

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
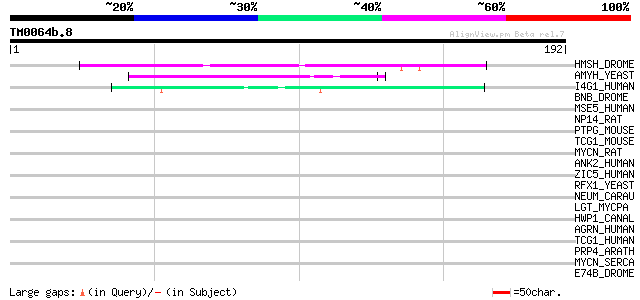
Query= TM0064b.8
(192 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
HMSH_DROME (Q03372) Muscle segmentation homeobox (Protein Drop) 45 1e-04
AMYH_YEAST (P08640) Glucoamylase S1/S2 precursor (EC 3.2.1.3) (G... 44 3e-04
I4G1_HUMAN (Q04637) Eukaryotic translation initiation factor 4 g... 42 9e-04
BNB_DROME (P29746) Bangles and beads protein 41 0.002
MSE5_HUMAN (Q00587) CDC42 effector protein 1 (Serum protein MSE55) 40 0.003
NP14_RAT (P41777) Nucleolar phosphoprotein p130 (Nucleolar 130 k... 39 0.010
PTPG_MOUSE (Q05909) Protein-tyrosine phosphatase gamma precursor... 38 0.013
TCG1_MOUSE (Q8CGF7) Transcription elongation regulator 1 (TATA b... 37 0.022
MYCN_RAT (Q63379) N-myc proto-oncogene protein 37 0.022
ANK2_HUMAN (Q01484) Ankyrin 2 (Brain ankyrin) (Ankyrin B) (Ankyr... 37 0.022
ZIC5_HUMAN (Q96T25) Zinc finger protein ZIC 5 (Zinc finger prote... 37 0.028
RFX1_YEAST (P48743) RFX-like DNA-binding protein RFX1 37 0.028
NEUM_CARAU (P17691) Neuromodulin (Axonal membrane protein GAP-43... 37 0.028
LGT_MYCPA (P60972) Prolipoprotein diacylglyceryl transferase (EC... 37 0.028
HWP1_CANAL (P46593) Hyphal wall protein 1 (Cell elongation prote... 37 0.028
AGRN_HUMAN (O00468) Agrin precursor 37 0.028
TCG1_HUMAN (O14776) Transcription elongation regulator 1 (TATA b... 37 0.037
PRP4_ARATH (O22212) Hypothetical Trp-Asp repeats containing prot... 37 0.037
MYCN_SERCA (P26014) N-myc proto-oncogene protein 37 0.037
E74B_DROME (P11536) Ecdysone-induced protein 74EF isoform B (ETS... 37 0.037
>HMSH_DROME (Q03372) Muscle segmentation homeobox (Protein Drop)
Length = 515
Score = 45.1 bits (105), Expect = 1e-04
Identities = 41/146 (28%), Positives = 61/146 (41%), Gaps = 9/146 (6%)
Query: 25 HASSRRGDHGAATQAVESSAPAATMSSPMVESSAPAASVPSTRGESSAMDATVLDPVVDL 84
HA + H AA + A AA + ++ A A S P++ SS +T L +
Sbjct: 199 HAHLLQAAHAAAAAHAQHQAMAAQLRQQQQQADARANSPPAST--SSTPSSTPLGSALGS 256
Query: 85 SDPVPTPPSPMVELSPYAIPQQPSSEETSGEESSSDHEREGSSEEEHILE-EELDAD--- 140
V + P+ SP + SE EE DHE + EE+ I++ E+++AD
Sbjct: 257 QGNVASTPAKNERHSP--LGSHTDSELEYDEEMLQDHEADHDEEEDSIVDIEDMNADDSP 314
Query: 141 -VVPEGAHGGDDDLIQRVAPFPGGLM 165
P+G G L P PG M
Sbjct: 315 RSTPDGLDGSGKSLESPHGPPPGSHM 340
>AMYH_YEAST (P08640) Glucoamylase S1/S2 precursor (EC 3.2.1.3)
(Glucan 1,4-alpha-glucosidase) (1,4-alpha-D-glucan
glucohydrolase)
Length = 1367
Score = 43.5 bits (101), Expect = 3e-04
Identities = 34/89 (38%), Positives = 39/89 (43%), Gaps = 3/89 (3%)
Query: 42 SSAPAATMSSPMVESSAPAASVPSTRGESSAMDATVLDPVVDLSDPVPTPPSPMVELSPY 101
SSAP T SS ESS+ A PS+ S+ S PVPTP S E S
Sbjct: 592 SSAPVPTPSSSTTESSSAPAPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSA 651
Query: 102 AIPQQPSSEETSGEESSSDHEREGSSEEE 130
+P PSS T E SS+ SS E
Sbjct: 652 PVP-TPSSSTT--ESSSAPVPTPSSSTTE 677
Score = 43.1 bits (100), Expect = 4e-04
Identities = 29/86 (33%), Positives = 36/86 (41%)
Query: 42 SSAPAATMSSPMVESSAPAASVPSTRGESSAMDATVLDPVVDLSDPVPTPPSPMVELSPY 101
SSAPA T SS ESS+ + +T S+ + S PVPTP S E S
