

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0065.18
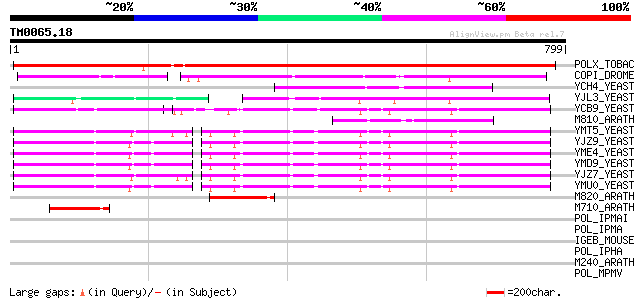
(799 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 835 0.0
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 360 1e-98
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 180 1e-44
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 162 2e-39
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 145 3e-34
M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810... 143 2e-33
YMT5_YEAST (Q04214) Transposon Ty1 protein B 135 4e-31
YJZ9_YEAST (P47100) Transposon Ty1 protein B 135 6e-31
YME4_YEAST (Q04711) Transposon Ty1 protein B 133 2e-30
YMD9_YEAST (Q03434) Transposon Ty1 protein B 132 3e-30
YJZ7_YEAST (P47098) Transposon Ty1 protein B 132 3e-30
YMU0_YEAST (Q04670) Transposon Ty1 protein B 130 1e-29
M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820... 80 2e-14
M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710... 77 2e-13
POL_IPMAI (P12894) Probable Pol polyprotein [Contains: Integrase... 43 0.004
POL_IPMA (P11368) Putative Pol polyprotein [Contains: Integrase ... 43 0.004
IGEB_MOUSE (P03975) IgE-binding protein 43 0.004
POL_IPHA (P04026) Putative Pol polyprotein [Contains: Integrase ... 42 0.005
M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240... 41 0.014
POL_MPMV (P07572) Pol polyprotein [Contains: Reverse transcripta... 40 0.032
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 835 bits (2157), Expect = 0.0
Identities = 430/793 (54%), Positives = 559/793 (70%), Gaps = 16/793 (2%)
Query: 6 VENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRTLNER 65
VE +T K+K L+SDNGGEY S+EF+++CS +GIR KT+ GTP+ NGVAERMNRT+ E+
Sbjct: 536 VERETGRKLKRLRSDNGGEYTSREFEEYCSSHGIRHEKTVPGTPQHNGVAERMNRTIVEK 595
Query: 66 ARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVFGCVS 125
R M + LPK FW +A+ TA YLINR PS+PL +++PE VW+ KEVS SHLKVFGC +
Sbjct: 596 VRSMLRMAKLPKSFWGEAVQTACYLINRSPSVPLAFEIPERVWTNKEVSYSHLKVFGCRA 655
Query: 126 YVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYKDRSS 185
+ + ++R KLD K+I C FIGYG + +GYR WD KK+IRSR+V F ES +
Sbjct: 656 FAHVPKEQRTKLDDKSIPCIFIGYGDEEFGYRLWDPVKKKVIRSRDVVFRESEVRTAADM 715
Query: 186 AESMS-----------SSKQLKLSERVALEEISESDVVKRNQINPENEDDEVEVELEQEP 234
+E + S+ S +E+SE I + DE E+E
Sbjct: 716 SEKVKNGIIPNFVTIPSTSNNPTSAESTTDEVSEQGEQPGEVIEQGEQLDEGVEEVEHP- 774
Query: 235 TTVIETPITQVRRSTRTPKAPQRYSPSLNYLLLTDAGEPEYFGEAMQGNDSIKWELAMKD 294
T E +RRS R P+ R PS Y+L++D EPE E + + + AM++
Sbjct: 775 -TQGEEQHQPLRRSER-PRVESRRYPSTEYVLISDDREPESLKEVLSHPEKNQLMKAMQE 832
Query: 295 EMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGSR-RYKARLVVKGFQQKQGIDFT 353
EM SLQKNGT+ L +LP+GK+ L+ +WV++LK++ D RYKARLVVKGF+QK+GIDF
Sbjct: 833 EMESLQKNGTYKLVELPKGKRPLKCKWVFKLKKDGDCKLVRYKARLVVKGFEQKKGIDFD 892
Query: 354 EIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGFEVLGTKNL 413
EIFSPVVKMT+IR ILS+ A+ +L +EQLDVK AFLHGDLEEEIYM QPEGFEV G K++
Sbjct: 893 EIFSPVVKMTSIRTILSLAASLDLEVEQLDVKTAFLHGDLEEEIYMEQPEGFEVAGKKHM 952
Query: 414 VCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKKFADS-YIILALYVDDM 472
VCKL+KSLYGLKQAPRQWY KF+ FM + + + D C + K+F+++ +IIL LYVDDM
Sbjct: 953 VCKLNKSLYGLKQAPRQWYMKFDSFMKSQTYLKTYSDPCVYFKRFSENNFIILLLYVDDM 1012
Query: 473 LIAGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVLKLSQEKYVEKLLDR 532
LI G + I +LK +S++F+MKDLGPA+QILGM+I R R+ L LSQEKY+E++L+R
Sbjct: 1013 LIVGKDKGLIAKLKGDLSKSFDMKDLGPAQQILGMKIVRERTSRKLWLSQEKYIERVLER 1072
Query: 533 FNVGDANTRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYASAVGSLMYAMVCTRPDIAHA 592
FN+ +A STPL HLK SKK P T EE+ M+ VPY+SAVGSLMYAMVCTRPDIAHA
Sbjct: 1073 FNMKNAKPVSTPLAGHLKLSKKMCPTTVEEKGNMAKVPYSSAVGSLMYAMVCTRPDIAHA 1132
Query: 593 VGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCFRRNNLTLQEFSDADLGGDSDGGKS 652
VGVVSRF+ NPGKEHWE VKWILRYL+G++ CLCF ++ L+ ++DAD+ GD D KS
Sbjct: 1133 VGVVSRFLENPGKEHWEAVKWILRYLRGTTGDCLCFGGSDPILKGYTDADMAGDIDNRKS 1192
Query: 653 TTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWLKSFLKELGKEQDVPP 712
+TGY+FT G A+SW+SKLQ VALSTTE+EY+A +E KEMIWLK FL+ELG Q
Sbjct: 1193 STGYLFTFSGGAISWQSKLQKCVALSTTEAEYIAATETGKEMIWLKRFLQELGLHQKEYV 1252
Query: 713 LFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSLLKILGSENPTDMLTKTVT 772
++ DSQS I L+KN ++H+R KHI ++YH+IRE++ DE L +LKI +ENP DMLTK V
Sbjct: 1253 VYCDSQSAIDLSKNSMYHARTKHIDVRYHWIREMVDDESLKVLKISTNENPADMLTKVVP 1312
Query: 773 ADNLRLCIASAGL 785
+ LC G+
Sbjct: 1313 RNKFELCKELVGM 1325
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 360 bits (923), Expect = 1e-98
Identities = 211/544 (38%), Positives = 307/544 (55%), Gaps = 27/544 (4%)
Query: 246 RRSTRTPKAPQ----RYSPSLNYLLLTDA----GEPEYFGEAMQGNDSIKWELAMKDEMT 297
RRS R PQ SLN ++L P F E +D WE A+ E+
Sbjct: 856 RRSERLKTKPQISYNEEDNSLNKVVLNAHTIFNDVPNSFDEIQYRDDKSSWEEAINTELN 915
