

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0065.1
(378 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
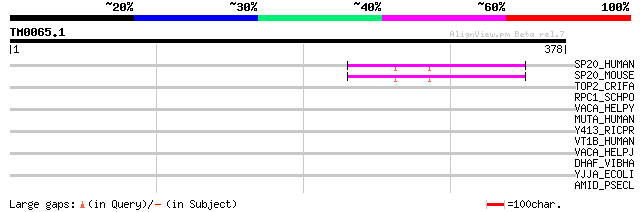
Score E
Sequences producing significant alignments: (bits) Value
SP20_HUMAN (Q8N0X7) Spartin (Trans-activated by hepatitis C viru... 47 1e-04
SP20_MOUSE (Q8R1X6) Spartin 44 9e-04
TOP2_CRIFA (P27570) DNA topoisomerase II (EC 5.99.1.3) 34 0.53
RPC1_SCHPO (O94666) DNA-directed RNA polymerase III largest subu... 34 0.70
VACA_HELPY (P55981) Vacuolating cytotoxin precursor 33 0.91
MUTA_HUMAN (P22033) Methylmalonyl-CoA mutase, mitochondrial prec... 33 1.6
Y413_RICPR (Q9ZDB9) Hypothetical protein RP413 32 2.0
VT1B_HUMAN (Q9UEU0) Vesicle transport through interaction with t... 31 4.5
VACA_HELPJ (Q9ZKW5) Vacuolating cytotoxin precursor 31 4.5
DHAF_VIBHA (Q56694) NADP-dependent fatty aldehyde dehydrogenase ... 31 5.9
YJJA_ECOLI (P18390) Hypothetical protein yjjA precursor (Protein... 30 7.7
AMID_PSECL (P27765) Amidase (EC 3.5.1.4) 30 7.7
>SP20_HUMAN (Q8N0X7) Spartin (Trans-activated by hepatitis C virus
core protein 1)
Length = 666
Score = 46.6 bits (109), Expect = 1e-04
Identities = 36/136 (26%), Positives = 63/136 (45%), Gaps = 15/136 (11%)
Query: 231 AMNENLKRVRQLTTMTEKLSKSLLDGVGIMS---GSVMAPVLKSQPGQAFLKMLPGE--- 284
A+ + L +Q T K+S+ L+DGV ++ G +AP +K + + L +
Sbjct: 472 AVTKGLYIAKQATGGAAKVSQFLVDGVCTVANCVGKELAPHVKKHGSKLVPESLKKDKDG 531
Query: 285 ---------VLLASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVF 335
V +S+ + V++ E A K ++ S + V +YG AGEAT H
Sbjct: 532 KSPLDGAMVVAASSVQGFSTVWQGLECAAKCIVNNVSAETVQTVRYKYGYNAGEATHHAV 591
Query: 336 ATAGHAANTAWNVSKI 351
+A + TA+N++ I
Sbjct: 592 DSAVNVGVTAYNINNI 607
>SP20_MOUSE (Q8R1X6) Spartin
Length = 671
Score = 43.5 bits (101), Expect = 9e-04
Identities = 35/136 (25%), Positives = 61/136 (44%), Gaps = 15/136 (11%)
Query: 231 AMNENLKRVRQLTTMTEKLSKSLLDGVGIMS---GSVMAPVLKSQPGQAFLKMLPGE--- 284
A+ L +Q T K+S+ L+DGV ++ G +AP +K + + L +
Sbjct: 476 AVTRGLYIAKQATGGAAKVSQLLVDGVCTVANCVGKELAPHVKKHGSKLVPESLKRDKDG 535
Query: 285 ---------VLLASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVF 335
V +S+ + V++ E A K ++ S + V +YG AGEAT +
Sbjct: 536 KSALDGAMVVAASSVQGFSTVWQGLECAAKCIVNNVSAETVQTVRYKYGHNAGEATHNAV 595
Query: 336 ATAGHAANTAWNVSKI 351
+A + TA+N+ I
Sbjct: 596 DSAINVGLTAYNIDNI 611
>TOP2_CRIFA (P27570) DNA topoisomerase II (EC 5.99.1.3)
Length = 1239
Score = 34.3 bits (77), Expect = 0.53
Identities = 37/122 (30%), Positives = 52/122 (42%), Gaps = 11/122 (9%)
Query: 264 VMAPVLKSQPGQAFLKML-PGEVLLASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNR 322
+ A + K+Q A LK P ++ L LD +K F+ E S+ Q R S
Sbjct: 1074 LQADIKKTQDSIAVLKQTTPVKMWLTDLDKFDKTFQEYERVLIHSI----QKEQRPASIT 1129