Sbjct: 607 SSAPAPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSA 666
Query: 102 AIPQQPSSEETSGEESSSDHEREGSS 127
+P SS S + E SS
Sbjct: 667 PVPTPSSSTTESSSAPVTSSTTESSS 692
Score = 42.0 bits (97), Expect = 9e-04
Identities = 33/89 (37%), Positives = 38/89 (42%), Gaps = 3/89 (3%)
Query: 42 SSAPAATMSSPMVESSAPAASVPSTRGESSAMDATVLDPVVDLSDPVPTPPSPMVELSPY 101
SSAP T SS ESS+ PS+ S+ S PVPTP S E S
Sbjct: 718 SSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSA 777
Query: 102 AIPQQPSSEETSGEESSSDHEREGSSEEE 130
+P PSS T E SS+ SS E
Sbjct: 778 PVP-TPSSSTT--ESSSAPVPTPSSSTTE 803
Score = 40.8 bits (94), Expect = 0.002
Identities = 29/105 (27%), Positives = 44/105 (41%), Gaps = 4/105 (3%)
Query: 27 SSRRGDHGAATQAVESSAPAATMSSPMVESSAPAASVPSTRGESSAMDATVLDPVVDLSD 86
SS + +A SS+ + S+P+ S+ ++S P T + + A V P ++
Sbjct: 657 SSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTE 716
Query: 87 ----PVPTPPSPMVELSPYAIPQQPSSEETSGEESSSDHEREGSS 127
PVPTP S E S +P SS S + E SS
Sbjct: 717 SSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSS 761
Score = 40.4 bits (93), Expect = 0.003
Identities = 28/83 (33%), Positives = 37/83 (43%), Gaps = 2/83 (2%)
Query: 42 SSAPAATMSSPMVESSAPAASVPSTRGESSAMDATVLDPVVDLSDPVPTPPSPMVELS-- 99
SSAP T SS ESS+ + +T S+ + S PVPTP S E S
Sbjct: 550 SSAPVPTPSSSTTESSSTPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSA 609
Query: 100 PYAIPQQPSSEETSGEESSSDHE 122
P P ++E +S +SS E
Sbjct: 610 PAPTPSSSTTESSSAPVTSSTTE 632
Score = 40.4 bits (93), Expect = 0.003
Identities = 27/86 (31%), Positives = 35/86 (40%)
Query: 42 SSAPAATMSSPMVESSAPAASVPSTRGESSAMDATVLDPVVDLSDPVPTPPSPMVELSPY 101
SSAP T SS ESS+ A PS+ S+ S PVPTP S E S
Sbjct: 508 SSAPVPTPSSSTTESSSAPAPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSST 567
Query: 102 AIPQQPSSEETSGEESSSDHEREGSS 127
+ + ++ + S E SS
Sbjct: 568 PVTSSTTESSSAPVPTPSSSTTESSS 593
Score = 38.1 bits (87), Expect = 0.013
Identities = 30/111 (27%), Positives = 43/111 (38%), Gaps = 1/111 (0%)
Query: 17 TEDRHRRLHASSRRGDHGAATQAVESSAPAATMSSPMVESSAPAASVPSTRGESSAMDAT 76
TE + +S+ T + S+ A SS SSAP + S+ ESS+ T
Sbjct: 415 TESSSAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVT 474
Query: 77 VLDPVVDLSDPVPTPPSPMVELSPYAIPQQPSSEETSGEESSSDHEREGSS 127
S PVPTP S E S + + ++ + S E SS
Sbjct: 475 S-STTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSS 524
Score = 37.7 bits (86), Expect = 0.016
Identities = 38/102 (37%), Positives = 43/102 (41%), Gaps = 17/102 (16%)
Query: 42 SSAPAATMSSPMVESS-------------APAASVPSTRGESSAMDATVLDPVVDLSDPV 88
SSAPA T SS ESS AP + S+ ESS+ T S PV
Sbjct: 523 SSAPAPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSTPVTS-STTESSSAPV 581
Query: 89 PTPPSPMVELSPYAIPQQPSSEETSGEESSSDHEREGSSEEE 130
PTP S E S +P PSS T E SS+ SS E
Sbjct: 582 PTPSSSTTESSSAPVP-TPSSSTT--ESSSAPAPTPSSSTTE 620
Score = 37.0 bits (84), Expect = 0.028
Identities = 27/78 (34%), Positives = 32/78 (40%), Gaps = 12/78 (15%)
Query: 42 SSAPAATMSSPMVESSAPAASVPSTRGESSAMDATVLDPVVDLSDPVPTPPSPMVELSPY 101
SSAP T SS ESS+ PS+ S+ S PVPTP S E S
Sbjct: 760 SSAPVPTPSSSTTESSSAPVPTPSSSTTESS------------SAPVPTPSSSTTESSVA 807