Query: 298 SLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGSR-RYKARLVVKGFQQKQGIDFTEIF 356
+ + N TW++TK PE K + +RWV+ +K G+ RYKARLV +GF QK ID+ E F
Sbjct: 916 AHKINNTWTITKRPENKNIVDSRWVFSVKYNELGNPIRYKARLVARGFTQKYQIDYEETF 975
Query: 357 SPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGFEVLGTKNLVCK 416
+PV ++++ R ILS+V NL + Q+DVK AFL+G L+EEIYM P+G + VCK
Sbjct: 976 APVARISSFRFILSLVIQYNLKVHQMDVKTAFLNGTLKEEIYMRLPQGISC--NSDNVCK 1033
Query: 417 LHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFV--KKFADSYIILALYVDDMLI 474
L+K++YGLKQA R W++ F + + F +D C ++ K + I + LYVDD++I
Sbjct: 1034 LNKAIYGLKQAARCWFEVFEQALKECEFVNSSVDRCIYILDKGNINENIYVLLYVDDVVI 1093
Query: 475 AGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVLKLSQEKYVEKLLDRFN 534
A +MT +N K+ + E F M DL K +G+RI E + LSQ YV+K+L +FN
Sbjct: 1094 ATGDMTRMNNFKRYLMEKFRMTDLNEIKHFIGIRI--EMQEDKIYLSQSAYVKKILSKFN 1151
Query: 535 VGDANTRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYASAVGSLMYAMVCTRPDIAHAVG 594
+ + N STPL + + + S DE+ P S +G LMY M+CTRPD+ AV
Sbjct: 1152 MENCNAVSTPLPSKINYELLNS---DED----CNTPCRSLIGCLMYIMLCTRPDLTTAVN 1204
Query: 595 VVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCFRRN---NLTLQEFSDADLGGDSDGGK 651
++SR+ S E W+ +K +LRYLKG+ M L F++N + + D+D G K
Sbjct: 1205 ILSRYSSKNNSELWQNLKRVLRYLKGTIDMKLIFKKNLAFENKIIGYVDSDWAGSEIDRK 1264
Query: 652 STTGYIFTL-GGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWLKSFLKELGKEQDV 710
STTGY+F + + W +K QN VA S+TE+EY+A+ EA +E +WLK L + + +
Sbjct: 1265 STTGYLFKMFDFNLICWNTKRQNSVAASSTEAEYMALFEAVREALWLKFLLTSINIKLEN 1324
Query: 711 P-PLFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSLLKILGSENPTDMLTK 769
P ++ D+Q I +A NP H R KHI +KYHF RE + + + L I D+ TK
Sbjct: 1325 PIKIYEDNQGCISIANNPSCHKRAKHIDIKYHFAREQVQNNVICLEYIPTENQLADIFTK 1384
Query: 770 TVTA 773
+ A
Sbjct: 1385 PLPA 1388
Score = 131 bits (330), Expect = 6e-30
Identities = 82/218 (37%), Positives = 118/218 (53%), Gaps = 6/218 (2%)
Query: 12 LKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRTLNERARCMRI 71
LK+ L DNG EY S E ++FC + GI T+ TP+ NGV+ERM RT+ E+AR M
Sbjct: 542 LKVVYLYIDNGREYLSNEMRQFCVKKGISYHLTVPHTPQLNGVSERMIRTITEKARTMVS 601
Query: 72 QSGLPKMFWVDAINTAAYLINRGPSIPL--DYQLPEEVWSGKEVSLSHLKVFGCVSYVLI 129
+ L K FW +A+ TA YLINR PS L + P E+W K+ L HL+VFG YV I
Sbjct: 602 GAKLDKSFWGEAVLTATYLINRIPSRALVDSSKTPYEMWHNKKPYLKHLRVFGATVYVHI 661
Query: 130 DSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYKDRSSAESM 189
+++ K D K+ K F+GY + G++ WD N+K I +R+V +E+ + R+
Sbjct: 662 -KNKQGKFDDKSFKSIFVGY--EPNGFKLWDAVNEKFIVARDVVVDETNMVNSRAVKFET 718
Query: 190 SSSKQLKLSERVALEEISESDVVKRNQINPENEDDEVE 227
K K SE S +++ N E D ++
Sbjct: 719 VFLKDSKESENKNFPNDSRK-IIQTEFPNESKECDNIQ 755
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 180 bits (457), Expect = 1e-44
Identities = 105/315 (33%), Positives = 167/315 (52%), Gaps = 9/315 (2%)
Query: 382 LDVKIAFLHGDLEEEIYMTQPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSN 441
+DV AFL+ ++E IY+ QP GF + V +L+ +YGLKQAP W + N +
Sbjct: 1 MDVDTAFLNSTMDEPIYVKQPPGFVNERNPDYVWELYGGMYGLKQAPLLWNEHINNTLKK 60
Query: 442 SGFNRCDMDHCCFVKKFADSYIILALYVDDMLIAGSNMTEINRLKQQMSENFEMKDLGPA 501
GF R + +H + + +D I + +YVDD+L+A + +R+KQ++++ + MKDLG
Sbjct: 61 IGFCRHEGEHGLYFRSTSDGPIYIGVYVDDLLVAAPSPKIYDRVKQELTKLYSMKDLGKV 120
Query: 502 KQILGMRISRNRSEGVLKLSQEKYVEKLLDRFNVGDANTRSTPLGNHLKFSKKQSPQTDE 561
+ LG+ I ++ + G + LS + Y+ K + TPL N + SP +
Sbjct: 121 DKFLGLNIHQS-TNGDITLSLQDYIAKAASESEINTFKLTQTPLCNSKPLFETTSPHLKD 179
Query: 562 EESYMSTVPYASAVGSLMYAMVCTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRYLKGS 621
PY S VG L++ RPDI++ V ++SRF+ P H E + +LRYL +
Sbjct: 180 ------ITPYQSIVGQLLFCANTGRPDISYPVSLLSRFLREPRAIHLESARRVLRYLYTT 233
Query: 622 SRMCLCFRR-NNLTLQEFSDADLGGDSDGGKSTTGYIFTLGGTAVSWKS-KLQNRVALST 679
MCL +R + + L + DA G D ST GY+ L G V+W S KL+ + + +
Sbjct: 234 RSMCLKYRSGSQVALTVYCDASHGAIHDLPHSTGGYVTLLAGAPVTWSSKKLKGVIPVPS 293
Query: 680 TESEYVAISEAAKEM 694
TE+EY+ SE E+
Sbjct: 294 TEAEYITASETVMEI 308
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 162 bits (411), Expect = 2e-39
Identities = 126/456 (27%), Positives = 219/456 (47%), Gaps = 23/456 (5%)
Query: 335 YKARLVVKGFQQKQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLE 394
YKAR+V +G Q ++ I + + I++ L I N+ ++ LD+ AFL+ LE
Sbjct: 1337 YKARIVCRGDTQSPDT-YSVITTESLNHNHIKIFLMIANNRNMFMKTLDINHAFLYAKLE 1395
Query: 395 EEIYMTQPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCF 454
EEIY+ P + V KL+K+LYGLKQ+P++W ++++ G D +
Sbjct: 1396 EEIYIPHPH------DRRCVVKLNKALYGLKQSPKEWNDHLRQYLNGIGLK--DNSYTPG 1447
Query: 455 VKKFADSYIILALYVDDMLIAGSNMTEINRLKQQMSENFEMKDLGPA------KQILGMR 508
+ + D +++A+YVDD +IA SN ++ ++ NFE+K G ILGM
Sbjct: 1448 LYQTEDKNLMIAVYVDDCVIAASNEQRLDEFINKLKSNFELKITGTLIDDVLDTDILGMD 1507
