

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0057b.8
(558 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
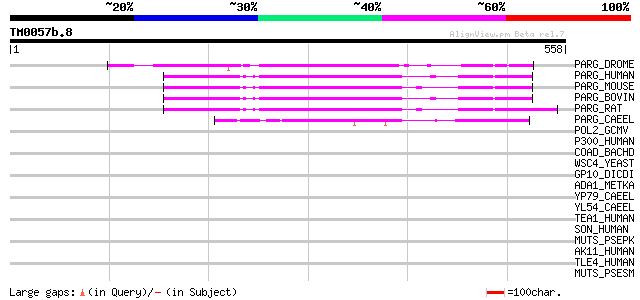
Score E
Sequences producing significant alignments: (bits) Value
PARG_DROME (O46043) Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143) 216 2e-55
PARG_HUMAN (Q86W56) Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143) 202 2e-51
PARG_MOUSE (O88622) Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143) 196 1e-49
PARG_BOVIN (O02776) Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143) 196 1e-49
PARG_RAT (Q9QYM2) Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143) 192 1e-48
PARG_CAEEL (Q867X0) Poly(ADP-ribose) glycohydrolase pme-3 (EC 3.... 119 2e-26
POL2_GCMV (P13026) RNA2 polyprotein (P2) [Contains: P2A protein ... 40 0.021
P300_HUMAN (Q09472) E1A-associated protein p300 (EC 2.3.1.48) 34 1.1
COAD_BACHD (Q9K9Q6) Phosphopantetheine adenylyltransferase (EC 2... 34 1.1
WSC4_YEAST (P38739) Cell wall integrity and stress response comp... 33 2.6
GP10_DICDI (Q06885) Glycoprotein gp100 precursor (P29F8) 33 2.6
ADA1_METKA (Q8TXF7) Acetyl-CoA decarbonylase/synthase complex al... 32 3.3
YP79_CAEEL (Q09439) Hypothetical protein B0228.9 in chromosome II 32 5.7
YL54_CAEEL (P34434) Hypothetical protein F44E2.4 in chromosome III 32 5.7
TEA1_HUMAN (P28347) Transcriptional enhancer factor TEF-1 (TEA d... 32 5.7
SON_HUMAN (P18583) SON protein (SON3) (Negative regulatory eleme... 32 5.7
MUTS_PSEPK (Q88ME7) DNA mismatch repair protein mutS 32 5.7
AK11_HUMAN (Q9UKA4) A-kinase anchor protein 11 (Protein kinase A... 32 5.7
TLE4_HUMAN (Q04727) Transducin-like enhancer protein 4 31 7.4
MUTS_PSESM (Q87XW6) DNA mismatch repair protein mutS 31 7.4
>PARG_DROME (O46043) Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143)
Length = 768
Score = 216 bits (549), Expect = 2e-55
Identities = 156/440 (35%), Positives = 219/440 (49%), Gaps = 91/440 (20%)
Query: 99 ESRKWFQEVVPALGILLLRLPSLLEAHYQNADRVLDEDGGLVRTGLRLLDSQEPGIVFLS 158
E+R +F++++P + L LRLP L+++ + LL + + LS
Sbjct: 204 ETRVFFEDLLPRIIRLALRLPDLIQSP------------------VPLLKHHKNASLSLS 245
Query: 159 QELIAALLVCSFFCLFPVNERYGKHLQSINFDEL-FGSLYDYYSQKQESKIQCIIHYFQR 217
Q+ I+ LL +F C FP + + F ++ F LY K++CI+HYF+R
Sbjct: 246 QQQISCLLANAFLCTFPRRNTLKRKSEYSTFPDINFNRLYQSTGPAVLEKLKCIMHYFRR 305
Query: 218 I------TSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVH--SSGLI 269
+ SN+P GVV+F R+ L + I WS S PL +H + G I
Sbjct: 306 VCPTERDASNVPTGVVTFVRRS-GLPEHLID--------WSQSAAPLGDVPLHVDAEGTI 356
Query: 270 EDQLSEAVEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGV 329