Query: 102 AIPQQPSSEETSGEESSS 119
+P SS + SS
Sbjct: 808 PVPTPSSSSNITSSAPSS 825
Score = 36.6 bits (83), Expect = 0.037
Identities = 33/123 (26%), Positives = 52/123 (41%), Gaps = 8/123 (6%)
Query: 7 SRSTSEDVEGTEDRHRRLHASSRRGDHGAATQAVESSAPAATMSSPMVESSAPAASVP-- 64
+ S+S V + +S + +A SS+ + S+P+ S+ ++S P
Sbjct: 427 TESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVP 486
Query: 65 ---STRGESSAMDATVLDPVVDLSDPVPTPPSPMVELS--PYAIPQQPSSEETSGEESSS 119
S+ ESS+ T S PVPTP S E S P P ++E +S +SS
Sbjct: 487 TPSSSTTESSSAPVT-SSTTESSSAPVPTPSSSTTESSSAPAPTPSSSTTESSSAPVTSS 545
Query: 120 DHE 122
E
Sbjct: 546 TTE 548
Score = 36.2 bits (82), Expect = 0.048
Identities = 23/93 (24%), Positives = 41/93 (43%), Gaps = 1/93 (1%)
Query: 36 ATQAVESSAPAATMSSPMVESSAPAASVPSTRGESSAMDATVLDPVVDLSD-PVPTPPSP 94
++ A +S+ + S+P+ S+ ++S P T + + A V + S PVPTP S
Sbjct: 405 SSSAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSSS 464
Query: 95 MVELSPYAIPQQPSSEETSGEESSSDHEREGSS 127
E S + + ++ + S E SS
Sbjct: 465 TTESSSAPVTSSTTESSSAPVPTPSSSTTESSS 497
Score = 36.2 bits (82), Expect = 0.048
Identities = 24/86 (27%), Positives = 34/86 (38%)
Query: 42 SSAPAATMSSPMVESSAPAASVPSTRGESSAMDATVLDPVVDLSDPVPTPPSPMVELSPY 101
SSAP T SS ESS+ + +T S+ + S P PTP S E S
Sbjct: 481 SSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPAPTPSSSTTESSSA 540
Query: 102 AIPQQPSSEETSGEESSSDHEREGSS 127
+ + ++ + S E SS
Sbjct: 541 PVTSSTTESSSAPVPTPSSSTTESSS 566
Score = 36.2 bits (82), Expect = 0.048
Identities = 30/100 (30%), Positives = 43/100 (43%), Gaps = 5/100 (5%)
Query: 42 SSAPAATMSSPMVESSAPAASVPSTRGESSAMDATVLDPVVDLSDPVPTPPSPMVELSPY 101
SSAP T SS ESS+ + +T S+ + ++ + S PVPTP S E S
Sbjct: 352 SSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTTE---SSSAPVPTPSSSTTESS-- 406
Query: 102 AIPQQPSSEETSGEESSSDHEREGSSEEEHILEEELDADV 141
+ P S+ E+S +S S+ E A V
Sbjct: 407 SAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPV 446
Score = 35.8 bits (81), Expect = 0.063
Identities = 27/112 (24%), Positives = 49/112 (43%), Gaps = 3/112 (2%)
Query: 35 AATQAVESSAPAATMSSPMVESSAPAASVPSTRGESSAMDATVLDPVVDLSDPVPTPPSP 94
+++ + SSAP++T S ESS+ PS+ S+ PVPTP S
Sbjct: 813 SSSSNITSSAPSSTPFSSSTESSSVPVPTPSSSTTESSSAPVSSSTTESSVAPVPTPSSS 872
Query: 95 --MVELSPYAIPQQPSSEE-TSGEESSSDHEREGSSEEEHILEEELDADVVP 143
+ +P +IP ++E ++G + + S+ E + + +VP
Sbjct: 873 SNITSSAPSSIPFSSTTESFSTGTTVTPSSSKYPGSQTETSVSSTTETTIVP 924
Score = 34.7 bits (78), Expect = 0.14
Identities = 25/87 (28%), Positives = 36/87 (40%), Gaps = 2/87 (2%)
Query: 41 ESSAPAATMSSPMVESSAPAASVPSTRGESSAMDATVLDPVVDLSDPVPTPPSPMVELSP 100
+++ P T SS ESS+ PS+ S+ S PVPTP S E S
Sbjct: 309 KTTTPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESS- 367
Query: 101 YAIPQQPSSEETSGEESSSDHEREGSS 127
+ P S+ E+S +S S+
Sbjct: 368 -SAPVTSSTTESSSAPVTSSTTESSSA 393
Score = 34.3 bits (77), Expect = 0.18
Identities = 24/116 (20%), Positives = 46/116 (38%)
Query: 7 SRSTSEDVEGTEDRHRRLHASSRRGDHGAATQAVESSAPAATMSSPMVESSAPAASVPST 66
+ S+S V + +S + +A SS+ + S+P+ S+ ++S P T
Sbjct: 364 TESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVT 423