Query: 509 ISRNRSEGVLKLSQEKYVEKLLDRFNVGDANTRSTPLGNHLKFS---KKQSPQTDEEESY 565
+ N+ G + L+ + ++ ++ ++N R + + + + KK Q EEE
Sbjct: 1508 LVYNKRLGTIDLTLKSFINRMDKKYNEELKKIRKSSIPHMSTYKIDPKKDVLQMSEEEFR 1567
Query: 566 MSTVPYASAVGSLMYAMVCTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRYLKGSSRMC 625
+ +G L Y R DI AV V+R ++ P + + + I++YL +
Sbjct: 1568 QGVLKLQQLLGELNYVRHKCRYDIEFAVKKVARLVNYPHERVFYMIYKIIQYLVRYKDIG 1627
Query: 626 LCFRRN---NLTLQEFSDADLGGDSDGGKSTTGYIFTLGGTAVSWKSKLQNRVALSTTES 682
+ + R+ + + +DA +G + D +S G I G + S +S+TE+
Sbjct: 1628 IHYDRDCNKDKKVIAITDASVGSEYD-AQSRIGVILWYGMNIFNVYSNKSTNRCVSSTEA 1686
Query: 683 EYVAISEAAKEMIWLKSFLKELGK-EQDVPPLFSDSQSVIFLAKNPVFHSRCKHIQMKYH 741
E AI E + LK LKELG+ + + + +DS+ I + K +K
Sbjct: 1687 ELHAIYEGYADSETLKVTLKELGEGDNNDIVMITDSKPAIQGLNRSYQQPKEKFTWIKTE 1746
Query: 742 FIRELISDEELSLLKILGSENPTDMLTKTVTADNLR 777
I+E I ++ + LLKI G N D+LTK V+A + +
Sbjct: 1747 IIKEKIKEKSIKLLKITGKGNIADLLTKPVSASDFK 1782
Score = 64.7 bits (156), Expect = 9e-10
Identities = 66/287 (22%), Positives = 117/287 (39%), Gaps = 15/287 (5%)
Query: 6 VENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRTLNER 65
VE Q K++ + SD G E+ + + +++ GI I T NG AER RT+
Sbjct: 682 VETQFDRKVREINSDRGTEFTNDQIEEYFISKGIHHILTSTQDHAANGRAERYIRTIITD 741
Query: 66 ARCMRIQSGLPKMFWVDAINTAA----YLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVF 121
A + QS L FW A+ +A YL ++ +LP + S + V++ +
Sbjct: 742 ATTLLRQSNLRVKFWEYAVTSATNIRNYLEHKSTG-----KLPLKAISRQPVTVRLMSFL 796
Query: 122 GCVSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYK 181
+I + KL P + + + YGY+F+ KI+ S N T +
Sbjct: 797 PFGEKGIIWNHNHKKLKPSGLPSIILCKDPNSYGYKFFIPSKNKIVTSDNYTIPNYTM-- 854
Query: 182 DRSSAESMSSSKQLKLSERVALEEISESDVVKRNQINPENEDDEVEVELEQEPTTVIETP 241
D + + +K + S EE V + EDD + ++ T E
Sbjct: 855 DGRVRNTQNINKSHQFSSDNDDEEDQIETVTNLCEALENYEDDNKPITRLEDLFT--EEE 912
Query: 242 ITQVRRSTRTPKAPQRYSPSLNYLL--LTDAGEPEYFGEAMQGNDSI 286
++Q+ + + P L+Y+ + ++G+ + E N+SI
Sbjct: 913 LSQIDSNAKYPSPSNNLEGDLDYVFSDVEESGDYDVESELSTTNNSI 959
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 145 bits (367), Expect = 3e-34
Identities = 146/594 (24%), Positives = 264/594 (43%), Gaps = 65/594 (10%)
Query: 222 EDDEVEVELEQEPTT-----VIETPITQVR----------RSTRTPKAPQRYSPSLNYLL 266
ED+E E+E+ ++ +E P ++ R +S + + RY ++ Y
Sbjct: 1198 EDNETEIEVSRDTWNNKNMRSLEPPRSKKRINLIAAIKGVKSIKPVRTTLRYDEAITYN- 1256
Query: 267 LTDAGEPEYFGEAMQGNDSIKWELAMKDEMTSLQKNGTWSLTKLPEG-----KKALQNRW 321
D E + + EA E++ L K TW K + KK + + +
Sbjct: 1257 -KDNKEKDRYVEAYH------------KEISQLLKMNTWDTNKYYDRNDIDPKKVINSMF 1303
Query: 322 VYRLKEESDGSRRYKARLVVKGFQQKQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQ 381
++ K DG+ +KAR V +G Q +++ S V + LSI + ++ Q
Sbjct: 1304 IFNKKR--DGT--HKARFVARGDIQHPDTYDSDMQSNTVHHYALMTSLSIALDNDYYITQ 1359
Query: 382 LDVKIAFLHGDLEEEIYMTQPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSN 441
LD+ A+L+ D++EE+Y+ P LG + + +L KSLYGLKQ+ WY+ ++ N
Sbjct: 1360 LDISSAYLYADIKEELYIRPPPH---LGLNDKLLRLRKSLYGLKQSGANWYETIKSYLIN 1416
Query: 442 SGFNRCDMDHCC-FVKKFADSYIILALYVDDMLIAGSNMTEINRLKQQMSENFEMK--DL 498
CDM + F +S + + L+VDDM++ ++ ++ + + ++ K +L
Sbjct: 1417 C----CDMQEVRGWSCVFKNSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINL 1472
Query: 499 GPAKQ-----ILGMRISRNRSEGVLKLSQEKYVEKLLDRFNVGDANTRSTPL---GNHLK 550
G + ILG+ I RS+ +KL EK + + L + NV N + L G
Sbjct: 1473 GESDNEIQYDILGLEIKYQRSK-YMKLGMEKSLTEKLPKLNV-PLNPKGKKLRAPGQPGH 1530
Query: 551 FSKKQSPQTDEEESYMSTVPYASAVGSLMYAMVCTRPDIAHAVGVVSRFMSNPGKEHWEC 610
+ + + DE+E +G Y R D+ + + +++ + P ++ +
Sbjct: 1531 YIDQDELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDM 1590
Query: 611 VKWILRYLKGSSRMCLCFRRNNLT-----LQEFSDADLGGDSDGGKSTTGYIFTLGGTAV 665
+++++ + L + +N T L SDA G + KS G IF L G +
Sbjct: 1591 TYELIQFMWDTRDKQLIWHKNKPTKPDNKLVAISDASYG-NQPYYKSQIGNIFLLNGKVI 1649
Query: 666 SWKSKLQNRVALSTTESEYVAISEAAKEMIWLKSFLKELGKEQDVPPLFSDSQSVIFLAK 725
KS + STTE+E A+SEA + L ++EL K+ + L +DS+S I + K
Sbjct: 1650 GGKSTKASLTCTSTTEAEIHAVSEAIPLLNNLSHLVQELNKKPIIKGLLTDSRSTISIIK 1709
Query: 726 NPVFHS-RCKHIQMKYHFIRELISDEELSLLKILGSENPTDMLTKTVTADNLRL 778
+ R + K +R+ +S L + I +N D++TK + +L
Sbjct: 1710 STNEEKFRNRFFGTKAMRLRDEVSGNNLYVYYIETKKNIADVMTKPLPIKTFKL 1763
Score = 62.8 bits (151), Expect = 4e-09
Identities = 53/232 (22%), Positives = 104/232 (43%), Gaps = 6/232 (2%)
Query: 6 VENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRTLNER 65
++NQ ++ ++ D G EY ++ KF + GI T +GVAER+NRTL
Sbjct: 718 IKNQFNARVLVIQMDRGSEYTNKTLHKFFTNRGITACYTTTADSRAHGVAERLNRTLLND 777
Query: 66 ARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVFGCVS 125
R + SGLP W A+ + + N S P + + + + ++ + FG
Sbjct: 778 CRTLLHCSGLPNHLWFSAVEFSTIIRNSLVS-PKNDKSARQHAGLAGLDITTILPFG--Q 834
Query: 126 YVLIDSDRRD-KLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRN-VTFNESVLYKDR 183
