

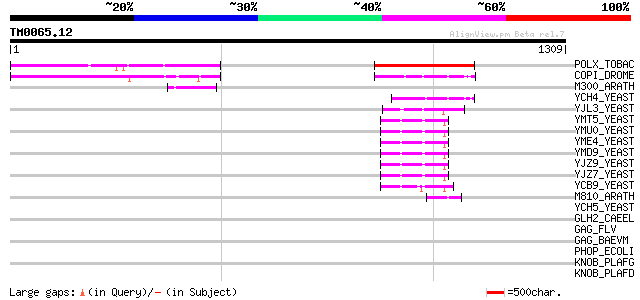
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0065.12
(1309 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 346 2e-94
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 142 6e-33
M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300... 101 1e-20
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 98 2e-19
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 84 2e-15
YMT5_YEAST (Q04214) Transposon Ty1 protein B 74 2e-12
YMU0_YEAST (Q04670) Transposon Ty1 protein B 74 3e-12
YME4_YEAST (Q04711) Transposon Ty1 protein B 74 3e-12
YMD9_YEAST (Q03434) Transposon Ty1 protein B 74 3e-12
YJZ9_YEAST (P47100) Transposon Ty1 protein B 73 6e-12
YJZ7_YEAST (P47098) Transposon Ty1 protein B 71 2e-11
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 71 2e-11
M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810... 55 2e-06
YCH5_YEAST (P25601) Transposon Ty5-1 16.0 kDa hypothetical protein 41 0.019
GLH2_CAEEL (Q966L9) ATP-dependent RNA helicase glh-2 (EC 3.6.1.-... 41 0.024
GAG_FLV (P10262) Gag polyprotein [Contains: Core protein p15; Co... 39 0.093
GAG_BAEVM (P03341) Gag polyprotein [Contains: Inner coat protein... 38 0.16
PHOP_ECOLI (P23836) Transcriptional regulatory protein phoP 37 0.27
KNOB_PLAFG (P09346) Knob-associated histidine-rich protein precu... 37 0.27
KNOB_PLAFD (P05229) Knob-associated histidine-rich protein (KAHR... 37 0.27
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 346 bits (888), Expect = 2e-94
Identities = 194/512 (37%), Positives = 305/512 (58%), Gaps = 25/512 (4%)
Query: 1 GAKFEVTRFYGTGNFGLWHRRVKDLLAQQSLQKALR--DEKPTDIATVDWNEMKEKAASL 58
G K+EV +F G F W RR++DLL QQ L K L +KP + DW ++ E+AAS
Sbjct: 3 GVKYEVAKFNGDNGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMKAEDWADLDERAASA 62
Query: 59 IRLCVSDDLMNHILDLTTPKDVWDKLESLYMSKTLMNKLFAKQRLYSLKMQEGGDLQAHV 118
IRL +SDD++N+I+D T + +W +LESLYMSKTL NKL+ K++LY+L M EG + +H+
Sbjct: 63 IRLHLSDDVVNNIIDEDTARGIWTRLESLYMSKTLTNKLYLKKQLYALHMSEGTNFLSHL 122
Query: 119 YAFNNILADLTRLRVKVDDEDKAIILLRSLPGSYDHLVTTLTYGKDSIKLDSISSTLLQH 178
FN ++ L L VK+++EDKAI+LL SLP SYD+L TT+ +GK +I+L ++S LL +
Sbjct: 123 NVFNGLITQLANLGVKIEEEDKAILLLNSLPSSYDNLATTILHGKTTIELKDVTSALLLN 182
Query: 179 AQRRQSVEEGGGSSGKCLFVKG-GQGRARGKGKAVDSGKKKRSKSKDR-KTAECYSCKQI 236
+ R+ E + G+ L +G G+ R SG + +SK++ + + CY+C Q
Sbjct: 183 EKMRKKPE----NQGQALITEGRGRSYQRSSNNYGRSGARGKSKNRSKSRVRNCYNCNQP 238
Query: 237 GHWKRDCPN-RPGK-----SSNSNSSANVVQSDGSC-----SEEDLLCVSSVKCTDAWVL 285
GH+KRDCPN R GK N +++A +VQ++ + EE+ + +S + WV+
Sbjct: 239 GHFKRDCPNPRKGKGETSGQKNDDNTAAMVQNNDNVVLFINEEEECMHLSGPE--SEWVV 296