ED+ ++VDFAN+YLGGG L GCVQEEIRF+I PEL+V LF + EA+ ++G
Sbjct: 357 EDEGIGLLQVDFANKYLGGGVLGHGCVQEEIRFVICPELLVGKLFTECLRPFEALVMLGA 416
Query: 330 ERFSSYTGYASSFRFSGNYVDERDVDTLGRRKTRIVAIDAL-CSPGMRQYRPKFLLREIN 388
ER+S+YTGYA SF +SGN+ D D+ GRR+T IVAIDAL + QYR + RE+N
Sbjct: 417 ERYSNYTGYAGSFEWSGNFEDSTPRDSSGRRQTAIVAIDALHFAQSHHQYREDLMERELN 476
Query: 389 KAFCGFLYGSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSNHEIRDSQNDYHRME 448
KA+ G H + T P
Sbjct: 477 KAYI----GFVHWM-----------------VTPPP------------------------ 491
Query: 449 QCNNIGVATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTFRLEALHNIDKVAHWI 508
GVATGNWGCGAFGGD +K ++Q + +Q RP +AYYTF + D W+
Sbjct: 492 -----GVATGNWGCGAFGGDSYLKALLQLMVCAQLGRP-LAYYTFGNVEFRD-DFHEMWL 544
Query: 509 L--SHRWTVGDLWNMLTEYS 526
L + TV LW++L YS
Sbjct: 545 LFRNDGTTVQQLWSILRSYS 564
>PARG_HUMAN (Q86W56) Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143)
Length = 976
Score = 202 bits (514), Expect = 2e-51
Identities = 136/373 (36%), Positives = 192/373 (51%), Gaps = 62/373 (16%)
Query: 155 VFLSQELIAALLVCSFFCLFPVNERYGKHLQSINFDELFGSLYDYYSQKQESKIQCIIHY 214
+ +SQE IA+LL +FFC FP K S D F L++ S ++ K++ + Y
Sbjct: 623 ITMSQEQIASLLANAFFCTFPRRNAKMKSEYSSYPDINFNRLFEGRSSRKPEKLKTLFCY 682
Query: 215 FQRITSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSSGLIEDQLS 274
F+R+T P G+V+F R+ LED +P W PL R V G IE+
Sbjct: 683 FRRVTEKKPTGLVTFTRQ--SLED------FPE---WERCEKPLTRLHVTYEGTIEENGQ 731
Query: 275 EAVEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGVERFSS 334
++VDFAN ++GGG G VQEEIRF+I+PELI+ LF + NE + I G E++S
Sbjct: 732 GMLQVDFANRFVGGGVTSAGLVQEEIRFLINPELIISRLFTEVLDHNECLIITGTEQYSE 791
Query: 335 YTGYASSFRFSGNYVDERDVDTLGRRKTRIVAIDAL-CSPGMRQYRPKFLLREINKAFCG 393
YTGYA ++R+S ++ D + D RR T IVAIDAL + Q+ P+ + RE+NKA+CG
Sbjct: 792 YTGYAETYRWSRSHEDGSERDDWQRRCTEIVAIDALHFRRYLDQFVPEKMRRELNKAYCG 851
Query: 394 FLYGSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSNHEIRDSQNDYHRMEQCNNI 453
FL P +SE N
Sbjct: 852 FL---------------------------RPGVSSE---------------------NLS 863
Query: 454 GVATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTF-RLEALHNIDKVAHWILSHR 512
VATGNWGCGAFGGD +K +IQ LAA+ A R + Y+TF E + +I + ++ +
Sbjct: 864 AVATGNWGCGAFGGDARLKALIQILAAAAAERD-VVYFTFGDSELMRDIYSMHIFLTERK 922
Query: 513 WTVGDLWNMLTEY 525
TVGD++ +L Y
Sbjct: 923 LTVGDVYKLLLRY 935
>PARG_MOUSE (O88622) Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143)
Length = 969
Score = 196 bits (499), Expect = 1e-49
Identities = 135/373 (36%), Positives = 188/373 (50%), Gaps = 62/373 (16%)
Query: 155 VFLSQELIAALLVCSFFCLFPVNERYGKHLQSINFDELFGSLYDYYSQKQESKIQCIIHY 214
V +SQE IA+LL +FFC FP K S D F L++ S ++ K++ + Y
