (ADF-4) (AtADF4)
Length = 139
Score = 224 bits (570), Expect = 6e-59
Identities = 110/139 (79%), Positives = 123/139 (88%), Gaps = 2/139 (1%)
Query: 1 MANAASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDKQ--VIVEKLGEPGQGYEDFTANL 58
MANAASGMAVHDDCKLRF+ELK KRT+RFIVYKIE+KQ VIVEK+GEP YEDF A+L
Sbjct: 1 MANAASGMAVHDDCKLRFLELKAKRTHRFIVYKIEEKQKQVIVEKVGEPILTYEDFAASL 60
Query: 59 PADECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIE 118
PADECRYA+YDF+++T N KS+IFFIAW PD ++VRSKMIYASSKDRFKRELDGIQ+E
Sbjct: 61 PADECRYAIYDFDFVTAENCQKSKIFFIAWCPDVAKVRSKMIYASSKDRFKRELDGIQVE 120
Query: 119 LQATDPTEMGLDVFKSRAN 137
LQATDPTEM LDV KSR N
Sbjct: 121 LQATDPTEMDLDVLKSRVN 139
>ADFX_ARATH (Q9LQ81) Actin-depolymerizing factor like At1g01750
(ADF-like)
Length = 140
Score = 208 bits (530), Expect = 2e-54
Identities = 98/139 (70%), Positives = 119/139 (85%), Gaps = 2/139 (1%)
Query: 1 MANAASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDK--QVIVEKLGEPGQGYEDFTANL 58
MAN+ASGM V D+CKL+F+ELK KR YRFIV+KI++K QV+++KLG P + YEDFT ++
Sbjct: 1 MANSASGMHVSDECKLKFLELKAKRNYRFIVFKIDEKAQQVMIDKLGNPEETYEDFTRSI 60
Query: 59 PADECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIE 118
P DECRYAVYD+++ T N KS+IFFIAWSPDTSRVRSKM+YASSKDRFKRELDGIQ+E
Sbjct: 61 PEDECRYAVYDYDFTTPENCQKSKIFFIAWSPDTSRVRSKMLYASSKDRFKRELDGIQVE 120
Query: 119 LQATDPTEMGLDVFKSRAN 137
LQATDP+EM LD+ K R N
Sbjct: 121 LQATDPSEMSLDIIKGRVN 139
>ADF_LILLO (P30175) Actin-depolymerizing factor (ADF)
Length = 139
Score = 206 bits (525), Expect = 9e-54
Identities = 96/138 (69%), Positives = 122/138 (87%), Gaps = 2/138 (1%)
Query: 1 MANAASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDK--QVIVEKLGEPGQGYEDFTANL 58
MAN++SGMAV D+CKL+FMELK KR +RFIV+KIE+K QV VE+LG+P + Y+DFT L
Sbjct: 1 MANSSSGMAVDDECKLKFMELKAKRNFRFIVFKIEEKVQQVTVERLGQPNESYDDFTECL 60
Query: 59 PADECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIE 118
P +ECRYAV+DF+++T+ N KS+IFFI+WSPDTSRVRSKM+YAS+KDRFKRELDGIQ+E
Sbjct: 61 PPNECRYAVFDFDFVTDENCQKSKIFFISWSPDTSRVRSKMLYASTKDRFKRELDGIQVE 120
Query: 119 LQATDPTEMGLDVFKSRA 136
LQATDP+EM +D+ K+RA
Sbjct: 121 LQATDPSEMSMDIIKARA 138
>ADF2_MAIZE (Q43694) Actin-depolymerizing factor 2 (ADF 2) (ZmABP2)
(ZmADF2)
Length = 139
Score = 196 bits (498), Expect = 1e-50
Identities = 89/139 (64%), Positives = 118/139 (84%), Gaps = 2/139 (1%)
Query: 1 MANAASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDK--QVIVEKLGEPGQGYEDFTANL 58
MAN++SG+AV D+CK++F +LK +R++RFIV++I+DK ++ V++LGEP QGY DFT +L
Sbjct: 1 MANSSSGLAVSDECKVKFRDLKARRSFRFIVFRIDDKDMEIKVDRLGEPNQGYGDFTDSL 60
Query: 59 PADECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIE 118
PADECRYA+YD ++ T N KS+IFF +WSPDT+R RSKM+YASSKDRF+RELDGIQ E