Query: 286 DSGCSYHMTPHREWFNSFKLGDFGYVYLGDDKPCIIKGMRQVKIALDDGRVRTLSQVRYV 345
D+ S+H TP R+ F + GDFG V +G+ I G+ + I + G L VR+V
Sbjct: 297 DTAASHHATPVRDLFCRYVAGDFGTVKMGNTSYSKIAGIGDICIKTNVGCTLVLKDVRHV 356
Query: 346 PEVTKNMISLGTLHENGYSFKSEENRDILRVSKGAMTVMRAKRTAGNIYKLLGGTVMGDV 405
P++ N+IS L +GY +S R++KG++ + + G +Y+ G++
Sbjct: 357 PDLRMNLISGIALDRDGY--ESYFANQKWRLTKGSLVIAKGV-ARGTLYRTNAEICQGEL 413
Query: 406 ASVETDDDATKMWHMRLGHLSERGMMQLHKRNLLKGVRSCTIGLCKYCVLGKQCRVRFKT 465
+ + D+ + +WH R+GH+SE+G+ L K++L+ + T+ C YC+ GKQ RV F+T
Sbjct: 414 NAAQ-DEISVDLWHKRMGHMSEKGLQILAKKSLISYAKGTTVKPCDYCLFGKQHRVSFQT 472
Query: 466 GQHKTKGILDYVHSDVWGPTKEPSVGGFRYFL 497
+ ILD V+SDV GP + S+GG +YF+
Sbjct: 473 SSERKLNILDLVYSDVCGPMEIESMGGNKYFV 504
Score = 259 bits (663), Expect = 3e-68
Identities = 131/236 (55%), Positives = 164/236 (68%)
Query: 861 KGIHSKRGLIMMRFFSPVVRHTSIRAVLALVASRDMHLEQMDVKTTFLHGNLEEQIYMEQ 920
KG K+G+ FSPVV+ TSIR +L+L AS D+ +EQ+DVKT FLHG+LEE+IYMEQ
Sbjct: 881 KGFEQKKGIDFDEIFSPVVKMTSIRTILSLAASLDLEVEQLDVKTAFLHGDLEEEIYMEQ 940
Query: 921 PEGFSETGDGRLVCKLKRSLYGLKQSPRQ*YKRFDSYMLRIGYRRCDYDCCVYVRSLDDG 980
PEGF G +VCKL +SLYGLKQ+PRQ Y +FDS+M Y + D CVY + +
Sbjct: 941 PEGFEVAGKKHMVCKLNKSLYGLKQAPRQWYMKFDSFMKSQTYLKTYSDPCVYFKRFSEN 1000
Query: 981 SFIFLLLYVDDMLIAANHLHNVNELKTKLGKEFDMKDLGAAKKILGMEIHRDRGTKKLWL 1040
+FI LLLYVDDMLI + +LK L K FDMKDLG A++ILGM+I R+R ++KLWL
Sbjct: 1001 NFIILLLYVDDMLIVGKDKGLIAKLKGDLSKSFDMKDLGPAQQILGMKIVRERTSRKLWL 1060
Query: 1041 SQKSYVEGVLSRFDMSKENPVSTPLANHFKLSLGQCSKTDSEIEGMSKIPYASAVG 1096
SQ+ Y+E VL RF+M PVSTPLA H KLS C T E M+K+PY+SAVG
Sbjct: 1061 SQEKYIERVLERFNMKNAKPVSTPLAGHLKLSKKMCPTTVEEKGNMAKVPYSSAVG 1116
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 142 bits (358), Expect = 6e-33
Identities = 87/240 (36%), Positives = 133/240 (55%), Gaps = 16/240 (6%)
Query: 861 KGIHSKRGLIMMRFFSPVVRHTSIRAVLALVASRDMHLEQMDVKTTFLHGNLEEQIYMEQ 920
+G K + F+PV R +S R +L+LV ++ + QMDVKT FL+G L+E+IYM
Sbjct: 961 RGFTQKYQIDYEETFAPVARISSFRFILSLVIQYNLKVHQMDVKTAFLNGTLKEEIYMRL 1020
Query: 921 PEGFSETGDGRLVCKLKRSLYGLKQSPRQ*YKRFDSYMLRIGYRRCDYDCCVYVRSLDDG 980
P+G S D VCKL +++YGLKQ+ R ++ F+ + + D C+Y+ LD G
Sbjct: 1021 PQGISCNSDN--VCKLNKAIYGLKQAARCWFEVFEQALKECEFVNSSVDRCIYI--LDKG 1076
Query: 981 SF---IFLLLYVDDMLIAANHLHNVNELKTKLGKEFDMKDLGAAKKILGMEIHRDRGTKK 1037
+ I++LLYVDD++IA + +N K L ++F M DL K +G+ I K
Sbjct: 1077 NINENIYVLLYVDDVVIATGDMTRMNNFKRYLMEKFRMTDLNEIKHFIGIRIEMQE--DK 1134
Query: 1038 LWLSQKSYVEGVLSRFDMSKENPVSTPLANHFKLSLGQCSKTDSEIEGMSKIPYASAVGC 1097
++LSQ +YV+ +LS+F+M N VSTPL + L +D + P S +GC
Sbjct: 1135 IYLSQSAYVKKILSKFNMENCNAVSTPLPSKINYEL---LNSDED----CNTPCRSLIGC 1187