V++++ D K+ P+ I + + + YGY + KK + + N V + D+
Sbjct: 835 PVIVNNHNPDSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQDKQSKLDQ 894
Query: 184 SSAESMSSSKQLKLSERVALEEISESDVVKRNQINPENEDD-EVEVELEQEP 234
+ ++++ L I +++ + N E++ D + E+E+ +P
Sbjct: 895 FNYDTLTFDDDLNRLTAHNQSFIEQNETEQSYDQNTESDHDYQSEIEINSDP 946
>M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810
(ORF240b)
Length = 240
Score = 143 bits (361), Expect = 2e-33
Identities = 90/233 (38%), Positives = 132/233 (56%), Gaps = 11/233 (4%)
Query: 465 LALYVDDMLIAGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVLKLSQEK 524
L LYVDD+L+ GS+ T +N L Q+S F MKDLGP LG++I + S L LSQ K
Sbjct: 3 LLLYVDDILLTGSSNTLLNMLIFQLSSTFSMKDLGPVHYFLGIQIKTHPSG--LFLSQTK 60
Query: 525 YVEKLLDRFNVGDANTRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYASAVGSLMYAMVC 584
Y E++L+ + D STPL L S + D + + S VG+L Y +
Sbjct: 61 YAEQILNNAGMLDCKPMSTPLPLKLNSSVSTAKYPDPSD-------FRSIVGALQY-LTL 112
Query: 585 TRPDIAHAVGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCFRRNN-LTLQEFSDADL 643
TRPDI++AV +V + M P ++ +K +LRY+KG+ L +N+ L +Q F D+D
Sbjct: 113 TRPDISYAVNIVCQRMHEPTLADFDLLKRVLRYVKGTIFHGLYIHKNSKLNVQAFCDSDW 172
Query: 644 GGDSDGGKSTTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIW 696
G + +STTG+ LG +SW +K Q V+ S+TE+EY A++ A E+ W
Sbjct: 173 AGCTSTRRSTTGFCTFLGCNIISWSAKRQPTVSRSSTETEYRALALTAAELTW 225
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 135 bits (340), Expect = 4e-31
Identities = 130/527 (24%), Positives = 236/527 (44%), Gaps = 36/527 (6%)
Query: 276 FGEAMQGNDSIK----WELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWV---YRLKEE 328
+ EA+ N IK + A E+ L K TW K + K+ R + + +
Sbjct: 807 YDEAITYNKDIKEKEKYIEAYHKEVNQLLKMKTWDTDKYYDRKEIDPKRVINSMFIFNRK 866
Query: 329 SDGSRRYKARLVVKGFQQKQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAF 388
DG+ +KAR V +G Q + + S V + LS+ N ++ QLD+ A+
Sbjct: 867 RDGT--HKARFVARGDIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAY 924
Query: 389 LHGDLEEEIYMTQPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCD 448
L+ D++EE+Y+ P LG + + +L KSLYGLKQ+ WY+ ++ +C
Sbjct: 925 LYADIKEELYIRPPPH---LGMNDKLIRLKKSLYGLKQSGANWYETIKSYL----IKQCG 977
Query: 449 MDHCC-FVKKFADSYIILALYVDDMLIAGSNMTEINRLKQQMSENFEMK--DLGPAKQ-- 503
M+ + F +S + + L+VDDM++ N+ R+ ++ ++ K +LG + +
Sbjct: 978 MEEVRGWSCVFENSQVTICLFVDDMVLFSKNLNSNKRIIDKLKMQYDTKIINLGESDEEI 1037
Query: 504 ---ILGMRISRNRSEGVLKLSQEKYVEKLLDRFNVGDANTRSTPL---GNHLKFSKKQSP 557
ILG+ I R + +KL E + + + + NV N + L G + +Q
Sbjct: 1038 QYDILGLEIKYQRGK-YMKLGMENSLTEKIPKLNV-PLNPKGRKLSAPGQPGLYIDQQEL 1095
Query: 558 QTDEEESYMSTVPYASAVGSLMYAMVCTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRY 617
+ +E++ M +G Y R D+ + + +++ + P K+ + ++++
Sbjct: 1096 ELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQF 1155
Query: 618 LKGSSRMCLCFRRNNLT-----LQEFSDADLGGDSDGGKSTTGYIFTLGGTAVSWKSKLQ 672
+ + L + ++ L SDA G + KS G I+ L G + KS
Sbjct: 1156 IWNTRDKQLIWHKSKPVKPTNKLVVISDASYG-NQPYYKSQIGNIYLLNGKVIGGKSTKA 1214
Query: 673 NRVALSTTESEYVAISEAAKEMIWLKSFLKELGKEQDVPPLFSDSQSVI-FLAKNPVFHS 731
+ STTE+E AISE+ + L ++EL K+ L +DS+S I + N
Sbjct: 1215 SLTCTSTTEAEIHAISESVPLLNNLSYLIQELDKKPITKGLLTDSKSTISIIISNNEEKF 1274
Query: 732 RCKHIQMKYHFIRELISDEELSLLKILGSENPTDMLTKTVTADNLRL 778
R + K +R+ +S L + I +N D++TK + +L
Sbjct: 1275 RNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMTKPLPIKTFKL 1321
Score = 70.9 bits (172), Expect = 1e-11
Identities = 66/276 (23%), Positives = 118/276 (41%), Gaps = 22/276 (7%)
Query: 6 VENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRTLNER 65
++NQ + ++ D G EY ++ KF +NGI T +GVAER+NRTL +
Sbjct: 295 IKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDD 354
Query: 66 ARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVFGCVS 125
R SGLP W AI + ++ + P + + + +S L FG
Sbjct: 355 CRTQLQCSGLPNHLWFSAIEFST-IVRNSLASPKSKKSARQHAGLAGLDISTLLPFG-QP 412
Query: 126 YVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTF---NESVLYKD 182
++ D + K+ P+ I + + + YGY + KK + + N ES L D
Sbjct: 413 VIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRL--D 470
Query: 183 RSSAESMSSSKQLKLSERVALEEISESDVVKRNQINPENEDD-EVEVELEQE-------- 233
+ + ++++ + L I+ +++ + N +N E++ D + ++EL E
Sbjct: 471 QFNYDALTFDEDLNRLTASYHSFIASNEIQQSNDLNIESDHDFQSDIELHPEQLRNVLSK 530
Query: 234 --PTTVIETPITQVRRSTRTPK----APQRYSPSLN 263
T P T S R K AP+ P+++
Sbjct: 531 AVSPTDSTPPSTHTEDSKRVSKTNIRAPREVDPNIS 566
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 135 bits (339), Expect = 6e-31
Identities = 129/527 (24%), Positives = 238/527 (44%), Gaps = 36/527 (6%)
Query: 276 FGEAMQGNDSIK----WELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWV---YRLKEE 328
+ EA+ N IK + A E+ L K TW + + K+ R + + ++
Sbjct: 1234 YDEAITYNKDIKEKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEIDPKRVINSMFIFNKK 1293
Query: 329 SDGSRRYKARLVVKGFQQKQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAF 388
DG+ +KAR V +G Q + + S V + LS+ N ++ QLD+ A+
Sbjct: 1294 RDGT--HKARFVARGDIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAY 1351
Query: 389 LHGDLEEEIYMTQPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCD 448
L+ D++EE+Y+ P LG + + +L KSLYGLKQ+ WY+ ++ +C
Sbjct: 1352 LYADIKEELYIRPPPH---LGMNDKLIRLKKSLYGLKQSGANWYETIKSYL----IQQCG 1404
Query: 449 MDHCC-FVKKFADSYIILALYVDDMLIAGSNMTEINRLKQQMSENFEMK--DLGPAKQ-- 503
M+ + F +S + + L+VDDM++ N+ R+ +++ ++ K +LG + +
Sbjct: 1405 MEEVRGWSCVFKNSQVTICLFVDDMVLFSKNLNSNKRIIEKLKMQYDTKIINLGESDEEI 1464
Query: 504 ---ILGMRISRNRSEGVLKLSQEKYVEKLLDRFNVGDANTRSTPL---GNHLKFSKKQSP 557
ILG+ I R + +KL E + + + + NV N + L G + +Q
Sbjct: 1465 QYDILGLEIKYQRGK-YMKLGMENSLTEKIPKLNV-PLNPKGRKLSAPGQPGLYIDQQEL 1522
Query: 558 QTDEEESYMSTVPYASAVGSLMYAMVCTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRY 617
+ +E++ M +G Y R D+ + + +++ + P K+ + ++++
Sbjct: 1523 ELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQF 1582
Query: 618 LKGSSRMCLCFRRNNLT-----LQEFSDADLGGDSDGGKSTTGYIFTLGGTAVSWKSKLQ 672
+ + L + ++ L SDA G + KS G I+ L G + KS
Sbjct: 1583 IWNTRDKQLIWHKSKPVKPTNKLVVISDASYG-NQPYYKSQIGNIYLLNGKVIGGKSTKA 1641
Query: 673 NRVALSTTESEYVAISEAAKEMIWLKSFLKELGKEQDVPPLFSDSQSVI-FLAKNPVFHS 731
+ STTE+E AISE+ + L ++EL K+ L +DS+S I + N
Sbjct: 1642 SLTCTSTTEAEIHAISESVPLLNNLSYLIQELDKKPITKGLLTDSKSTISIIISNNEEKF 1701
Query: 732 RCKHIQMKYHFIRELISDEELSLLKILGSENPTDMLTKTVTADNLRL 778
R + K +R+ +S L + I +N D++TK + +L
Sbjct: 1702 RNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMTKPLPIKTFKL 1748
Score = 72.0 bits (175), Expect = 6e-12
Identities = 64/277 (23%), Positives = 122/277 (43%), Gaps = 24/277 (8%)
Query: 6 VENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRTLNER 65
++NQ + ++ D G EY ++ KF +NGI T +GVAER+NRTL +
Sbjct: 722 IKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDD 781
Query: 66 ARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVFGCVS 125
R SGLP W AI + ++ + P + + + +S L FG
Sbjct: 782 CRTQLQCSGLPNHLWFSAIEFST-IVRNSLASPKSKKSARQHAGLAGLDISTLLPFG-QP 839
Query: 126 YVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRN-------------- 171
++ D + K+ P+ I + + + YGY + KK + + N
Sbjct: 840 VIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRLDQF 899
Query: 172 ----VTFNESVLYKDRSSAESMSSSKQLKLSERVALEEISESDVVKRNQINPENEDDEVE 227
+TF+E L + +S +S +S +++ S+ + +E S+ D +++PE + +
Sbjct: 900 NYDALTFDED-LNRLTASYQSFIASNEIQQSDDLNIE--SDHDFQSDIELHPEQPRNVLS 956
Query: 228 VELEQEPTTVIETPITQVRRSTRTP-KAPQRYSPSLN 263
+ +T T +R ++T +AP+ P+++
Sbjct: 957 KAVSPTDSTPPSTHTEDSKRVSKTNIRAPREVDPNIS 993
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 133 bits (335), Expect = 2e-30
Identities = 129/527 (24%), Positives = 234/527 (43%), Gaps = 36/527 (6%)
Query: 276 FGEAMQGNDSIK----WELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWV---YRLKEE 328
+ EA+ N IK + A E+ L K TW K + K+ R + + +
Sbjct: 807 YDEAITYNKDIKEKEKYIEAYHKEVNQLLKMNTWDTDKYYDRKEIDPKRVINSMFIFNRK 866
Query: 329 SDGSRRYKARLVVKGFQQKQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAF 388
DG+ +KAR V +G Q + + S V + LS+ N ++ QLD+ A+
Sbjct: 867 RDGT--HKARFVARGDIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAY 924
Query: 389 LHGDLEEEIYMTQPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCD 448
L+ D++EE+Y+ P LG + + +L KSLYGLKQ+ WY+ ++ +C
Sbjct: 925 LYADIKEELYIRPPPH---LGMNDKLIRLKKSLYGLKQSGANWYETIKSYL----IKQCG 977
Query: 449 MDHCC-FVKKFADSYIILALYVDDMLIAGSNMTEINRLKQQMSENFEMK--DLGPAKQ-- 503
M+ + F +S + + L+VDDM++ ++ ++ + + ++ K +LG +
Sbjct: 978 MEEVRGWSCVFKNSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEI 1037
Query: 504 ---ILGMRISRNRSEGVLKLSQEKYVEKLLDRFNVGDANTRSTPL---GNHLKFSKKQSP 557
ILG+ I R + +KL E + + + + NV N + L G + +
Sbjct: 1038 QYDILGLEIKYQRGK-YMKLGMENSLTEKIPKLNV-PLNPKGRKLSAPGQPGLYIDQDEL 1095
Query: 558 QTDEEESYMSTVPYASAVGSLMYAMVCTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRY 617
+ DE+E +G Y R D+ + + +++ + P ++ + ++++
Sbjct: 1096 EIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQF 1155
Query: 618 LKGSSRMCLCFRRNNLT-----LQEFSDADLGGDSDGGKSTTGYIFTLGGTAVSWKSKLQ 672
+ + L + +N T L SDA G + KS G I+ L G + KS
Sbjct: 1156 MWDTRDKQLIWHKNKPTEPDNKLVAISDASYG-NQPYYKSQIGNIYLLNGKVIGGKSTKA 1214
Query: 673 NRVALSTTESEYVAISEAAKEMIWLKSFLKELGKEQDVPPLFSDSQSVI-FLAKNPVFHS 731
+ STTE+E AISE+ + L ++EL K+ L +DS+S I + N
Sbjct: 1215 SLTCTSTTEAEIHAISESVPLLNNLSHLVQELNKKPITKGLLTDSKSTISIIISNNEEKF 1274
Query: 732 RCKHIQMKYHFIRELISDEELSLLKILGSENPTDMLTKTVTADNLRL 778
R + K +R+ +S L + I +N D++TK + +L
Sbjct: 1275 RNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMTKPLPIKTFKL 1321
Score = 72.0 bits (175), Expect = 6e-12
Identities = 64/277 (23%), Positives = 122/277 (43%), Gaps = 24/277 (8%)