Sbjct: 61 PADECRYAIYDLDFTTVENCQKSKIFFFSWSPDTARTRSKMLYASSKDRFRRELDGIQCE 120
Query: 119 LQATDPTEMGLDVFKSRAN 137
+QATDP+EM LD+ KSR N
Sbjct: 121 IQATDPSEMSLDIVKSRTN 139
>ADF1_MAIZE (P46251) Actin-depolymerizing factor 1 (ADF 1) (ZmABP1)
(ZmADF1)
Length = 139
Score = 194 bits (493), Expect = 5e-50
Identities = 88/139 (63%), Positives = 119/139 (85%), Gaps = 2/139 (1%)
Query: 1 MANAASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDK--QVIVEKLGEPGQGYEDFTANL 58
MAN++SG+AV+D+CK++F ELK++RT+RFIV++I+D ++ V++LGEP QGY DFT +L
Sbjct: 1 MANSSSGLAVNDECKVKFRELKSRRTFRFIVFRIDDTDMEIKVDRLGEPNQGYGDFTDSL 60
Query: 59 PADECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIE 118
PA+ECRYA+YD ++ T N KS+IFF +WSPDT+R RSKM+YASSKDRF+RELDGIQ E
Sbjct: 61 PANECRYAIYDLDFTTIENCQKSKIFFFSWSPDTARTRSKMLYASSKDRFRRELDGIQCE 120
Query: 119 LQATDPTEMGLDVFKSRAN 137
+QATDP+EM LD+ +SR N
Sbjct: 121 IQATDPSEMSLDIVRSRTN 139
>ADF_BRANA (P30174) Actin depolymerizing factor (ADF) (Fragment)
Length = 126
Score = 190 bits (483), Expect = 7e-49
Identities = 88/125 (70%), Positives = 110/125 (87%), Gaps = 1/125 (0%)
Query: 12 DDCKLRFMELKTKRTYRFIVYKIEDKQVIVEKLGEPGQGYEDFTANLPADECRYAVYDFE 71
D+CKL+F+ELK KR +RFI+++I+ +QV+VEKLG P + Y+DFTA+LPADECRYAV+DF+
Sbjct: 2 DNCKLKFLELK-KRIFRFIIFRIDGQQVVVEKLGNPQETYDDFTASLPADECRYAVFDFD 60
Query: 72 YLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIELQATDPTEMGLDV 131
+ T N KS+IFFIAWSPD+SRVR KM+YASSKDRFKRELDGIQ+ELQATDP+EM D+
Sbjct: 61 FTTNENCQKSKIFFIAWSPDSSRVRMKMVYASSKDRFKRELDGIQVELQATDPSEMSFDI 120
Query: 132 FKSRA 136
KSRA
Sbjct: 121 IKSRA 125
>ADF_VITVI (Q8SAG3) Actin-depolymerizing factor (ADF)
Length = 143
Score = 172 bits (437), Expect = 1e-43
Identities = 77/137 (56%), Positives = 111/137 (80%), Gaps = 2/137 (1%)
Query: 3 NAASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDK--QVIVEKLGEPGQGYEDFTANLPA 60
NA+SGM V D K F+ELK K+ +R++++KI++K +V+VEK G P + +++F A LP
Sbjct: 7 NASSGMGVADHSKNTFLELKRKKVHRYVIFKIDEKKKEVVVEKTGGPAESFDEFAAALPE 66
Query: 61 DECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIELQ 120
++CRYAVYDF+++T N KS+IFFIAWSPD+SR+R+KM+YA+SK+RF+RELDG+ E+Q
Sbjct: 67 NDCRYAVYDFDFVTSENCQKSKIFFIAWSPDSSRIRAKMLYATSKERFRRELDGVHYEIQ 126
Query: 121 ATDPTEMGLDVFKSRAN 137
ATDPTEM L+V + RA+
Sbjct: 127 ATDPTEMDLEVLRERAH 143
>ADF6_ARATH (Q9ZSK2) Actin-depolymerizing factor 6 (ADF-6) (AtADF6)
Length = 146
Score = 166 bits (419), Expect = 2e-41
Identities = 75/137 (54%), Positives = 107/137 (77%), Gaps = 2/137 (1%)
Query: 3 NAASGMAVHDDCKLRFMELKTKRTYRFIVYKIED--KQVIVEKLGEPGQGYEDFTANLPA 60
NA SGM V D+ K F+EL+ K+T+R++V+KI++ K+V+VEK G P + Y+DF A+LP
Sbjct: 10 NAISGMGVADESKTTFLELQRKKTHRYVVFKIDESKKEVVVEKTGNPTESYDDFLASLPD 69
Query: 61 DECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIELQ 120