Query: 67 RGESSAMDATVLDPVVDLSDPVPTPPSPMVELSPYAIPQQPSSEETSGEESSSDHE 122
+ + A V + S T + +P P ++E +S +SS E
Sbjct: 424 SSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTE 479
Score = 34.3 bits (77), Expect = 0.18
Identities = 26/86 (30%), Positives = 34/86 (39%), Gaps = 12/86 (13%)
Query: 42 SSAPAATMSSPMVESSAPAASVPSTRGESSAMDATVLDPVVDLSDPVPTPPSPMVELSPY 101
SSAP T SS ESS+ PS+ S+ S PVPTP S E S
Sbjct: 703 SSAPVPTPSSSTTESSSAPVPTPSSSTTESS------------SAPVPTPSSSTTESSSA 750
Query: 102 AIPQQPSSEETSGEESSSDHEREGSS 127
+ + ++ + S E SS
Sbjct: 751 PVTSSTTESSSAPVPTPSSSTTESSS 776
Score = 33.5 bits (75), Expect = 0.31
Identities = 28/86 (32%), Positives = 36/86 (41%), Gaps = 14/86 (16%)
Query: 42 SSAPAATMSSPMVESSAPAASVPSTRGESSAMDATVLDPVVDLSDPVPTPPSPMVELSPY 101
SSAP T SS ESS+ PS+ S+ S PVPTP S E S
Sbjct: 634 SSAPVPTPSSSTTESSSAPVPTPSSSTTESS------------SAPVPTPSSSTTESS-- 679
Query: 102 AIPQQPSSEETSGEESSSDHEREGSS 127
+ P S+ E+S +S S+
Sbjct: 680 SAPVTSSTTESSSAPVTSSTTESSSA 705
Score = 33.1 bits (74), Expect = 0.41
Identities = 34/88 (38%), Positives = 40/88 (44%), Gaps = 9/88 (10%)
Query: 42 SSAPAATMSSPMVE-SSAPAASVPSTRGESSAMDATVLDPV----VDLSDPVPTPPSPMV 96
SSAP T SS E SSAP + S+ ESS A V P + S P TP S
Sbjct: 775 SSAPVPTPSSSTTESSSAPVPTPSSSTTESSV--APVPTPSSSSNITSSAPSSTPFSSST 832
Query: 97 ELS--PYAIPQQPSSEETSGEESSSDHE 122
E S P P ++E +S SSS E
Sbjct: 833 ESSSVPVPTPSSSTTESSSAPVSSSTTE 860
Score = 32.7 bits (73), Expect = 0.53
Identities = 25/86 (29%), Positives = 33/86 (38%), Gaps = 12/86 (13%)
Query: 42 SSAPAATMSSPMVESSAPAASVPSTRGESSAMDATVLDPVVDLSDPVPTPPSPMVELSPY 101
SSAP T SS ESS+ PS+ S+ S P PTP S E S
Sbjct: 577 SSAPVPTPSSSTTESSSAPVPTPSSSTTESS------------SAPAPTPSSSTTESSSA 624
Query: 102 AIPQQPSSEETSGEESSSDHEREGSS 127
+ + ++ + S E SS
Sbjct: 625 PVTSSTTESSSAPVPTPSSSTTESSS 650
>I4G1_HUMAN (Q04637) Eukaryotic translation initiation factor 4
gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G) (p220)
Length = 1600
Score = 42.0 bits (97), Expect = 9e-04
Identities = 41/147 (27%), Positives = 59/147 (39%), Gaps = 21/147 (14%)
Query: 36 ATQAVESSAPAATMSS----PMVESSAPAASVPSTRGESSAMDATVLDPVVDLSDPVPTP 91
A+ VE P + S P VESS A P+ ES A P +L + P+P
Sbjct: 352 ASHTVEIHEPNGMVPSEDLEPEVESSPELAPPPACPSESPVPIAPTAQPE-ELLNGAPSP 410
Query: 92 PSPMVELSPYAIPQQ--------------PSSEETSGEESSSDHEREGSSEEEHILEEEL 137
P+ V+LSP + P++ PS+ + ++S + E EEE E E
Sbjct: 411 PA--VDLSPVSEPEEQAKEVTASVAPPTIPSATPATAPSATSPAQEEEMEEEEEEEEGEA 468
Query: 138 DADVVPEGAHGGDDDLIQRVAPFPGGL 164
E GG++ L P P L
Sbjct: 469 GEAGEAESEKGGEELLPPESTPIPANL 495
>BNB_DROME (P29746) Bangles and beads protein
Length = 442
Score = 40.8 bits (94), Expect = 0.002
Identities = 40/166 (24%), Positives = 64/166 (38%), Gaps = 15/166 (9%)
Query: 8 RSTSEDVEGTE--DRHRRLHASSRRGDHGAATQAVESSAPAA-TMSSPMVESSAPAASVP 64
+S++ DVE + L ++ A +A + A T + P VE+ A
Sbjct: 101 KSSAPDVETAPAIPEKKTLPEEAKPAQENAPVEAEKKQEKTARTEAEPTVEAQPQATKAI 160
Query: 65 STRGESSAMDATVLDPVVDLSDPV-----------PTPPSPMVELSPYAIPQQPSSEETS 113
E+ A +A V VVD P P P+ + +P+QP+ +E
Sbjct: 161 EQAPEAPAANAEVQKQVVDEVKPQEPKIDAKSAEEPAIPAVVAAEKETPVPEQPARQERI 220