Query: 323 YGEEAGEATEHVF---ATAGHAANTAWNVSKIRKAINPVT---PASTAGGALRNSIKNRN 376
GEE + A A AA++++ V R P + P T GGAL + RN
Sbjct: 1130 GGEEVPALRQPPLMLEAPAKGAASSSYRVHICRYEEPPASKRKPEDTYGGALSSGGSTRN 1189
Query: 377 VG 378
VG
Sbjct: 1190 VG 1191
>RPC1_SCHPO (O94666) DNA-directed RNA polymerase III largest subunit
(EC 2.7.7.6)
Length = 1405
Score = 33.9 bits (76), Expect = 0.70
Identities = 33/128 (25%), Positives = 55/128 (42%), Gaps = 8/128 (6%)
Query: 236 LKRVRQLTTMTEKLSKSLLDGVGIMSGSVMAPVLKSQPGQAFLKMLPGEVLLASLDAVNK 295
L VR+ + L + +D + M G APV K + + +++ E L S V++
Sbjct: 976 LASVRERRDLAPMLEEPDMDDLDDMEGDEFAPVAKRKSVENIIRVT--EKQLRSF--VDR 1031
Query: 296 VFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVFATAGHAANTAWNVSKIRKAI 355
+E A+ + +A + + GE + T F AG AA T V +I++ I
Sbjct: 1032 CWEKYMRAKVEPGTAVGAIGAQSI----GEPGTQMTLKTFHFAGVAAQTTLGVPRIKEII 1087
Query: 356 NPVTPAST 363
N ST
Sbjct: 1088 NAAKTIST 1095
>VACA_HELPY (P55981) Vacuolating cytotoxin precursor
Length = 1290
Score = 33.5 bits (75), Expect = 0.91
Identities = 19/65 (29%), Positives = 30/65 (45%), Gaps = 1/65 (1%)
Query: 202 QTILTDNKNNGLVRQSVSNNKTADATKK-NAMNENLKRVRQLTTMTEKLSKSLLDGVGIM 260
QT+L D+ + G RQ + N T + TK+ NA L + L T L L I+
Sbjct: 913 QTLLIDSHDAGYARQMIDNTSTGEITKQLNAATTTLNNIASLEHKTSSLQTLSLSNAMIL 972
Query: 261 SGSVM 265
+ ++
Sbjct: 973 NSRLV 977
>MUTA_HUMAN (P22033) Methylmalonyl-CoA mutase, mitochondrial
precursor (EC 5.4.99.2) (MCM)
Length = 750
Score = 32.7 bits (73), Expect = 1.6
Identities = 38/164 (23%), Positives = 69/164 (41%), Gaps = 24/164 (14%)
Query: 171 LAKAIAGGTGQIVKGIFMCSNAYTNQVQSGGQTILTDNKNNGLVRQSVSNNKTADATKKN 230
+AKA+A G ++ I C+ ++ SG + I+ NK +V + + +N
Sbjct: 455 MAKAVAEGIPKL--RIEECAARRQARIDSGSEVIVGVNKYQLEKEDAVEVLAIDNTSVRN 512
Query: 231 AMNENLKRVRQLT--TMTEKLSKSLLDGVGIMSGSVMAPVLKSQPGQAFLKMLPGEVLLA 288
E LK+++ + E +L + G+++A + + + + GE+
Sbjct: 513 RQIEKLKKIKSSRDQALAEHCLAALTECAASGDGNILALAVDASRARCTV----GEIT-- 566
Query: 289 SLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATE 332
DA+ KVF +A RMVS Y +E GE+ E
Sbjct: 567 --DALKKVF------------GEHKANDRMVSGAYRQEFGESKE 596
>Y413_RICPR (Q9ZDB9) Hypothetical protein RP413
Length = 219
Score = 32.3 bits (72), Expect = 2.0
Identities = 20/56 (35%), Positives = 28/56 (49%), Gaps = 4/56 (7%)
Query: 185 GIFM---CSNAYTNQVQSGGQTILTDNKN-NGLVRQSVSNNKTADATKKNAMNENL 236
GIF+ C++ + N Q L D K G R+ V NNK D KKN + +N+
Sbjct: 11 GIFLLSSCTDNFRNYFQRSANNKLLDIKGAKGGKRKPVYNNKYIDLAKKNILEDNI 66
>VT1B_HUMAN (Q9UEU0) Vesicle transport through interaction with
t-SNAREs homolog 1B (Vesicle transport v-SNARE protein
Vti1-like 1) (Vti1-rp1)
Length = 232
Score = 31.2 bits (69), Expect = 4.5
Identities = 23/88 (26%), Positives = 42/88 (47%), Gaps = 6/88 (6%)