Query: 6 VENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRTLNER 65
++NQ + ++ D G EY ++ KF +NGI T +GVAER+NRTL +
Sbjct: 295 IKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDD 354
Query: 66 ARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVFGCVS 125
R SGLP W AI + ++ + P + + + +S L FG
Sbjct: 355 CRTQLQCSGLPNHLWFSAIEFST-IVRNSLASPKSKKSARQHAGLAGLDISTLLPFG-QP 412
Query: 126 YVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRN-------------- 171
++ D + K+ P+ I + + + YGY + KK + + N
Sbjct: 413 VIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRLDQF 472
Query: 172 ----VTFNESVLYKDRSSAESMSSSKQLKLSERVALEEISESDVVKRNQINPENEDDEVE 227
+TF+E L + +S +S +S +++ S+ + +E S+ D +++PE + +
Sbjct: 473 NYDALTFDED-LNRLTASYQSFIASNEIQQSDDLNIE--SDHDFQSDIELHPEQPRNVLS 529
Query: 228 VELEQEPTTVIETPITQVRRSTRTP-KAPQRYSPSLN 263
+ +T T +R ++T +AP+ P+++
Sbjct: 530 KAVSPTDSTPPSTHTEDSKRVSKTNIRAPREVDPNIS 566
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 132 bits (333), Expect = 3e-30
Identities = 128/527 (24%), Positives = 237/527 (44%), Gaps = 36/527 (6%)
Query: 276 FGEAMQGNDSIK----WELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWV---YRLKEE 328
+ EA+ N IK + A E+ L K TW + + K+ R + + ++
Sbjct: 807 YDEAITYNKDIKEKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEIDPKRVINSMFIFNKK 866
Query: 329 SDGSRRYKARLVVKGFQQKQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAF 388
DG+ +KAR V +G Q + + S V + LS+ N ++ QLD+ A+
Sbjct: 867 RDGT--HKARFVARGDIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAY 924
Query: 389 LHGDLEEEIYMTQPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCD 448
L+ D++EE+Y+ P LG + + +L KSLYGLKQ+ WY+ ++ +C
Sbjct: 925 LYADIKEELYIRPPPH---LGMNDKLIRLKKSLYGLKQSGANWYETIKSYL----IKQCG 977
Query: 449 MDHCC-FVKKFADSYIILALYVDDMLIAGSNMTEINRLKQQMSENFEMK--DLGPAKQ-- 503
M+ + F +S + + L+VDDM++ ++ ++ + + ++ K +LG +
Sbjct: 978 MEEVRGWSCVFKNSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEI 1037
Query: 504 ---ILGMRISRNRSEGVLKLSQEKYVEKLLDRFNVGDANTRSTPL---GNHLKFSKKQSP 557
ILG+ I R + +KL E + + + + NV N + L G + +
Sbjct: 1038 QYDILGLEIKYQRGK-YMKLGMENSLTEKIPKLNV-PLNPKGRKLSAPGQPGLYIDQDEL 1095
Query: 558 QTDEEESYMSTVPYASAVGSLMYAMVCTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRY 617
+ DE+E +G Y R D+ + + +++ + P ++ + ++++
Sbjct: 1096 EIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQF 1155
Query: 618 LKGSSRMCLCFRRNNLT-----LQEFSDADLGGDSDGGKSTTGYIFTLGGTAVSWKSKLQ 672
+ + L + +N T L SDA G + KS G I+ L G + KS
Sbjct: 1156 MWDTRDKQLIWHKNKPTEPDNKLVAISDASYG-NQPYYKSQIGNIYLLNGKVIGGKSTKA 1214
Query: 673 NRVALSTTESEYVAISEAAKEMIWLKSFLKELGKEQDVPPLFSDSQSVIFLAKNPVFHS- 731
+ STTE+E AISE+ + L ++EL K+ + L +DS+S I + K+
Sbjct: 1215 SLTCTSTTEAEIHAISESVPLLNNLSYLIQELNKKPIIKGLLTDSRSTISIIKSTNEEKF 1274
Query: 732 RCKHIQMKYHFIRELISDEELSLLKILGSENPTDMLTKTVTADNLRL 778
R + K +R+ +S L + I +N D++TK + +L
Sbjct: 1275 RNRFFGTKAMRLRDEVSGNNLYVYYIETKKNIADVMTKPLPIKTFKL 1321
Score = 72.0 bits (175), Expect = 6e-12
Identities = 64/277 (23%), Positives = 122/277 (43%), Gaps = 24/277 (8%)
Query: 6 VENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRTLNER 65
++NQ + ++ D G EY ++ KF +NGI T +GVAER+NRTL +
Sbjct: 295 IKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDD 354
Query: 66 ARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVFGCVS 125
R SGLP W AI + ++ + P + + + +S L FG
Sbjct: 355 CRTQLQCSGLPNHLWFSAIEFST-IVRNSLASPKSKKSARQHAGLAGLDISTLLPFG-QP 412
Query: 126 YVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRN-------------- 171
++ D + K+ P+ I + + + YGY + KK + + N
Sbjct: 413 VIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRLDQF 472
Query: 172 ----VTFNESVLYKDRSSAESMSSSKQLKLSERVALEEISESDVVKRNQINPENEDDEVE 227
+TF+E L + +S +S +S +++ S+ + +E S+ D +++PE + +
Sbjct: 473 NYDALTFDED-LNRLTASYQSFIASNEIQQSDDLNIE--SDHDFQSDIELHPEQPRNVLS 529
Query: 228 VELEQEPTTVIETPITQVRRSTRTP-KAPQRYSPSLN 263
+ +T T +R ++T +AP+ P+++
Sbjct: 530 KAVSPTDSTPPSTHTEDSKRVSKTNIRAPREVDPNIS 566
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 132 bits (333), Expect = 3e-30
Identities = 129/527 (24%), Positives = 236/527 (44%), Gaps = 36/527 (6%)
Query: 276 FGEAMQGNDSIK----WELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWV---YRLKEE 328
+ EA+ N IK + A E+ L K TW + + K+ R + + ++
Sbjct: 1234 YDEAITYNKDIKEKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEIDPKRVINSMFIFNKK 1293
Query: 329 SDGSRRYKARLVVKGFQQKQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAF 388
DG+ +KAR V +G Q T + S V + LS+ N ++ QLD+ A+
Sbjct: 1294 RDGT--HKARFVARGDIQHPDTYDTGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAY 1351
Query: 389 LHGDLEEEIYMTQPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCD 448
L+ D++EE+Y+ P LG + + +L KS YGLKQ+ WY+ ++ +C
Sbjct: 1352 LYADIKEELYIRPPPH---LGMNDKLIRLKKSHYGLKQSGANWYETIKSYL----IKQCG 1404
