

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
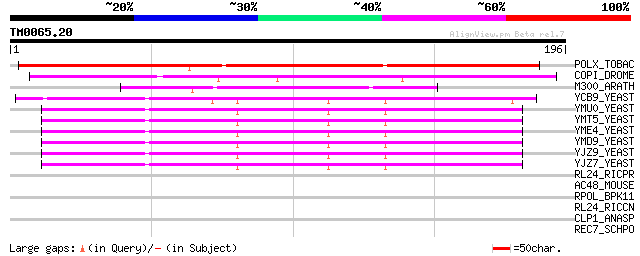
Query= TM0065.20
(196 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 166 3e-41
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 86 5e-17
M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300... 78 1e-14
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 58 1e-08
YMU0_YEAST (Q04670) Transposon Ty1 protein B 45 1e-04
YMT5_YEAST (Q04214) Transposon Ty1 protein B 45 1e-04
YME4_YEAST (Q04711) Transposon Ty1 protein B 45 1e-04
YMD9_YEAST (Q03434) Transposon Ty1 protein B 45 1e-04
YJZ9_YEAST (P47100) Transposon Ty1 protein B 45 1e-04
YJZ7_YEAST (P47098) Transposon Ty1 protein B 43 4e-04
RL24_RICPR (Q9ZCR6) 50S ribosomal protein L24 31 1.6
AC48_MOUSE (Q9R0X4) Acyl coenzyme A thioester hydrolase 2, mitoc... 31 1.6
RPOL_BPK11 (P18147) DNA-directed RNA polymerase (EC 2.7.7.6) 31 2.1
RL24_RICCN (Q92GX7) 50S ribosomal protein L24 31 2.1
CLP1_ANASP (Q8YXH5) ATP-dependent Clp protease proteolytic subun... 29 6.0
REC7_SCHPO (P36625) Meiotic recombination protein rec7 29 7.9
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 166 bits (420), Expect = 3e-41
Identities = 83/188 (44%), Positives = 126/188 (66%), Gaps = 6/188 (3%)
Query: 4 GTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYM- 62
G L +VRHVP ++ NLIS LD +GY + F W++TKG+LV+A+G RG+LY
Sbjct: 345 GCTLVLKDVRHVPDLRMNLISGIALDRDGYESYFANQKWRLTKGSLVIAKGVARGTLYRT 404
Query: 63 ---VAEEDMIAVTEAVNSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLGVCEHCILG 119
+ + ++ A + + S +WH+R+GHMSEKG++I+A K +S K + C++C+ G
Sbjct: 405 NAEICQGELNAAQDEI-SVDLWHKRMGHMSEKGLQILAKKSLISYAKGTTVKPCDYCLFG 463
Query: 120 KQRKVSFSKAGRKSKLEKLKLVHTDVWGPAPVKSLGGSRYYVTFIDDSTRKV*VYFLKNK 179
KQ +VSF + + KL L LV++DV GP ++S+GG++Y+VTFIDD++RK+ VY LK K
Sbjct: 464 KQHRVSFQTSSER-KLNILDLVYSDVCGPMEIESMGGNKYFVTFIDDASRKLWVYILKTK 522
Query: 180 SDVFSVFK 187
VF VF+
Sbjct: 523 DQVFQVFQ 530
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 85.9 bits (211), Expect = 5e-17
Identities = 59/195 (30%), Positives = 96/195 (48%), Gaps = 11/195 (5%)
Query: 8 TLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMVAEED 67
TL +V NL+S+ +L + G F ++K L+V K G L V +
Sbjct: 344 TLEDVLFCKEAAGNLMSVKRLQEAGMSIEFDKSGVTISKNGLMVV--KNSGMLNNVPVIN 401
Query: 68 MIAVT---EAVNSSSIWHQRLGHMSEKGM-----KIMASKGKMSNLKHVDLGVCEHCILG 119
A + + N+ +WH+R GH+S+ + K M S + N + +CE C+ G
Sbjct: 402 FQAYSINAKHKNNFRLWHERFGHISDGKLLEIKRKNMFSDQSLLNNLELSCEICEPCLNG 461
Query: 120 KQRKVSFSKAGRKSKLEK-LKLVHTDVWGPAPVKSLGGSRYYVTFIDDSTRKV*VYFLKN 178