++CRYAVYDF+++T N KS+IFF AWSP TS +R+K++Y++SKD+ REL GI E+Q
Sbjct: 70 NDCRYAVYDFDFVTSENCQKSKIFFFAWSPSTSGIRAKVLYSTSKDQLSRELQGIHYEIQ 129
Query: 121 ATDPTEMGLDVFKSRAN 137
ATDPTE+ L+V + RAN
Sbjct: 130 ATDPTEVDLEVLRERAN 146
>ADF3_MAIZE (Q41764) Actin-depolymerizing factor 3 (ADF 3) (ZmABP3)
(ZmADF3)
Length = 139
Score = 164 bits (414), Expect = 7e-41
Identities = 74/138 (53%), Positives = 111/138 (79%), Gaps = 2/138 (1%)
Query: 1 MANAASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDK--QVIVEKLGEPGQGYEDFTANL 58
MANA SG+AV+D+C L+F EL++KR +RFI +K++DK +++V+++G+ Y+DFT +L
Sbjct: 1 MANARSGVAVNDECMLKFGELQSKRLHRFITFKMDDKFKEIVVDQVGDRATSYDDFTNSL 60
Query: 59 PADECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIE 118
P ++CRYA+YDF+++T +V KSRIF+I WSP +++V+SKM+YASS +FK L+GIQ+E
Sbjct: 61 PENDCRYAIYDFDFVTAEDVQKSRIFYILWSPSSAKVKSKMLYASSNQKFKSGLNGIQVE 120
Query: 119 LQATDPTEMGLDVFKSRA 136
LQATD +E+ LD K RA
Sbjct: 121 LQATDASEISLDEIKDRA 138
>ADF5_ARATH (Q9ZNT3) Actin-depolymerizing factor 5 (ADF-5) (AtADF5)
Length = 143
Score = 158 bits (399), Expect = 4e-39
Identities = 72/135 (53%), Positives = 103/135 (75%), Gaps = 2/135 (1%)
Query: 4 AASGMAVHDDCKLRFMELKTKRTYRFIVYKIEDK--QVIVEKLGEPGQGYEDFTANLPAD 61
A +GM V D+C FM++K K+ +R+IV+KIE+K +V V+K+G G+ Y D +LP D
Sbjct: 8 ATTGMRVTDECTSSFMDMKWKKVHRYIVFKIEEKSRKVTVDKVGGAGESYHDLEDSLPVD 67
Query: 62 ECRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIELQA 121
+CRYAV+DF+++T N KS+IFFIAWSP+ S++R+K++YA+SKD +R L+GI ELQA
Sbjct: 68 DCRYAVFDFDFVTVDNCRKSKIFFIAWSPEASKIRAKILYATSKDGLRRVLEGIHYELQA 127
Query: 122 TDPTEMGLDVFKSRA 136
TDPTEMG D+ + RA
Sbjct: 128 TDPTEMGFDIIQDRA 142
>ACTP_ACACA (P37167) Actophorin
Length = 137
Score = 125 bits (313), Expect = 4e-29
Identities = 58/133 (43%), Positives = 91/133 (67%), Gaps = 3/133 (2%)
Query: 6 SGMAVHDDCKLRFMELKTKRTYRFIVYKIE--DKQVIVEKLGEPGQGYEDFTANLPADEC 63
SG+AV DDC +F ELK +R++ +K+ + +V+VE +G P YEDF + LP +C
Sbjct: 1 SGIAVSDDCVQKFNELKLGHQHRYVTFKMNASNTEVVVEHVGGPNATYEDFKSQLPERDC 60
Query: 64 RYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIELQATD 123
RYA++D+E+ +G +++I FI W+PD++ ++SKM+Y S+KD K++L GIQ+E+QATD
Sbjct: 61 RYAIFDYEFQVDGG-QRNKITFILWAPDSAPIKSKMMYTSTKDSIKKKLVGIQVEVQATD 119
Query: 124 PTEMGLDVFKSRA 136
E+ D RA
Sbjct: 120 AAEISEDAVSERA 132
>COFI_YEAST (Q03048) Cofilin
Length = 143
Score = 111 bits (277), Expect = 5e-25
Identities = 59/134 (44%), Positives = 82/134 (61%), Gaps = 2/134 (1%)
Query: 6 SGMAVHDDCKLRFMELKTKRTYRFIVYKIED-KQVIVEKLGEPGQGYEDFTANLPADECR 64
SG+AV D+ F +LK + Y+FI++ + D K IV K Y+ F LP ++C
Sbjct: 4 SGVAVADESLTAFNDLKLGKKYKFILFGLNDAKTEIVVKETSTDPSYDAFLEKLPENDCL 63
Query: 65 YAVYDFEYLTEGNVPK-SRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIELQATD 123