Query: 114 GEESSSDHEREGSSEEEHILEEELDADVVPEGAHGGDDDLIQRVAP 159
E D +++ + EE E PE A D + IQ +AP
Sbjct: 221 NEIEQKDAKKDAAVAEEPAKAAEATPTAAPEAATKSDSN-IQVIAP 265
>MSE5_HUMAN (Q00587) CDC42 effector protein 1 (Serum protein MSE55)
Length = 391
Score = 40.0 bits (92), Expect = 0.003
Identities = 20/63 (31%), Positives = 29/63 (45%)
Query: 39 AVESSAPAATMSSPMVESSAPAASVPSTRGESSAMDATVLDPVVDLSDPVPTPPSPMVEL 98
A E+ APAA +P + PAA+ P+T A A P + P PP+P
Sbjct: 221 AAETPAPAANPPAPTANPTGPAANPPATTANPPAPAANPSAPAATPTGPAANPPAPAASS 280
Query: 99 SPY 101
+P+
Sbjct: 281 TPH 283
>NP14_RAT (P41777) Nucleolar phosphoprotein p130 (Nucleolar 130 kDa
protein) (140 kDa nucleolar phosphoprotein) (Nopp140)
(Nucleolar and coiled-body phosphoprotein 1)
Length = 704
Score = 38.5 bits (88), Expect = 0.010
Identities = 31/109 (28%), Positives = 50/109 (45%), Gaps = 6/109 (5%)
Query: 28 SRRGDHGAATQAVESSAPAATMSS---PMVESSAPAA-SVPSTRGESSAMDATVLDPVVD 83
SR+ D ++ + SS AT S P +A AA S+P+ + + D++
Sbjct: 427 SRKADSSSSEEESSSSEEEATKKSVTTPKARVTAKAAPSLPAKQAPRAGGDSSSDSESSS 486
Query: 84 LSDPVPTPPSPMVE--LSPYAIPQQPSSEETSGEESSSDHEREGSSEEE 130
+ TPP P + + A+P+ ++ + E SSS E SSEEE
Sbjct: 487 SEEEKKTPPKPPAKKKAAGAAVPKPTPVKKAAAESSSSSSSSEDSSEEE 535
Score = 36.2 bits (82), Expect = 0.048
Identities = 31/130 (23%), Positives = 52/130 (39%), Gaps = 8/130 (6%)
Query: 5 KRSRSTSEDVEGTEDRHRRLHASSRRGDHGAATQAVESSAPAATMSSPMVESSAPAASVP 64
K++ + ++ + + D +SS A V + ESS+ ++
Sbjct: 320 KKAAAQTQPADSSADSSEESDSSSEEEKKTPAKTVVSKTPAKPAPVKKKAESSSDSSDSD 379
Query: 65 STRGESSAMDATVLDPVVDLSDPVPTPPSPMVELSPYAIPQQPSSE----ETSGEESSSD 120
S+ E+ A + LS P TP P + A P+QP+ ++ +SSS
Sbjct: 380 SSEDEAPAKPVSATKS--PLSKPAVTPKPPAAKA--VATPKQPAGSGQKPQSRKADSSSS 435
Query: 121 HEREGSSEEE 130
E SSEEE
Sbjct: 436 EEESSSSEEE 445
Score = 34.7 bits (78), Expect = 0.14
Identities = 34/133 (25%), Positives = 57/133 (42%), Gaps = 13/133 (9%)
Query: 3 GEKRSRSTSEDVEGTEDRHRRLHASSRRGDHGAATQAVESSAPAATMSSPMVESSAPAAS 62
G+ S S+S ++D A++ Q V + AP ++P +SS
Sbjct: 222 GKAGSSSSSSSSSSSDDSEEEKKAAAPLKKTAPKKQVV-AKAPVKVTAAPTQKSS----- 275
Query: 63 VPSTRGESSAMDATVLDPVVDLSDPVPTPPSPMVELSPYAIPQQ---PSSEETSGEESSS 119
S+ SS + P+ + P + P P V LS ++ Q ++ +T +SS+
Sbjct: 276 --SSEDSSSEEEEEQKKPMKKKAGPYSSVPPPSVSLSKKSVGAQSPKKAAAQTQPADSSA 333
Query: 120 D--HEREGSSEEE 130
D E + SSEEE
Sbjct: 334 DSSEESDSSSEEE 346
Score = 29.6 bits (65), Expect = 4.5
Identities = 29/118 (24%), Positives = 43/118 (35%), Gaps = 4/118 (3%)
Query: 7 SRSTSEDVEGTEDRHRRLHASSRRGDHGAATQAVESSAPAATMSSPMVESSAPAASVPST 66
S S+ E E E++ ++ ++ A +AV P SS S+ P T
Sbjct: 128 SSSSEESSEEEEEKDKKKKPVQQKAVKPQA-KAVRPP-PKKAESSESESDSSSEDEAPQT 185
Query: 67 RGESSAMDATVLDPVVDLSDPVPTPPSPMVELSPYAIPQQPSSEETSGEESSSDHERE 124
+ +A AT P P P + P A + S +S SSSD E
Sbjct: 186 QKPKAA--ATAAKAPTKAQTKAPAKPGPPAKAQPKAANGKAGSSSSSSSSSSSDDSEE 241
>PTPG_MOUSE (Q05909) Protein-tyrosine phosphatase gamma precursor
(EC 3.1.3.48) (R-PTP-gamma)
Length = 1442
Score = 38.1 bits (87), Expect = 0.013
Identities = 36/129 (27%), Positives = 53/129 (40%), Gaps = 10/129 (7%)
Query: 2 KGEKRSRSTSEDVEGTEDRHRRLHASSRRGDHGAATQAVESSAPAATMSSP-MVESSAPA 60