Score = 139 bits (350), Expect = 5e-32
Identities = 126/514 (24%), Positives = 229/514 (44%), Gaps = 31/514 (6%)
Query: 2 AKFEVTRFYGTGNFGLWHRRVKDLLAQQSLQKALRDEKPTDIATVDWNEMKEKAASLIRL 61
AK + F G + +W R++ LLA+Q + K + P ++ W + + A S I
Sbjct: 4 AKRNIKPFDGE-KYAIWKFRIRALLAEQDVLKVVDGLMPNEVDD-SWKKAERCAKSTIIE 61
Query: 62 CVSDDLMNHILDLTTPKDVWDKLESLYMSKTLMNKLFAKQRLYSLKMQEGGDLQAHVYAF 121
+SD +N T + + + L+++Y K+L ++L ++RL SLK+ L +H + F
Sbjct: 62 YLSDSFLNFATSDITARQILENLDAVYERKSLASQLALRKRLLSLKLSSEMSLLSHFHIF 121
Query: 122 NNILADLTRLRVKVDDEDKAIILLRSLPGSYDHLVTTL-TYGKDSIKLDSISSTLL-QHA 179
+ ++++L K+++ DK LL +LP YD ++T + T ++++ L + + LL Q
Sbjct: 122 DELISELLAAGAKIEEMDKISHLLITLPSCYDGIITAIETLSEENLTLAFVKNRLLDQEI 181
Query: 180 QRRQSVEEGGGSSGKCLFVKGGQGRARGKGKAVDSGKKKRSKSKDRKTAECYSCKQIGHW 239
+ + + + K + KK K + +C+ C + GH
Sbjct: 182 KIKNDHNDTSKKVMNAIVHNNNNTYKNNLFKNRVTKPKKIFKGNSKYKVKCHHCGREGHI 241
Query: 240 KRDCPNRPGKSSNSNSSANVVQSDGSCSEEDLLCVSSVKCTD-----AWVLDSGCSYHMT 294
K+DC + +N N N Q + S V V T +VLDSG S H+
Sbjct: 242 KKDCFHYKRILNNKNKE-NEKQVQTATSHGIAFMVKEVNNTSVMDNCGFVLDSGASDHLI 300
Query: 295 PHREWF-NSFKLGDFGYVYLGDDKPCIIKGMRQVKIALDDGRVRTLSQVRYVPEVTKNMI 353
+ +S ++ + + I R + +D + TL V + E N++
Sbjct: 301 NDESLYTDSVEVVPPLKIAVAKQGEFIYATKRGIVRLRNDHEI-TLEDVLFCKEAAGNLM 359
Query: 354 SLGTLHENGYSFKSEENRDILRVSKGAMTVMRAKRTAGNIYKLLGGTVMGDVASVET-DD 412
S+ L E G S E ++ + +SK + V++ N+ + S+
Sbjct: 360 SVKRLQEAGMSI--EFDKSGVTISKNGLMVVKNSGMLNNV-----PVINFQAYSINAKHK 412
Query: 413 DATKMWHMRLGHLSERGMMQLHKRNLLKGVR-------SCTIGLCKYCVLGKQCRVRFKT 465
+ ++WH R GH+S+ ++++ ++N+ SC I C+ C+ GKQ R+ FK
Sbjct: 413 NNFRLWHERFGHISDGKLLEIKRKNMFSDQSLLNNLELSCEI--CEPCLNGKQARLPFKQ 470
Query: 466 GQHKT--KGILDYVHSDVWGPTKEPSVGGFRYFL 497
+ KT K L VHSDV GP ++ YF+
Sbjct: 471 LKDKTHIKRPLFVVHSDVCGPITPVTLDDKNYFV 504
>M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300
(ORF145a) (ORF1451)
Length = 145
Score = 101 bits (252), Expect = 1e-20
Identities = 49/116 (42%), Positives = 68/116 (58%), Gaps = 1/116 (0%)
Query: 373 ILRVSKGAMTVMRAKRTAGNIYKLLGGTVMGDVASVETDDDATKMWHMRLGHLSERGMMQ 432
+L+V KG T+++ R ++Y L G G+ ET D T++WH RL H+S+RGM
Sbjct: 28 VLKVLKGCRTILKGNRH-DSLYILQGSVETGESNLAETAKDETRLWHSRLAHMSQRGMEL 86
Query: 433 LHKRNLLKGVRSCTIGLCKYCVLGKQCRVRFKTGQHKTKGILDYVHSDVWGPTKEP 488
L K+ L + ++ C+ C+ GK RV F TGQH TK LDYVHSD+WG P
Sbjct: 87 LVKKGFLDSSKVSSLKFCEDCIYGKTHRVNFSTGQHTTKNPLDYVHSDLWGAPSVP 142
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 97.8 bits (242), Expect = 2e-19
Identities = 65/196 (33%), Positives = 104/196 (52%), Gaps = 8/196 (4%)
Query: 901 MDVKTTFLHGNLEEQIYMEQPEGFSETGDGRLVCKLKRSLYGLKQSPRQ*YKRFDSYMLR 960