KQ ++ F + K+ +++ L +VH+DV GP +L Y+V F+D T Y +K
Sbjct: 462 KQARLPFKQLKDKTHIKRPLFVVHSDVCGPITPVTLDDKNYFVIFVDQFTHYCVTYLIKY 521
Query: 179 KSDVFSVFKGTTKKT 193
KSDVFS+F+ K+
Sbjct: 522 KSDVFSMFQDFVAKS 536
>M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300
(ORF145a) (ORF1451)
Length = 145
Score = 78.2 bits (191), Expect = 1e-14
Identities = 43/117 (36%), Positives = 65/117 (54%), Gaps = 7/117 (5%)
Query: 40 GAWKVTKGNLVVARGKKRGSLYMV-----AEEDMIAVTEAVNSSSIWHQRLGHMSEKGMK 94
G KV KG + +G + SLY++ E +A T A + + +WH RL HMS++GM+
Sbjct: 27 GVLKVLKGCRTILKGNRHDSLYILQGSVETGESNLAET-AKDETRLWHSRLAHMSQRGME 85
Query: 95 IMASKGKMSNLKHVDLGVCEHCILGKQRKVSFSKAGRKSKLEKLKLVHTDVWGPAPV 151
++ KG + + K L CE CI GK +V+FS G+ + L VH+D+WG V
Sbjct: 86 LLVKKGFLDSSKVSSLKFCEDCIYGKTHRVNFS-TGQHTTKNPLDYVHSDLWGAPSV 141
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 58.2 bits (139), Expect = 1e-08
Identities = 51/210 (24%), Positives = 92/210 (43%), Gaps = 28/210 (13%)
Query: 3 NGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYM 62
NGT ++ + H P I +L+S+ +L ++ F + + G V+A K G Y
Sbjct: 504 NGTKTSIKAL-HTPNIAYDLLSLSELANQNITACFTRNTLERSDGT-VLAPIVKHGDFYW 561
Query: 63 VAEEDMIA------VTEAVNSSS--------IWHQRLGHMSEKGMKIMASKGKMSNLKHV 108
++++ +I VN S + H+ LGH + + ++ K ++ LK
Sbjct: 562 LSKKYLIPSHISKLTINNVNKSKSVNKYPYPLIHRMLGHANFRSIQKSLKKNAVTYLKES 621
Query: 109 DLG-------VCEHCILGKQRKVSFSKAGR---KSKLEKLKLVHTDVWGPAPVKSLGGSR 158
D+ C C++GK K K R + E + +HTD++GP
Sbjct: 622 DIEWSNASTYQCPDCLIGKSTKHRHVKGSRLKYQESYEPFQYLHTDIFGPVHHLPKSAPS 681
Query: 159 YYVTFIDDSTRKV*VYFL--KNKSDVFSVF 186
Y+++F D+ TR VY L + + + +VF
Sbjct: 682 YFISFTDEKTRFQWVYPLHDRREESILNVF 711
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 45.1 bits (105), Expect = 1e-04
Identities = 40/194 (20%), Positives = 80/194 (40%), Gaps = 25/194 (12%)
Query: 12 VRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMVAEEDMIAV 71
V H P I +L+S+ +L F + + G V+A K G Y V+++ ++
Sbjct: 89 VLHTPNIAYDLLSLNELAAVDITACFTKNVLERSDGT-VLAPIVKYGDFYWVSKKYLLPS 147
Query: 72 TEAVNSSS--------------IWHQRLGHMSEKGMKIMASKGKMSNLKHVDLG------ 111
+V + + H+ L H + + ++ ++ D+
Sbjct: 148 NISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAID 207
Query: 112 -VCEHCILGKQRKVSFSKAGR---KSKLEKLKLVHTDVWGPAPVKSLGGSRYYVTFIDDS 167
C C++GK K K R ++ E + +HTD++GP Y+++F D++
Sbjct: 208 YQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDET 267
Query: 168 TRKV*VYFLKNKSD 181
T+ VY L ++ +
Sbjct: 268 TKFRWVYPLHDRRE 281
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 45.1 bits (105), Expect = 1e-04