+GEK +S SED E + + + + + +APA T SSP +
Sbjct: 579 EGEKEEKSESEDGEREHEEEEKDSEKKEKSEATHTAAESDRTAPAPTPSSPHRTAAEGGH 638
Query: 61 ASVPSTRGESSAMD-------ATVLDPVVDLSDPV-PTPPSPMVELSPYAIPQQPSSEET 112
++P R + SA A LDP+VD + V PT S P+ PS +
Sbjct: 639 QTIPGRRQDHSAPATDQPGHVAPDLDPLVDTATQVPPTATEEHYSGSDPRRPEMPSKKPM 698
Query: 113 S-GEESSSD 120
S G+ S D
Sbjct: 699 SRGDRFSED 707
>TCG1_MOUSE (Q8CGF7) Transcription elongation regulator 1 (TATA
box-binding protein-associated factor 2S) (Transcription
factor CA150) (p144) (Formin-binding protein 28) (FBP
28)
Length = 1100
Score = 37.4 bits (85), Expect = 0.022
Identities = 26/89 (29%), Positives = 38/89 (42%), Gaps = 18/89 (20%)
Query: 34 GAATQAVESSAPAATMSSPMVESSAPAA------------SVPSTRGESSAMDATVLDPV 81
GA T S APA + S+P S+ A S P+T+ ++ + +V P
Sbjct: 259 GAPTPTTSSPAPAVSTSTPTSTPSSTTATTTTATSVAQTVSTPTTQDQTPSSAVSVATPT 318
Query: 82 VDLSDPVPTP------PSPMVELSPYAIP 104
V +S P PT P P + P A+P
Sbjct: 319 VSVSAPAPTATPVQTVPQPHPQTLPPAVP 347
>MYCN_RAT (Q63379) N-myc proto-oncogene protein
Length = 462
Score = 37.4 bits (85), Expect = 0.022
Identities = 40/126 (31%), Positives = 52/126 (40%), Gaps = 11/126 (8%)
Query: 22 RRLHASSRRGDHGAATQAVESSAPAATMSSPMVESSAP-----AASVPSTRGESSAMDAT 76
R L S+ G GA T + S PAA P V P +ASVP+ + A A
Sbjct: 161 RALGGSASAGRTGA-TLPTDLSHPAAECVDPAVVFPFPVNKRESASVPAAPTSAPATSAV 219
Query: 77 VLDPVVDLSDPVPTPPSPMVELSPYAIPQQPSSEETSGEESSSDHEREGSSEEEHILEEE 136
V V PV P A + + TSGE++ SD + E EE+ EEE
Sbjct: 220 VTSVSVPAVAPVAAPARGS---GRPANSGEHKALSTSGEDTLSDSDDEDDEEEDE--EEE 274
Query: 137 LDADVV 142
+D V
Sbjct: 275 IDVVTV 280
>ANK2_HUMAN (Q01484) Ankyrin 2 (Brain ankyrin) (Ankyrin B) (Ankyrin,
nonerythroid)
Length = 3924
Score = 37.4 bits (85), Expect = 0.022
Identities = 35/140 (25%), Positives = 53/140 (37%), Gaps = 10/140 (7%)
Query: 2 KGEKRSRSTSEDVEGTEDRHRRLHASSRRGDHGAATQAVESSAPAATMSSPMVESSAPAA 61
K E+ S S+ D + + D H S ES P S SS+ +
Sbjct: 2583 KQEESSSSSDPDADCSVDVDEPKHTGSGED---------ESGVPVLVTSESRKVSSSSES 2633
Query: 62 SVPSTRGESSAMDATVLDPVVDLSDPVPTPPSPMVELSPYAIPQQP-SSEETSGEESSSD 120
+ + A + +PV+ + P P P S SP + QP S++ + + +
Sbjct: 2634 EPELAQLKKGADSGLLPEPVIRVQPPSPLPSSMDSNSSPEEVQFQPVVSKQYTFKMNEDT 2693
Query: 121 HEREGSSEEEHILEEELDAD 140
E G SEEE E L D
Sbjct: 2694 QEEPGKSEEEKDSESHLAED 2713
>ZIC5_HUMAN (Q96T25) Zinc finger protein ZIC 5 (Zinc finger protein
of the cerebellum 5)
Length = 639
Score = 37.0 bits (84), Expect = 0.028
Identities = 28/105 (26%), Positives = 39/105 (36%), Gaps = 7/105 (6%)
Query: 44 APAATMSSPMVESSAPAASVPSTRGESSAMDATVLDPVVDLSDPVPTPPSPMVELSPYAI 103
APAA + + + A S P G S A + P L P P+PP P P A+
Sbjct: 93 APAAAARAAALVAHPGAGSYPCGGGSSGAQPSAPPPPAPPLP-PTPSPPPPPPPPPPPAL 151
Query: 104 PQQPSSEETSGEESSSDHEREGSSEEEHILEEELDADVVPEGAHG 148
++ G S H R + +L +L A HG
Sbjct: 152 SGYTTTNSGGGGSSGKGHSR------DFVLRRDLSATAPAAAMHG 190
>RFX1_YEAST (P48743) RFX-like DNA-binding protein RFX1
Length = 811
Score = 37.0 bits (84), Expect = 0.028
Identities = 25/73 (34%), Positives = 40/73 (54%), Gaps = 4/73 (5%)
Query: 79 DPVVDLSDPVPTPPSPMVE--LSPYAIPQQPSSEETSG-EESSSDHEREGSSEEEHILEE 135