Query: 449 MDHCC-FVKKFADSYIILALYVDDMLIAGSNMTEINRLKQQMSENFEMK--DLGPAKQ-- 503
M+ + F +S + + L+VDDM++ ++ ++ + + ++ K +LG +
Sbjct: 1405 MEEVRGWSCVFKNSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEI 1464
Query: 504 ---ILGMRISRNRSEGVLKLSQEKYVEKLLDRFNVGDANTRSTPL---GNHLKFSKKQSP 557
ILG+ I R + +KL E + + + + NV N + L G + +
Sbjct: 1465 QYDILGLEIKYQRGK-YMKLGMENSLTEKIPKLNV-PLNPKGRKLSAPGQPGLYIDQDEL 1522
Query: 558 QTDEEESYMSTVPYASAVGSLMYAMVCTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRY 617
+ DE+E +G Y R D+ + + +++ + P ++ + ++++
Sbjct: 1523 EIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQF 1582
Query: 618 LKGSSRMCLCFRRNNLT-----LQEFSDADLGGDSDGGKSTTGYIFTLGGTAVSWKSKLQ 672
+ + L + +N T L SDA G + KS G IF L G + KS
Sbjct: 1583 MWDTRDKQLIWHKNKPTEPDNKLVAISDASYG-NQPYYKSQIGNIFLLNGKVIGGKSTKA 1641
Query: 673 NRVALSTTESEYVAISEAAKEMIWLKSFLKELGKEQDVPPLFSDSQSVIFLAKNPVFHS- 731
+ STTE+E AISE+ + L ++EL K+ + L +DS+S I + K+
Sbjct: 1642 SLTCTSTTEAEIHAISESVPLLNNLSYLIQELNKKPIIKGLLTDSRSTISIIKSTNEEKF 1701
Query: 732 RCKHIQMKYHFIRELISDEELSLLKILGSENPTDMLTKTVTADNLRL 778
R + K +R+ +S L + I +N D++TK + +L
Sbjct: 1702 RNRFFGTKAMRLRDEVSGNNLYVYYIETKKNIADVMTKPLPIKTFKL 1748
Score = 72.8 bits (177), Expect = 3e-12
Identities = 66/276 (23%), Positives = 121/276 (42%), Gaps = 22/276 (7%)
Query: 6 VENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRTLNER 65
++NQ + ++ D G EY ++ KF +NGI T +GVAER+NRTL +
Sbjct: 722 IKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDD 781
Query: 66 ARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVFGCVS 125
R SGLP W AI + ++ + P + + + +S L FG
Sbjct: 782 CRTQLQCSGLPNHLWFSAIEFST-IVRNSLASPKSKKSARQHAGLAGLDISTLLPFG-QP 839
Query: 126 YVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTF---NESVLYKD 182
++ D + K+ P+ I + + + YGY + KK + + N ES L D
Sbjct: 840 VIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRL--D 897
Query: 183 RSSAESMSSSKQLKLSERVALEEISESDVVKRNQINPENEDD-EVEVELE-QEPTTVIE- 239
+ + ++++ + L I+ +++ + N +N E++ D + ++EL ++P V+
Sbjct: 898 QFNYDALTFDEDLNRLTASYQSFIASNEIQESNDLNIESDHDFQSDIELHPEQPRNVLSK 957
Query: 240 --------TPITQVRRSTRTPK----APQRYSPSLN 263
P T S R K AP+ P+++
Sbjct: 958 AVSPTDSTPPSTHTEDSKRVSKTNIRAPREVDPNIS 993
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 130 bits (327), Expect = 1e-29
Identities = 127/527 (24%), Positives = 234/527 (44%), Gaps = 36/527 (6%)
Query: 276 FGEAMQGNDSIK----WELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWV---YRLKEE 328
+ EA+ N IK + A E+ L K TW + + K+ R + + +
Sbjct: 807 YDEAITYNKDIKEKEKYIQAYHKEVNQLLKMKTWDTDRYYDRKEIDPKRVINSMFIFNRK 866
Query: 329 SDGSRRYKARLVVKGFQQKQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAF 388
DG+ +KAR V +G Q + S V + LS+ N ++ QLD+ A+
Sbjct: 867 RDGT--HKARFVARGDIQHPDTYDPGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAY 924
Query: 389 LHGDLEEEIYMTQPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCD 448
L+ D++EE+Y+ P LG + + +L KSLYGLKQ+ WY+ ++ +C
Sbjct: 925 LYADIKEELYIRPPPH---LGMNDKLIRLKKSLYGLKQSGANWYETIKSYL----IKQCG 977
Query: 449 MDHCC-FVKKFADSYIILALYVDDMLIAGSNMTEINRLKQQMSENFEMK--DLGPAKQ-- 503
M+ + F +S + + L+VDDM++ ++ ++ + + ++ K +LG +
Sbjct: 978 MEEVRGWSCVFKNSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEI 1037
Query: 504 ---ILGMRISRNRSEGVLKLSQEKYVEKLLDRFNVGDANTRSTPL---GNHLKFSKKQSP 557
ILG+ I R + +KL E + + + + NV N + L G + +Q
Sbjct: 1038 QYDILGLEIKYQRGK-YMKLGMENSLTEKIPKLNV-PLNPKGRKLSAPGQPGLYIDQQEL 1095
Query: 558 QTDEEESYMSTVPYASAVGSLMYAMVCTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRY 617
+ +E++ M +G Y R D+ + + +++ + P K+ + ++++
Sbjct: 1096 ELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQF 1155
Query: 618 LKGSSRMCLCFRRNNLT-----LQEFSDADLGGDSDGGKSTTGYIFTLGGTAVSWKSKLQ 672
+ + L + ++ L SDA G + KS G I+ L G + KS
Sbjct: 1156 IWNTRDKQLIWHKSKPVKPTNKLVVISDASYG-NQPYYKSQIGNIYLLNGKVIGGKSTKA 1214
Query: 673 NRVALSTTESEYVAISEAAKEMIWLKSFLKELGKEQDVPPLFSDSQSVI-FLAKNPVFHS 731
+ STTE+E AISE+ + L ++EL K+ L +DS+S I + N
Sbjct: 1215 SLTCTSTTEAEIHAISESVPLLNNLSHLVQELNKKPITKGLLTDSKSTISIIISNNEEKF 1274
Query: 732 RCKHIQMKYHFIRELISDEELSLLKILGSENPTDMLTKTVTADNLRL 778
R + K +R+ +S L + I +N D++TK + +L
Sbjct: 1275 RNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMTKPLPIKTFKL 1321
Score = 72.0 bits (175), Expect = 6e-12
Identities = 64/277 (23%), Positives = 122/277 (43%), Gaps = 24/277 (8%)
Query: 6 VENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRTLNER 65
++NQ + ++ D G EY ++ KF +NGI T +GVAER+NRTL +
Sbjct: 295 IKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDD 354
Query: 66 ARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVFGCVS 125
R SGLP W AI + ++ + P + + + +S L FG
Sbjct: 355 CRTQLQCSGLPNHLWFSAIEFST-IVRNSLASPKSKKSARQHAGLAGLDISTLLPFG-QP 412
Query: 126 YVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRN-------------- 171