MDV T FL+ ++E IY++QP GF + V +L +YGLKQ+P + ++ + +
Sbjct: 1 MDVDTAFLNSTMDEPIYVKQPPGFVNERNPDYVWELYGGMYGLKQAPLLWNEHINNTLKK 60
Query: 961 IGYRRCDYDCCVYVRSLDDGSFIFLLLYVDDMLIAANHLHNVNELKTKLGKEFDMKDLGA 1020
IG+ R + + +Y RS DG I++ +YVDD+L+AA + +K +L K + MKDLG
Sbjct: 61 IGFCRHEGEHGLYFRSTSDGP-IYIGVYVDDLLVAAPSPKIYDRVKQELTKLYSMKDLGK 119
Query: 1021 AKKILGMEIHRDRGTKKLWLSQKSYVEGVLSRFDMSKENPVSTPLANHFKLSLGQCSKTD 1080
K LG+ IH+ + LS + Y+ S +++ TPL N L T
Sbjct: 120 VDKFLGLNIHQST-NGDITLSLQDYIAKAASESEINTFKLTQTPLCNSKPL----FETTS 174
Query: 1081 SEIEGMSKIPYASAVG 1096
++ ++ PY S VG
Sbjct: 175 PHLKDIT--PYQSIVG 188
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 84.3 bits (207), Expect = 2e-15
Identities = 56/201 (27%), Positives = 102/201 (49%), Gaps = 17/201 (8%)
Query: 879 VRHTSIRAVLALVASRDMHLEQMDVKTTFLHGNLEEQIYMEQPEGFSETGDGRLVCKLKR 938
+ H I+ L + +R+M ++ +D+ FL+ LEE+IY+ P D R V KL +
Sbjct: 1361 LNHNHIKIFLMIANNRNMFMKTLDINHAFLYAKLEEEIYIPHPH------DRRCVVKLNK 1414
Query: 939 SLYGLKQSPRQ*YKRFDSYMLRIGYRRCDYDCCVYVRSLDDGSFIFLLLYVDDMLIAANH 998
+LYGLKQSP++ Y+ IG + Y +Y + + + +YVDD +IAA++
Sbjct: 1415 ALYGLKQSPKEWNDHLRQYLNGIGLKDNSYTPGLY---QTEDKNLMIAVYVDDCVIAASN 1471
Query: 999 LHNVNELKTKLGKEFDMKDLGAA------KKILGMEIHRDRGTKKLWLSQKSYVEGVLSR 1052
++E KL F++K G ILGM++ ++ + L+ KS++ + +
Sbjct: 1472 EQRLDEFINKLKSNFELKITGTLIDDVLDTDILGMDLVYNKRLGTIDLTLKSFINRMDKK 1531
Query: 1053 F--DMSKENPVSTPLANHFKL 1071
+ ++ K S P + +K+
Sbjct: 1532 YNEELKKIRKSSIPHMSTYKI 1552
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 74.3 bits (181), Expect = 2e-12
Identities = 48/167 (28%), Positives = 90/167 (53%), Gaps = 16/167 (9%)
Query: 876 SPVVRHTSIRAVLALVASRDMHLEQMDVKTTFLHGNLEEQIYMEQPEGFSETGDGRLVCK 935
S V H ++ L+L + ++ Q+D+ + +L+ +++E++Y+ P G + +
Sbjct: 893 SNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPP---PHLGMNDKLIR 949
Query: 936 LKRSLYGLKQSPRQ*YKRFDSYMLR-IGYRRCDYDCCVYVRSLDDGSFIFLLLYVDDMLI 994
LK+SLYGLKQS Y+ SY+++ G CV+ + S + + L+VDDM++
Sbjct: 950 LKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVF-----ENSQVTICLFVDDMVL 1004
Query: 995 AANHLHNVNELKTKLGKEFDMK--DLGAAKK-----ILGMEIHRDRG 1034
+ +L++ + KL ++D K +LG + + ILG+EI RG
Sbjct: 1005 FSKNLNSNKRIIDKLKMQYDTKIINLGESDEEIQYDILGLEIKYQRG 1051
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 73.9 bits (180), Expect = 3e-12
Identities = 47/167 (28%), Positives = 86/167 (51%), Gaps = 16/167 (9%)
Query: 876 SPVVRHTSIRAVLALVASRDMHLEQMDVKTTFLHGNLEEQIYMEQPEGFSETGDGRLVCK 935
S V H ++ L+L + ++ Q+D+ + +L+ +++E++Y+ P G + +
Sbjct: 893 SNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPP---PHLGMNDKLIR 949
Query: 936 LKRSLYGLKQSPRQ*YKRFDSYMLR-IGYRRCDYDCCVYVRSLDDGSFIFLLLYVDDMLI 994