YA+YDFEY GN K S+I F WSPDT+ VRSKM+YASSKD +R L+G+ ++Q TD
Sbjct: 64 YAIYDFEYEINGNEGKRSKIVFFTWSPDTAPVRSKMVYASSKDALRRALNGVSTDVQGTD 123
Query: 124 PTEMGLDVFKSRAN 137
+E+ D R +
Sbjct: 124 FSEVSYDSVLERVS 137
>COFI_DICDI (P54706) Cofilin
Length = 137
Score = 107 bits (266), Expect = 1e-23
Identities = 53/125 (42%), Positives = 81/125 (64%), Gaps = 3/125 (2%)
Query: 5 ASGMAVHDDCKLRFMELKTKRTYRFIVYKIED--KQVIVEKLGEPGQGYEDFTANLPADE 62
+SG+A+ +C F +LK R Y I+Y+I D K++IV+ G +++FT LP +E
Sbjct: 2 SSGIALAPNCVSTFNDLKLGRKYGGIIYRISDDSKEIIVDSTLPAGCSFDEFTKCLPENE 61
Query: 63 CRYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIELQAT 122
CRY V D++Y EG KS+I F+AW PDT+ ++ KM+ SSKD ++ GIQ+E+Q T
Sbjct: 62 CRYVVLDYQYKEEG-AQKSKICFVAWCPDTANIKKKMMATSSKDSLRKACVGIQVEIQGT 120
Query: 123 DPTEM 127
D +E+
Sbjct: 121 DASEV 125
>COFI_SCHPO (P78929) Cofilin
Length = 137
Score = 102 bits (255), Expect = 2e-22
Identities = 50/127 (39%), Positives = 83/127 (64%), Gaps = 4/127 (3%)
Query: 6 SGMAVHDDCKLRFMELKTKRTYRFIVYKIEDK--QVIVEKLGEPGQGYEDFTANLPADEC 63
SG+ V +C F ELK ++ R++V+K+ D +++VEK + ++ F +LP +C
Sbjct: 4 SGVKVSPECLEAFQELKLGKSLRYVVFKMNDTKTEIVVEKKSTD-KDFDTFLGDLPEKDC 62
Query: 64 RYAVYDFEYLTEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGIQIELQATD 123
RYA+YDFE+ G +++I FI+WSPD + ++SKM+Y+SSKD +R GI ++QATD
Sbjct: 63 RYAIYDFEF-NLGEGVRNKIIFISWSPDVAPIKSKMVYSSSKDTLRRAFTGIGTDIQATD 121
Query: 124 PTEMGLD 130
+E+ +
Sbjct: 122 FSEVAYE 128
>CADF_DROME (P45594) Cofilin/actin depolymerizing factor homolog
(D61 protein) (Twinstar protein)
Length = 148
Score = 98.2 bits (243), Expect = 5e-21
Identities = 49/140 (35%), Positives = 84/140 (60%), Gaps = 9/140 (6%)
Query: 5 ASGMAVHDDCKLRFMELKTKRTYRFIVYKIED-KQVIVEKLGEPGQGYEDFTANLPA--- 60
ASG+ V D CK + E+K + +R++++ I D KQ+ VE + + Y+ F ++
Sbjct: 2 ASGVTVSDVCKTTYEEIKKDKKHRYVIFYIRDEKQIDVETVADRNAEYDQFLEDIQKCGP 61
Query: 61 DECRYAVYDFEYL-----TEGNVPKSRIFFIAWSPDTSRVRSKMIYASSKDRFKRELDGI 115
ECRY ++DFEY+ T + K ++F ++W PDT++V+ KM+Y+SS D K+ L G+
Sbjct: 62 GECRYGLFDFEYMHQCQGTSESSKKQKLFLMSWCPDTAKVKKKMLYSSSFDALKKSLVGV 121
Query: 116 QIELQATDPTEMGLDVFKSR 135
Q +QATD +E + + +
Sbjct: 122 QKYIQATDLSEASREAVEEK 141
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,468,100
Number of Sequences: 164201
Number of extensions: 594602
Number of successful extensions: 1369
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 50
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 1273
Number of HSP's gapped (non-prelim): 63
length of query: 137
length of database: 59,974,054
effective HSP length: 98
effective length of query: 39
effective length of database: 43,882,356
effective search space: 1711411884
effective search space used: 1711411884
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0066.6