Query: 285 VLLASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVFATAGHAANT 344
+LL +++N+ AT++ ++ AT T + + EE GE + + T NT
Sbjct: 131 MLLQGTESLNR---ATQSIERSHRIATE---TDQIGSEIIEELGEQRDQLERTKSRLVNT 184
Query: 345 AWNVSKIRKAINPVTPASTAGGALRNSI 372
+ N+SK RK + ++ T L + I
Sbjct: 185 SENLSKSRKILRSMSRKVTTNKLLLSII 212
>VACA_HELPJ (Q9ZKW5) Vacuolating cytotoxin precursor
Length = 1288
Score = 31.2 bits (69), Expect = 4.5
Identities = 19/65 (29%), Positives = 30/65 (45%), Gaps = 1/65 (1%)
Query: 202 QTILTDNKNNGLVRQSVSNNKTADATKK-NAMNENLKRVRQLTTMTEKLSKSLLDGVGIM 260
QT+L D+ + G RQ + N T + TK+ NA + L V L L L I+
Sbjct: 910 QTLLIDSHDAGYARQMIDNTSTGEITKQLNAATDALNNVASLEHKQSGLQTLSLSNAMIL 969
Query: 261 SGSVM 265
+ ++
Sbjct: 970 NSRLV 974
>DHAF_VIBHA (Q56694) NADP-dependent fatty aldehyde dehydrogenase (EC
1.2.1.4)
Length = 510
Score = 30.8 bits (68), Expect = 5.9
Identities = 26/97 (26%), Positives = 41/97 (41%), Gaps = 4/97 (4%)
Query: 284 EVLLASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRY--GEEAGEATEH--VFATAG 339
E+L SL A E Q + + A R+V N + G E G A H + +
Sbjct: 399 EMLAGSLTATIHATEEDYPQVSQLIPRLEEIAGRLVFNGWPTGVEVGYAMVHGGPYPAST 458
Query: 340 HAANTAWNVSKIRKAINPVTPASTAGGALRNSIKNRN 376
H+A+T+ I + + PV + L +S+K N
Sbjct: 459 HSASTSVGAEAIHRWLRPVAYQALPESLLPDSLKAEN 495
>YJJA_ECOLI (P18390) Hypothetical protein yjjA precursor (Protein
P-18)
Length = 164
Score = 30.4 bits (67), Expect = 7.7
Identities = 21/66 (31%), Positives = 34/66 (50%), Gaps = 3/66 (4%)
Query: 194 TNQVQSGGQTILTDNKNNGL-VRQSVSNNKTADAT-KKNAMNENLKRVRQLTTMTEKLSK 251
TN + SG Q + DN NN + Q + K A T +N N+ L+++ L + +K
Sbjct: 57 TNLLSSGNQALSADNMNNAAGILQYCAKQKLASVTDAENIKNQVLEKL-GLNSEEQKEDT 115
Query: 252 SLLDGV 257
+ LDG+
Sbjct: 116 NYLDGI 121
>AMID_PSECL (P27765) Amidase (EC 3.5.1.4)
Length = 505
Score = 30.4 bits (67), Expect = 7.7
Identities = 18/58 (31%), Positives = 29/58 (49%), Gaps = 1/58 (1%)
Query: 120 GVTFPEQCYGSMGLLESFLKDHSCFSDLKTGKKGGLDWEQFAPRVEDYNHFLAKAIAG 177
GV E +G + ++D++ G G LD Q AP+V+DY +L K ++G
Sbjct: 220 GVMAIEATIDHVGPITGNVRDNALMLQAMAGADG-LDPRQAAPQVDDYCSYLEKGVSG 276
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.313 0.130 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,380,701
Number of Sequences: 164201
Number of extensions: 1730104
Number of successful extensions: 3491
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 3487
Number of HSP's gapped (non-prelim): 14
length of query: 378
length of database: 59,974,054
effective HSP length: 112
effective length of query: 266
effective length of database: 41,583,542
effective search space: 11061222172
effective search space used: 11061222172
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0065.1