LK+SLYGLKQS Y+ SY+++ G CV+ S + + L+VDDM++
Sbjct: 950 LKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVF-----KNSQVTICLFVDDMIL 1004
Query: 995 AANHLHNVNELKTKLGKEFDMKDLGAAKK-------ILGMEIHRDRG 1034
+ L+ ++ T L K++D K + + ILG+EI RG
Sbjct: 1005 FSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRG 1051
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 73.9 bits (180), Expect = 3e-12
Identities = 47/167 (28%), Positives = 86/167 (51%), Gaps = 16/167 (9%)
Query: 876 SPVVRHTSIRAVLALVASRDMHLEQMDVKTTFLHGNLEEQIYMEQPEGFSETGDGRLVCK 935
S V H ++ L+L + ++ Q+D+ + +L+ +++E++Y+ P G + +
Sbjct: 893 SNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPP---PHLGMNDKLIR 949
Query: 936 LKRSLYGLKQSPRQ*YKRFDSYMLR-IGYRRCDYDCCVYVRSLDDGSFIFLLLYVDDMLI 994
LK+SLYGLKQS Y+ SY+++ G CV+ S + + L+VDDM++
Sbjct: 950 LKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVF-----KNSQVTICLFVDDMIL 1004
Query: 995 AANHLHNVNELKTKLGKEFDMKDLGAAKK-------ILGMEIHRDRG 1034
+ L+ ++ T L K++D K + + ILG+EI RG
Sbjct: 1005 FSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRG 1051
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 73.9 bits (180), Expect = 3e-12
Identities = 47/167 (28%), Positives = 86/167 (51%), Gaps = 16/167 (9%)
Query: 876 SPVVRHTSIRAVLALVASRDMHLEQMDVKTTFLHGNLEEQIYMEQPEGFSETGDGRLVCK 935
S V H ++ L+L + ++ Q+D+ + +L+ +++E++Y+ P G + +
Sbjct: 893 SNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPP---PHLGMNDKLIR 949
Query: 936 LKRSLYGLKQSPRQ*YKRFDSYMLR-IGYRRCDYDCCVYVRSLDDGSFIFLLLYVDDMLI 994
LK+SLYGLKQS Y+ SY+++ G CV+ S + + L+VDDM++
Sbjct: 950 LKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVF-----KNSQVTICLFVDDMIL 1004
Query: 995 AANHLHNVNELKTKLGKEFDMKDLGAAKK-------ILGMEIHRDRG 1034
+ L+ ++ T L K++D K + + ILG+EI RG
Sbjct: 1005 FSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRG 1051
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 72.8 bits (177), Expect = 6e-12
Identities = 48/167 (28%), Positives = 89/167 (52%), Gaps = 16/167 (9%)
Query: 876 SPVVRHTSIRAVLALVASRDMHLEQMDVKTTFLHGNLEEQIYMEQPEGFSETGDGRLVCK 935
S V H ++ L+L + ++ Q+D+ + +L+ +++E++Y+ P G + +
Sbjct: 1320 SNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPP---PHLGMNDKLIR 1376
Query: 936 LKRSLYGLKQSPRQ*YKRFDSYML-RIGYRRCDYDCCVYVRSLDDGSFIFLLLYVDDMLI 994
LK+SLYGLKQS Y+ SY++ + G CV+ S + + L+VDDM++
Sbjct: 1377 LKKSLYGLKQSGANWYETIKSYLIQQCGMEEVRGWSCVF-----KNSQVTICLFVDDMVL 1431
Query: 995 AANHLHNVNELKTKLGKEFDMK--DLGAAKK-----ILGMEIHRDRG 1034
+ +L++ + KL ++D K +LG + + ILG+EI RG
Sbjct: 1432 FSKNLNSNKRIIEKLKMQYDTKIINLGESDEEIQYDILGLEIKYQRG 1478
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 71.2 bits (173), Expect = 2e-11