DP+ LS P P+ PSP V SP+++ ++ S S ++SS+D+ E E +H E
Sbjct: 382 DPISPLSSPSPSSPSPQVPNVSSPFSLNRKSLSRTGSPVKQSSNDNPNEPELESQHPNET 441
Query: 136 ELD-ADVVPEGAH 147
E + D +P A+
Sbjct: 442 EANKLDSLPPAAN 454
>NEUM_CARAU (P17691) Neuromodulin (Axonal membrane protein GAP-43)
(PP46) (B-50) (Protein F1) (Calmodulin-binding protein
P-57)
Length = 213
Score = 37.0 bits (84), Expect = 0.028
Identities = 33/142 (23%), Positives = 52/142 (36%), Gaps = 15/142 (10%)
Query: 6 RSRSTSEDVEGTEDRHRRLHASSRRGDHGAAT---QAVESSAPAATMSSPMVESSAPAAS 62
R + ED +G D A + + + + VE S A S P + ++PAA
Sbjct: 49 RKKMKDEDKDGENDTAPDESAETEEKEERVSPSEEKPVEVSTETAEESKPAEQPNSPAAE 108
Query: 63 VPSTRGESSAMDATVLDPVVDLSDPVPTPPSPMVELSPYAIPQQPSSEETSGEESSSDHE 122
P T SA T PT +L P++ + + + ++ E
Sbjct: 109 APPTAATDSAPSDT------------PTKEEAQEQLQDAEEPKETENTAAADDITTQKEE 156
Query: 123 REGSSEEEHILEEELDADVVPE 144
+ EEE EEE VP+
Sbjct: 157 EKEEEEEEEEEEEEAKRADVPD 178
>LGT_MYCPA (P60972) Prolipoprotein diacylglyceryl transferase (EC
2.4.99.-)
Length = 427
Score = 37.0 bits (84), Expect = 0.028
Identities = 26/118 (22%), Positives = 42/118 (35%), Gaps = 14/118 (11%)
Query: 41 ESSAPAATMSSPMVESSAPAASVPSTRGESSAMDATVLDPVVDLSDPVPTPPSPMVELSP 100
E + P + E++ + P+ E++ DAT P V P S E +
Sbjct: 294 EPAEPEPATVAATTEAATEGVAAPADGAEAAGADATAQRPEESAEPDVEKPESEETEAAE 353
Query: 101 YAI--------------PQQPSSEETSGEESSSDHEREGSSEEEHILEEELDADVVPE 144
A P++P +EE + E G + E+ + EE A PE
Sbjct: 354 EASEPEAEEPEAPEAEEPEEPETEEPEADSGEEPEEESGEAPEQLVAEEPEPAPQQPE 411
>HWP1_CANAL (P46593) Hyphal wall protein 1 (Cell elongation protein
2)
Length = 634
Score = 37.0 bits (84), Expect = 0.028
Identities = 38/132 (28%), Positives = 56/132 (41%), Gaps = 26/132 (19%)
Query: 34 GAATQAVESSAPAATMSSPMVESSAPAASVPS-TRGESSAMDATVLDPVVDLSDPVPTPP 92
G + AV S AT S+P+ E + PA S PS GE+S P V SD T
Sbjct: 446 GETSPAVPKSDVPATESAPVPEMT-PAGSQPSIPAGETS--------PAVPKSDVPATES 496
Query: 93 SPMVELSPYAIPQQPSSEETS-----------GEESSSDHEREGSSEEEHILEEELD--- 138
+P E++P +P++ ++S G ES+ SS + +L E
Sbjct: 497 APAPEMTPAGTETKPAAPKSSAPATEPSPVAPGTESAPAGPGASSSPKSSVLASETSPIA 556
Query: 139 --ADVVPEGAHG 148
A+ P G+ G
Sbjct: 557 PGAETAPAGSSG 568
Score = 32.7 bits (73), Expect = 0.53
Identities = 27/90 (30%), Positives = 36/90 (40%), Gaps = 9/90 (10%)
Query: 40 VESSAPAATMSSPMVES-----SAPAASVPSTR----GESSAMDATVLDPVVDLSDPVPT 90
V S T P+ E+ S P AS+P G S+M A P V SD T
Sbjct: 401 VTSEETVYTTFCPLTENTPGTDSTPEASIPPMETIPAGSESSMPAGETSPAVPKSDVPAT 460
Query: 91 PPSPMVELSPYAIPQQPSSEETSGEESSSD 120
+P+ E++P + ETS SD
Sbjct: 461 ESAPVPEMTPAGSQPSIPAGETSPAVPKSD 490
>AGRN_HUMAN (O00468) Agrin precursor
Length = 2045
Score = 37.0 bits (84), Expect = 0.028
Identities = 27/90 (30%), Positives = 44/90 (48%), Gaps = 6/90 (6%)
Query: 36 ATQAVESSAPAATMSSPMVESSAPAASVPSTRGESSAMDATVLDPVVDLSDPVPTPPSPM 95
A A + P A S+ +++ P +S P T ++++ T + PV+ + PT PSP
Sbjct: 996 ALPAPPGALPLAPSSTAHSQTTPPPSSRPRT---TASVPRTTVWPVLTVP---PTAPSPA 1049
Query: 96 VELSPYAIPQQPSSEETSGEESSSDHEREG 125
L A + S++ +S EE S D E G
Sbjct: 1050 PSLVASAFGESGSTDGSSDEELSGDQEASG 1079
>TCG1_HUMAN (O14776) Transcription elongation regulator 1 (TATA