++ D + K+ P+ I + + + YGY + KK + + N
Sbjct: 413 VIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRLDQF 472
Query: 172 ----VTFNESVLYKDRSSAESMSSSKQLKLSERVALEEISESDVVKRNQINPENEDDEVE 227
+TF+E L + +S +S +S +++ S+ + +E S+ D +++PE + +
Sbjct: 473 NYDALTFDED-LNRLTASYQSFIASNEIQQSDDLNIE--SDHDFQSDIELHPEQPRNVLS 529
Query: 228 VELEQEPTTVIETPITQVRRSTRTP-KAPQRYSPSLN 263
+ +T T +R ++T +AP+ P+++
Sbjct: 530 KAVSPTDSTPPSTHTEDSKRVSKTNIRAPREVDPNIS 566
>M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820
(ORF170)
Length = 170
Score = 80.1 bits (196), Expect = 2e-14
Identities = 43/95 (45%), Positives = 61/95 (63%), Gaps = 3/95 (3%)
Query: 288 WELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGSR-RYKARLVVKGFQQ 346
W AM++E+ +L +N TW L P + L +WV++ K SDG+ R KARLV KGF Q
Sbjct: 40 WCQAMQEELDALSRNKTWILVPPPVNQNILGCKWVFKTKLHSDGTLDRLKARLVAKGFHQ 99
Query: 347 KQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQ 381
++GI F E +SPVV+ TIR IL++ A+ L + Q
Sbjct: 100 EEGIYFVETYSPVVRTATIRTILNV--AQQLEVGQ 132
>M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710
(ORF120)
Length = 120
Score = 76.6 bits (187), Expect = 2e-13
Identities = 38/86 (44%), Positives = 55/86 (63%), Gaps = 3/86 (3%)
Query: 58 MNRTLNERARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSH 117
MNRT+ E+ R M + GLPK F DA NTA ++IN+ PS +++ +P+EVW + S+
Sbjct: 1 MNRTIIEKVRSMLCECGLPKTFRADAANTAVHIINKYPSTAINFHVPDEVWFQSVPTYSY 60
Query: 118 LKVFGCVSYVLIDSDRRDKLDPKAIK 143
L+ FGCV+Y+ D KL P+A K
Sbjct: 61 LRRFGCVAYIHCD---EGKLKPRAKK 83
>POL_IPMAI (P12894) Probable Pol polyprotein [Contains: Integrase
(IN); Reverse transcriptase/ribonuclease H (EC 2.7.7.49)
(EC 3.1.26.4) (RT)]
Length = 814
Score = 42.7 bits (99), Expect = 0.004
Identities = 19/50 (38%), Positives = 31/50 (62%)
Query: 13 KIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRTL 62
K + LK+DNG Y SQ+F++FC + + + + P+ G+ ER +RTL
Sbjct: 622 KPRLLKTDNGPAYTSQKFQQFCRQMDVTHLTGLPYNPQGQGIVERAHRTL 671
>POL_IPMA (P11368) Putative Pol polyprotein [Contains: Integrase
(IN); Reverse transcriptase/ribonuclease H (EC 2.7.7.49)
(EC 3.1.26.4) (RT)]
Length = 867
Score = 42.7 bits (99), Expect = 0.004
Identities = 19/50 (38%), Positives = 31/50 (62%)
Query: 13 KIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRTL 62
K + LK+DNG Y SQ+F++FC + + + + P+ G+ ER +RTL
Sbjct: 704 KPRLLKTDNGPAYTSQKFQQFCRQMDVTHLTGLPYNPQGQGIVERAHRTL 753
>IGEB_MOUSE (P03975) IgE-binding protein
Length = 557
Score = 42.7 bits (99), Expect = 0.004
Identities = 19/50 (38%), Positives = 31/50 (62%)
Query: 13 KIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRTL 62
K + LK+DNG Y SQ+F++FC + + + + P+ G+ ER +RTL
Sbjct: 434 KPRLLKTDNGPAYTSQKFQQFCRQMDVTHLTGLPYNPQGQGIVERAHRTL 483
>POL_IPHA (P04026) Putative Pol polyprotein [Contains: Integrase
(IN); Reverse transcriptase/ribonuclease H (EC 2.7.7.49)
(EC 3.1.26.4) (RT)]
Length = 863
Score = 42.4 bits (98), Expect = 0.005
Identities = 23/68 (33%), Positives = 36/68 (52%), Gaps = 6/68 (8%)
Query: 1 GWKTEVENQTCLKIKS------LKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGV 54
G K Q CL+ S +K+DNG Y SQ+F++FC + + + + P+ G+
Sbjct: 638 GEKASYVIQHCLEAWSAWGKPRIKTDNGPAYTSQKFRQFCRQMDVTHLTGLPYNPQGQGI 697
Query: 55 AERMNRTL 62
ER +RTL
Sbjct: 698 VERAHRTL 705
>M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240
(ORF111a)
Length = 111
Score = 40.8 bits (94), Expect = 0.014
Identities = 24/76 (31%), Positives = 40/76 (52%), Gaps = 1/76 (1%)
Query: 582 MVCTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCF-RRNNLTLQEFSD 640
+ TRPD+ AV +S+F S + V +L Y+KG+ L + ++L L+ F+D
Sbjct: 3 LTITRPDLTFAVNRLSQFSSASRTAQMQAVYKVLHYVKGTVGQGLFYSATSDLQLKAFAD 62
Query: 641 ADLGGDSDGGKSTTGY 656
+D D +S TG+
Sbjct: 63 SDWASCPDTRRSVTGF 78
>POL_MPMV (P07572) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)]
Length = 867
Score = 39.7 bits (91), Expect = 0.032
Identities = 19/48 (39%), Positives = 29/48 (59%)
Query: 15 KSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRTL 62
K +K+DNG Y S+ F++FCS I+ I I P+ G+ ER + +L
Sbjct: 709 KQIKTDNGPGYTSKNFQEFCSTLQIKHITGIPYNPQGQGIVERAHLSL 756
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 92,681,909
Number of Sequences: 164201
Number of extensions: 3990853
Number of successful extensions: 11303
Number of sequences better than 10.0: 110
Number of HSP's better than 10.0 without gapping: 82
Number of HSP's successfully gapped in prelim test: 28
Number of HSP's that attempted gapping in prelim test: 11109
Number of HSP's gapped (non-prelim): 171
length of query: 799
length of database: 59,974,054
effective HSP length: 118
effective length of query: 681
effective length of database: 40,598,336
effective search space: 27647466816
effective search space used: 27647466816
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0065.18