Identities = 46/167 (27%), Positives = 85/167 (50%), Gaps = 16/167 (9%)
Query: 876 SPVVRHTSIRAVLALVASRDMHLEQMDVKTTFLHGNLEEQIYMEQPEGFSETGDGRLVCK 935
S V H ++ L+L + ++ Q+D+ + +L+ +++E++Y+ P G + +
Sbjct: 1320 SNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPP---PHLGMNDKLIR 1376
Query: 936 LKRSLYGLKQSPRQ*YKRFDSYMLR-IGYRRCDYDCCVYVRSLDDGSFIFLLLYVDDMLI 994
LK+S YGLKQS Y+ SY+++ G CV+ S + + L+VDDM++
Sbjct: 1377 LKKSHYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVF-----KNSQVTICLFVDDMIL 1431
Query: 995 AANHLHNVNELKTKLGKEFDMKDLGAAKK-------ILGMEIHRDRG 1034
+ L+ ++ T L K++D K + + ILG+EI RG
Sbjct: 1432 FSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRG 1478
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 71.2 bits (173), Expect = 2e-11
Identities = 49/184 (26%), Positives = 89/184 (47%), Gaps = 24/184 (13%)
Query: 876 SPVVRHTSIRAVLALVASRDMHLEQMDVKTTFLHGNLEEQIYMEQPEGFSETGDGRLVCK 935
S V H ++ L++ D ++ Q+D+ + +L+ +++E++Y+ P G + +
Sbjct: 1335 SNTVHHYALMTSLSIALDNDYYITQLDISSAYLYADIKEELYIRPP---PHLGLNDKLLR 1391
Query: 936 LKRSLYGLKQSPRQ*YKRFDSYMLRIGYRRCDYD-----CCVYVRSLDDGSFIFLLLYVD 990
L++SLYGLKQS Y+ SY++ CD CV+ S + + L+VD
Sbjct: 1392 LRKSLYGLKQSGANWYETIKSYLINC----CDMQEVRGWSCVF-----KNSQVTICLFVD 1442
Query: 991 DMLIAANHLHNVNELKTKLGKEFDMKDLGAAKK-------ILGMEIHRDRGTKKLWLSQK 1043
DM++ + L+ ++ T L K++D K + + ILG+EI R +K
Sbjct: 1443 DMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRSKYMKLGMEK 1502
Query: 1044 SYVE 1047
S E
Sbjct: 1503 SLTE 1506
>M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810
(ORF240b)
Length = 240
Score = 54.7 bits (130), Expect = 2e-06
Identities = 33/83 (39%), Positives = 46/83 (54%), Gaps = 2/83 (2%)
Query: 983 IFLLLYVDDMLIAANHLHNVNELKTKLGKEFDMKDLGAAKKILGMEIHRDRGTKKLWLSQ 1042
++LLLYVDD+L+ + +N L +L F MKDLG LG++I L+LSQ
Sbjct: 1 MYLLLYVDDILLTGSSNTLLNMLIFQLSSTFSMKDLGPVHYFLGIQIKTH--PSGLFLSQ 58
Query: 1043 KSYVEGVLSRFDMSKENPVSTPL 1065
Y E +L+ M P+STPL
Sbjct: 59 TKYAEQILNNAGMLDCKPMSTPL 81
>YCH5_YEAST (P25601) Transposon Ty5-1 16.0 kDa hypothetical protein
Length = 146
Score = 41.2 bits (95), Expect = 0.019
Identities = 37/121 (30%), Positives = 56/121 (45%), Gaps = 17/121 (14%)
Query: 247 PGKSSNSNSSANVV--------QSDGSCSEEDLLCV--SSVKCTDA--WVLDSGCSYHMT 294
P ++ +SSA+V Q+ G + + LC+ S+V T + W+ D+GC+ HM
Sbjct: 31 PNDKTSRSSSASVAIPDYETQGQTAGQITPKSWLCMLSSTVPATKSSEWIFDTGCTSHMC 90
Query: 295 PHREWFNSFKLGDFGYVYLGDDKPCIIKGMRQVKIALDDGRVRTLSQVRYVPEVTKNMIS 354
R F+SF G I G V I G V+ L V YVP++ N+IS
Sbjct: 91 HDRSIFSSFTRSSRKDFVRGVGGSIPIMGSGTVNI----GTVQ-LHDVSYVPDLPVNLIS 145
Query: 355 L 355
+
Sbjct: 146 V 146