box-binding protein-associated factor 2S) (Transcription
factor CA150)
Length = 1098
Score = 36.6 bits (83), Expect = 0.037
Identities = 24/86 (27%), Positives = 40/86 (45%), Gaps = 7/86 (8%)
Query: 35 AATQAVESSAPAATMSSPMVESS-APAASVPSTRGESSAMDATVLDPVVDLS------DP 87
A + + SS P++T S+ +S A S P+T+ ++ + +V P V +S P
Sbjct: 269 AVSTSTSSSTPSSTTSTTTTATSVAQTVSTPTTQDQTPSSAVSVATPTVSVSTPARTATP 328
Query: 88 VPTPPSPMVELSPYAIPQQPSSEETS 113
V T P P + P A+P T+
Sbjct: 329 VQTVPQPHPQTLPPAVPHSVPQPTTA 354
>PRP4_ARATH (O22212) Hypothetical Trp-Asp repeats containing protein
At2g41500
Length = 554
Score = 36.6 bits (83), Expect = 0.037
Identities = 32/106 (30%), Positives = 47/106 (44%), Gaps = 7/106 (6%)
Query: 56 SSAPAASVPSTRGESSAMDATVLDPVVDLSDPVPTPPSPMVELSPYAIP---QQPSSEET 112
++A ++ P + SS + + PVV S P P P PM+ P A P + P S+
Sbjct: 13 ATAQISAPPVLQDASSLPGFSAIPPVVPPSFPPPMAPIPMMPHPPVARPPTFRPPVSQNG 72
Query: 113 SGEESSSDHEREGSSEEEHILEEELDADVVPEGAHGGDDDLIQRVA 158
+ S SD E S++EHI E V D L++R A
Sbjct: 73 GVKTSDSDSE----SDDEHIEISEESKQVRERQEKALQDLLVKRRA 114
>MYCN_SERCA (P26014) N-myc proto-oncogene protein
Length = 427
Score = 36.6 bits (83), Expect = 0.037
Identities = 35/121 (28%), Positives = 52/121 (42%), Gaps = 14/121 (11%)
Query: 27 SSRRGDHGAATQAVESSAPAATMSSPMVESSAPAASVPSTRGESSAMDATVLDPVVDLSD 86
S+R A T+ +++ PAA P S P VP+ G + +DP V
Sbjct: 133 SAREKLERAVTEKLQNKTPAAPPPPPGTAGSPP---VPARSGRAVP---ECVDPAVVFPF 186
Query: 87 PVPTPPSPMVE-----LSPYAIPQQPSSEETSGEESSSDHEREGSSEEEHILEEELDADV 141
PV +P E + SS +SG+++ SD E + EEE +EE + DV
Sbjct: 187 PVNKREAPAAEGLRRRRRARGDSRASSSSSSSGDDTLSDSEDDEDEEEE---DEEEEIDV 243
Query: 142 V 142
V
Sbjct: 244 V 244
>E74B_DROME (P11536) Ecdysone-induced protein 74EF isoform B
(ETS-related protein E74B)
Length = 883
Score = 36.6 bits (83), Expect = 0.037
Identities = 33/134 (24%), Positives = 56/134 (41%), Gaps = 10/134 (7%)
Query: 33 HGAATQAVESSAPAATMSSPMVESSAPAASVPSTRGESSAMDATVLDPVVDLSDPVPTPP 92
H A+QA S ++ SS SS+ + S+ S+ SS AT P S PT P
Sbjct: 13 HNFASQAAASLVNVSSSSSSSSSSSSSSLSLSSSSSSSSLSSAT---PTPVASPVTPTSP 69
Query: 93 SPMVELSPYAIPQQPSSEETSGEESSSDHEREGSSEEEHILEEELD-----ADVVPEGAH 147
P A P P+ E + + + + + +++ +LE+ +V GA
Sbjct: 70 PPAAAAPAEASP--PAGAELQEDGQQAKTQEDPTMKDQDMLEKTRQEVKDPVNVEEPGAI 127
Query: 148 GGDDDLIQRVAPFP 161
+ ++ R +P P
Sbjct: 128 VDTESVMARQSPSP 141
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.307 0.125 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,246,167
Number of Sequences: 164201
Number of extensions: 1054092
Number of successful extensions: 8148
Number of sequences better than 10.0: 450
Number of HSP's better than 10.0 without gapping: 108
Number of HSP's successfully gapped in prelim test: 348
Number of HSP's that attempted gapping in prelim test: 6897
Number of HSP's gapped (non-prelim): 1171
length of query: 192
length of database: 59,974,054
effective HSP length: 104
effective length of query: 88
effective length of database: 42,897,150
effective search space: 3774949200
effective search space used: 3774949200
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0064b.8