Identities = 40/194 (20%), Positives = 80/194 (40%), Gaps = 25/194 (12%)
Query: 12 VRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMVAEEDMIAV 71
V H P I +L+S+ +L F + + G V+A K G Y V+++ ++
Sbjct: 89 VLHTPNIAYDLLSLNELAAVDITACFTKNVLERSDGT-VLAPIVKYGDFYWVSKKYLLPS 147
Query: 72 TEAVNSSS--------------IWHQRLGHMSEKGMKIMASKGKMSNLKHVDLG------ 111
+V + + H+ L H + + ++ ++ D+
Sbjct: 148 NISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAID 207
Query: 112 -VCEHCILGKQRKVSFSKAGR---KSKLEKLKLVHTDVWGPAPVKSLGGSRYYVTFIDDS 167
C C++GK K K R ++ E + +HTD++GP Y+++F D++
Sbjct: 208 YQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDET 267
Query: 168 TRKV*VYFLKNKSD 181
T+ VY L ++ +
Sbjct: 268 TKFRWVYPLHDRRE 281
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 45.1 bits (105), Expect = 1e-04
Identities = 40/194 (20%), Positives = 80/194 (40%), Gaps = 25/194 (12%)
Query: 12 VRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMVAEEDMIAV 71
V H P I +L+S+ +L F + + G V+A K G Y V+++ ++
Sbjct: 89 VLHTPNIAYDLLSLNELAAVDITACFTKNVLERSDGT-VLAPIVKYGDFYWVSKKYLLPS 147
Query: 72 TEAVNSSS--------------IWHQRLGHMSEKGMKIMASKGKMSNLKHVDLG------ 111
+V + + H+ L H + + ++ ++ D+
Sbjct: 148 NISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAID 207
Query: 112 -VCEHCILGKQRKVSFSKAGR---KSKLEKLKLVHTDVWGPAPVKSLGGSRYYVTFIDDS 167
C C++GK K K R ++ E + +HTD++GP Y+++F D++
Sbjct: 208 YQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDET 267
Query: 168 TRKV*VYFLKNKSD 181
T+ VY L ++ +
Sbjct: 268 TKFRWVYPLHDRRE 281
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 45.1 bits (105), Expect = 1e-04
Identities = 40/194 (20%), Positives = 80/194 (40%), Gaps = 25/194 (12%)
Query: 12 VRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMVAEEDMIAV 71
V H P I +L+S+ +L F + + G V+A K G Y V+++ ++
Sbjct: 89 VLHTPNIAYDLLSLNELAAVDITACFTKNVLERSDGT-VLAPIVKYGDFYWVSKKYLLPS 147
Query: 72 TEAVNSSS--------------IWHQRLGHMSEKGMKIMASKGKMSNLKHVDLG------ 111
+V + + H+ L H + + ++ ++ D+
Sbjct: 148 NISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDRSSAID 207
Query: 112 -VCEHCILGKQRKVSFSKAGR---KSKLEKLKLVHTDVWGPAPVKSLGGSRYYVTFIDDS 167
C C++GK K K R ++ E + +HTD++GP Y+++F D++
Sbjct: 208 YQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDET 267
Query: 168 TRKV*VYFLKNKSD 181
T+ VY L ++ +
Sbjct: 268 TKFRWVYPLHDRRE 281
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 45.1 bits (105), Expect = 1e-04
Identities = 40/194 (20%), Positives = 80/194 (40%), Gaps = 25/194 (12%)