>GLH2_CAEEL (Q966L9) ATP-dependent RNA helicase glh-2 (EC 3.6.1.-)
(Germline helicase-2)
Length = 974
Score = 40.8 bits (94), Expect = 0.024
Identities = 28/98 (28%), Positives = 41/98 (41%), Gaps = 21/98 (21%)
Query: 188 GGGSSGKCLFVKGGQGRARGKGKA-VDSGKKKRS----------------KSKDRKTAEC 230
GGG+SG F GG G G D G++ + K+R+ C
Sbjct: 225 GGGNSGGSGFGSGGNSNGFGSGGGGQDRGERNNNCFNCQQPGHRSNDCPEPKKEREPRVC 284
Query: 231 YSCKQIGHWKRDCPN----RPGKSSNSNSSANVVQSDG 264
Y+C+Q GH RDCP R G++ + S+ +G
Sbjct: 285 YNCQQPGHNSRDCPEERKPREGRNGFTGGSSGFGGGNG 322
Score = 38.9 bits (89), Expect = 0.093
Identities = 28/87 (32%), Positives = 40/87 (45%), Gaps = 13/87 (14%)
Query: 188 GGGSSGKCLFVKGGQGRARGKGK-----AVDSGKKKRS---KSKDRKTAECYSCKQIGHW 239
GG S+G F GG G+ RG+ G + K+R+ CY+C+Q GH
Sbjct: 351 GGNSNG---FGSGGGGQDRGERNNNCFNCQQPGHRSNDCPEPKKEREPRVCYNCQQPGHN 407
Query: 240 KRDCP--NRPGKSSNSNSSANVVQSDG 264
RDCP +P + N +S +DG
Sbjct: 408 SRDCPEERKPREGRNGFTSGFGGGNDG 434
Score = 35.8 bits (81), Expect = 0.79
Identities = 24/85 (28%), Positives = 35/85 (40%), Gaps = 9/85 (10%)
Query: 188 GGGSSGKCLFVKGGQGRARGKGKAVDSGKKKRSKSKDRKTAECYSCKQIGHWKRDCPNRP 247
G G++G+ F GG G G + G + + + C++C+Q GH DCP P
Sbjct: 336 GSGNNGESGFGSGGFG-----GNSNGFGSGGGGQDRGERNNNCFNCQQPGHRSNDCP-EP 389
Query: 248 GKSSNSNSSANVVQ---SDGSCSEE 269
K N Q + C EE
Sbjct: 390 KKEREPRVCYNCQQPGHNSRDCPEE 414
Score = 32.7 bits (73), Expect = 6.7
Identities = 21/67 (31%), Positives = 30/67 (44%), Gaps = 14/67 (20%)
Query: 188 GGGSSGKCLFVKGGQGRARGKGKAVDSGKKK--RSKSKDRKTAEC-------YSCKQIGH 238
GGG+ G G G A G G + G K K + ++AEC ++C + GH
Sbjct: 429 GGGNDGGF-----GGGNAEGFGNNEERGPMKCFNCKGEGHRSAECPEPPRGCFNCGEQGH 483
Query: 239 WKRDCPN 245
+CPN
Sbjct: 484 RSNECPN 490
>GAG_FLV (P10262) Gag polyprotein [Contains: Core protein p15; Core
protein p12; Core protein p30; Core protein p10]
Length = 580
Score = 38.9 bits (89), Expect = 0.093
Identities = 14/31 (45%), Positives = 20/31 (64%)
Query: 229 ECYSCKQIGHWKRDCPNRPGKSSNSNSSANV 259
+C CK+ GHW RDCP RP K +++ N+
Sbjct: 548 QCAYCKEKGHWVRDCPKRPRKKPANSTLLNL 578
>GAG_BAEVM (P03341) Gag polyprotein [Contains: Inner coat protein
p12; Core protein p15; Core shell protein p30;
Nucleoprotein p10]
Length = 537
Score = 38.1 bits (87), Expect = 0.16
Identities = 16/38 (42%), Positives = 23/38 (60%), Gaps = 2/38 (5%)
Query: 210 KAVDSGKKKRSKSKDRKTAECYSCKQIGHWKRDCPNRP 247
+A SG+ +R D+ +C CK+ GHW +DCP RP
Sbjct: 485 RAGKSGETRRRPKVDKD--QCAYCKERGHWIKDCPKRP 520
>PHOP_ECOLI (P23836) Transcriptional regulatory protein phoP
Length = 223
Score = 37.4 bits (85), Expect = 0.27
Identities = 40/155 (25%), Positives = 68/155 (43%), Gaps = 15/155 (9%)