Query: 12 VRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMVAEEDMIAV 71
V H P I +L+S+ +L F + + G V+A K G Y V+++ ++
Sbjct: 516 VLHTPNIAYDLLSLNELAAVDITACFTKNVLERSDGT-VLAPIVKYGDFYWVSKKYLLPS 574
Query: 72 TEAVNSSS--------------IWHQRLGHMSEKGMKIMASKGKMSNLKHVDLG------ 111
+V + + H+ L H + + ++ ++ D+
Sbjct: 575 NISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAID 634
Query: 112 -VCEHCILGKQRKVSFSKAGR---KSKLEKLKLVHTDVWGPAPVKSLGGSRYYVTFIDDS 167
C C++GK K K R ++ E + +HTD++GP Y+++F D++
Sbjct: 635 YQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDET 694
Query: 168 TRKV*VYFLKNKSD 181
T+ VY L ++ +
Sbjct: 695 TKFRWVYPLHDRRE 708
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 43.1 bits (100), Expect = 4e-04
Identities = 39/194 (20%), Positives = 79/194 (40%), Gaps = 25/194 (12%)
Query: 12 VRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMVAEEDMIAV 71
V H P I +L+S+ +L F + + G V+A + G Y V++ ++
Sbjct: 516 VLHTPNIAYDLLSLNELAAVDITACFTKNVLERSDGT-VLAPIVQYGDFYWVSKRYLLPS 574
Query: 72 TEAVNSSS--------------IWHQRLGHMSEKGMKIMASKGKMSNLKHVDLG------ 111
+V + + H+ L H + + ++ ++ D+
Sbjct: 575 NISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAID 634
Query: 112 -VCEHCILGKQRKVSFSKAGR---KSKLEKLKLVHTDVWGPAPVKSLGGSRYYVTFIDDS 167
C C++GK K K R ++ E + +HTD++GP Y+++F D++
Sbjct: 635 YQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDET 694
Query: 168 TRKV*VYFLKNKSD 181
T+ VY L ++ +
Sbjct: 695 TKFRWVYPLHDRRE 708
>RL24_RICPR (Q9ZCR6) 50S ribosomal protein L24
Length = 113
Score = 31.2 bits (69), Expect = 1.6
Identities = 24/71 (33%), Positives = 35/71 (48%), Gaps = 5/71 (7%)
Query: 43 KVTKGN-LVVARGK---KRGSLYMVAEEDMIAVTEAVNSSSIWHQRLGHMSEKGMKIMAS 98
KV KG+ +V+ GK K+G + V E+ V VN H + MSE G+ S
Sbjct: 5 KVKKGDEVVIITGKYKGKKGKVLKVFPEENTVVVSGVNLVKK-HTKPNKMSEGGIITQES 63
Query: 99 KGKMSNLKHVD 109
+SN+ H+D
Sbjct: 64 PIHISNIAHID 74
>AC48_MOUSE (Q9R0X4) Acyl coenzyme A thioester hydrolase 2,
mitochondrial precursor (EC 3.1.2.-) (48 kDa acyl-CoA
thioester hydrolase) (p48) (Mt-ACT48) (Protein U8)
(MTE-2)
Length = 439
Score = 31.2 bits (69), Expect = 1.6
Identities = 28/108 (25%), Positives = 42/108 (37%), Gaps = 25/108 (23%)
Query: 42 WKVTKGNLVVARGKKRGSLYMVAEEDMI-----AVTEAVNSSSIWHQRLGHMSEKGMKIM 96
W + KG L RG +GS Y ++E I + E V S++W + M E+ +
Sbjct: 9 WTLNKGLLTHGRGLSQGSQYKISEPLHIHQVRDKLREIVGVSTVWRDHVKAMEERKLL-- 66
Query: 97 ASKGKMSNLKHVDLGVCEHCILGKQRKVSFSKAGRKSKLEKLKLVHTD 144
H L K +KV + R S +E L + TD
Sbjct: 67 ------------------HSFLPKSQKVLPPRKMRDSYIEVLLPLGTD 96
>RPOL_BPK11 (P18147) DNA-directed RNA polymerase (EC 2.7.7.6)
Length = 906
Score = 30.8 bits (68), Expect = 2.1
Identities = 22/78 (28%), Positives = 36/78 (45%), Gaps = 10/78 (12%)
Query: 36 TFGGGAWKVTKGNLVVARGKKRGSLYMVAEEDMIAVTEAVN--SSSIWHQRLGHMSEKGM 93
T GGG W V + L + R + +L A+ M V +AVN ++ W +
Sbjct: 304 TVGGGYWSVGRRPLALVRTHSKKALRRYADVHMPEVYKAVNLAQNTPW--------KVNK 355
Query: 94 KIMASKGKMSNLKHVDLG 111
K++A ++ N KH +G
Sbjct: 356 KVLAVVNEIVNWKHCPVG 373
>RL24_RICCN (Q92GX7) 50S ribosomal protein L24
Length = 109
Score = 30.8 bits (68), Expect = 2.1
Identities = 24/71 (33%), Positives = 34/71 (47%), Gaps = 5/71 (7%)
Query: 43 KVTKGN-LVVARGK---KRGSLYMVAEEDMIAVTEAVNSSSIWHQRLGHMSEKGMKIMAS 98
KV KG+ +VV GK K+G + V ED + VN H + MSE G+
Sbjct: 5 KVKKGDEVVVITGKHKGKKGKILKVFPEDSKVIVSGVNVVKK-HTKSNQMSEGGIITKEL 63
Query: 99 KGKMSNLKHVD 109
+SN+ H+D
Sbjct: 64 PIHISNIAHID 74
>CLP1_ANASP (Q8YXH5) ATP-dependent Clp protease proteolytic subunit
1 (EC 3.4.21.92) (Endopeptidase Clp 1)
Length = 204
Score = 29.3 bits (64), Expect = 6.0
Identities = 25/103 (24%), Positives = 44/103 (42%), Gaps = 7/103 (6%)
Query: 87 HMSEKGMKIMASKGKMSNLKHVDLGVCEHCI-LGKQRKVSFSKAGRKSKLEKLKLVHTDV 145
+++ G + A G +KH+ VC C L AG K K ++ L H+ +
Sbjct: 62 YINSPGGSVTAGMGIFDTMKHIRPDVCTICTGLAASMGAFLLSAGAKGK--RMSLPHSRI 119
Query: 146 WGPAPVKSLGGSRYYVTFIDDSTRKV*VYFLKNKSDVFSVFKG 188
P LGG++ T I+ R++ +Y + +D + G
Sbjct: 120 MIHQP---LGGAQGQATDIEIQAREI-LYHKRRLNDYLAEHTG 158
>REC7_SCHPO (P36625) Meiotic recombination protein rec7
Length = 339
Score = 28.9 bits (63), Expect = 7.9
Identities = 25/101 (24%), Positives = 45/101 (43%), Gaps = 6/101 (5%)
Query: 58 GSLYMVAEEDMIAVTEAVNSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLGVCEHCI 117
G L + E++I +++N +WHQ + EK + ++ S+ N++ V +
Sbjct: 40 GLLQIRQHEELI---QSINLLDLWHQLIPGTKEKCLTLL-SRAPCMNIRAFTNNVMKRFQ 95
Query: 118 LGKQRKVSFSKAGRKSKLEKLKLVHTDVWGPAPVKSLGGSR 158
+ V + KA K + EKL LV D + K S+
Sbjct: 96 VKFPSDVHYMKA--KVEFEKLGLVFKDAKSSSEKKQFNNSQ 134
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.136 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,596,149
Number of Sequences: 164201
Number of extensions: 868518
Number of successful extensions: 1826
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1809
Number of HSP's gapped (non-prelim): 18
length of query: 196
length of database: 59,974,054
effective HSP length: 105
effective length of query: 91
effective length of database: 42,732,949
effective search space: 3888698359
effective search space used: 3888698359
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0065.20