Query: 37 DEKPTDIATVDWNEMKEKAASLIRLCVSDDLMNHILDLTTPKDVWDKLESL------YMS 90
+E DIA VD E SLIR S+D+ IL LT + DK+E L Y++
Sbjct: 41 NEHIPDIAIVDLGLPDEDGLSLIRRWRSNDVSLPILVLTARESWQDKVEVLSAGADDYVT 100
Query: 91 KTL-MNKLFAKQRLYSLKMQEGGDLQAHVYAFNNILADLTRLRVKVDDEDKAIILLRSLP 149
K + ++ A+ + M+ L + V + DL+R + ++DE +++
Sbjct: 101 KPFHIEEVMARMQAL---MRRNSGLASQVISLPPFQVDLSRRELSINDE-----VIKLTA 152
Query: 150 GSYDHLVTTLTYGKDSIKLDSISSTLLQHAQRRQS 184
Y + T + + DS+ L A+ R+S
Sbjct: 153 FEYTIMETLIRNNGKVVSKDSLMLQLYPDAELRES 187
>KNOB_PLAFG (P09346) Knob-associated histidine-rich protein
precursor (KAHRP) (KP)
Length = 634
Score = 37.4 bits (85), Expect = 0.27
Identities = 27/90 (30%), Positives = 39/90 (43%), Gaps = 7/90 (7%)
Query: 180 QRRQSVEEGGGSSGKCLFVKGGQGRARGKGKAVDSGKKKRSKSKDRKTAECYSCKQIGHW 239
++ S ++ G+ G+ G + + K K D KKK K KD + AE S K H
Sbjct: 358 EKHHSSKKHEGNDGE-----GEKKKKSKKHKDHDGEKKKSKKHKDNEDAE--SVKSKKHK 410
Query: 240 KRDCPNRPGKSSNSNSSANVVQSDGSCSEE 269
DC + K N A V+S S E+
Sbjct: 411 SHDCEKKKSKKHKDNEDAESVKSKKSVKEK 440
Score = 33.1 bits (74), Expect = 5.1
Identities = 22/69 (31%), Positives = 34/69 (48%), Gaps = 7/69 (10%)
Query: 208 KGKAVDSGKKKRSKSKDRKTAECY----SCKQIG---HWKRDCPNRPGKSSNSNSSANVV 260
K K+ D KKK K KD + AE S K+ G + K+ C + + + + N +
Sbjct: 408 KHKSHDCEKKKSKKHKDNEDAESVKSKKSVKEKGEKHNGKKPCSKKTNEENKNKEKTNNL 467
Query: 261 QSDGSCSEE 269
+SDGS + E
Sbjct: 468 KSDGSKAHE 476
>KNOB_PLAFD (P05229) Knob-associated histidine-rich protein (KAHRP)
(Fragment)
Length = 277
Score = 37.4 bits (85), Expect = 0.27
Identities = 27/90 (30%), Positives = 39/90 (43%), Gaps = 7/90 (7%)
Query: 180 QRRQSVEEGGGSSGKCLFVKGGQGRARGKGKAVDSGKKKRSKSKDRKTAECYSCKQIGHW 239
++ S ++ G+ G+ G + + K K D KKK K KD + AE S K H
Sbjct: 173 EKHHSSKKHEGNDGE-----GEKKKKSKKHKDHDGEKKKSKKHKDNEDAE--SVKSKKHK 225
Query: 240 KRDCPNRPGKSSNSNSSANVVQSDGSCSEE 269
DC + K N A V+S S E+
Sbjct: 226 SHDCEKKKSKKHKDNEDAESVKSKKSVKEK 255
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.337 0.146 0.474
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 139,440,617
Number of Sequences: 164201
Number of extensions: 5577193
Number of successful extensions: 19260
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 45
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 19105
Number of HSP's gapped (non-prelim): 129
length of query: 1309
length of database: 59,974,054
effective HSP length: 122
effective length of query: 1187
effective length of database: 39,941,532
effective search space: 47410598484
effective search space used: 47410598484
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 72 (32.3 bits)
Lotus: description of TM0065.12