Sbjct: 616 VTMSQEQIASLLANAFFCTFPRRNAKMKSEYSSYPDINFNRLFEGRSSRKPEKLKTLFCY 675
Query: 215 FQRITSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSSGLIEDQLS 274
F+R+T P G+V+F R+ LED +P W PL R V G IE
Sbjct: 676 FRRVTEKKPTGLVTFTRQ--SLED------FPE---WERCEKPLTRLHVTYEGTIEGNGR 724
Query: 275 EAVEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGVERFSS 334
++VDFAN ++GGG G VQEEIRF+I+PELIV LF + NE + I G E++S
Sbjct: 725 GMLQVDFANRFVGGGVTGAGLVQEEIRFLINPELIVSRLFTEVLDHNECLIITGTEQYSE 784
Query: 335 YTGYASSFRFSGNYVDERDVDTLGRRKTRIVAIDAL-CSPGMRQYRPKFLLREINKAFCG 393
YTGYA ++R++ ++ D + D RR T IVAIDAL + Q+ P+ + RE+NKA+CG
Sbjct: 785 YTGYAETYRWARSHEDGSEKDDWQRRCTEIVAIDALHFRRYLDQFVPEKVRRELNKAYCG 844
Query: 394 FLYGSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSNHEIRDSQNDYHRMEQCNNI 453
FL G PS N
Sbjct: 845 FL-------------RPGVPSE-----------------------------------NLS 856
Query: 454 GVATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTF-RLEALHNIDKVAHWILSHR 512
VATGNWGCGAFGGD +K +IQ LAA+ A R + Y+TF E + +I + ++ +
Sbjct: 857 AVATGNWGCGAFGGDARLKALIQILAAAAAERD-VVYFTFGDSELMRDIYSMHTFLTERK 915
Query: 513 WTVGDLWNMLTEY 525
VG ++ +L Y
Sbjct: 916 LDVGKVYKLLLRY 928
>PARG_BOVIN (O02776) Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143)
Length = 977
Score = 196 bits (499), Expect = 1e-49
Identities = 134/373 (35%), Positives = 190/373 (50%), Gaps = 62/373 (16%)
Query: 155 VFLSQELIAALLVCSFFCLFPVNERYGKHLQSINFDELFGSLYDYYSQKQESKIQCIIHY 214
+ +SQE IA+LL +FFC FP K S D F L++ S ++ K++ + Y
Sbjct: 624 ITMSQEQIASLLANAFFCTFPRRNAKMKSEYSSYPDINFNRLFEGRSSRKPEKLKTLFCY 683
Query: 215 FQRITSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSSGLIEDQLS 274
F+R+T P G+V+F R+ LED +P W L R V G IE
Sbjct: 684 FRRVTEKKPTGLVTFTRQ--SLED------FPE---WERCEKLLTRLHVTYEGTIEGNGQ 732
Query: 275 EAVEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGVERFSS 334
++VDFAN ++GGG G VQEEIRF+I+PELIV LF + NE + I G E++S
Sbjct: 733 GMLQVDFANRFVGGGVTSAGLVQEEIRFLINPELIVSRLFTEVLDHNECLIITGTEQYSE 792
Query: 335 YTGYASSFRFSGNYVDERDVDTLGRRKTRIVAIDAL-CSPGMRQYRPKFLLREINKAFCG 393
YTGYA ++R++ ++ D + D RR T IVAIDAL + Q+ P+ + RE+NKA+CG
Sbjct: 793 YTGYAETYRWARSHEDRSERDDWQRRTTEIVAIDALHFRRYLDQFVPEKIRRELNKAYCG 852
Query: 394 FLYGSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSNHEIRDSQNDYHRMEQCNNI 453
FL P +SE N
Sbjct: 853 FL---------------------------RPGVSSE---------------------NLS 864
Query: 454 GVATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTF-RLEALHNIDKVAHWILSHR 512
VATGNWGCGAFGGD +K +IQ LAA+ A R + Y+TF E + +I + ++ +
Sbjct: 865 AVATGNWGCGAFGGDARLKALIQILAAAVAERD-VVYFTFGDSELMRDIYSMHTFLTERK 923
Query: 513 WTVGDLWNMLTEY 525
TVG+++ +L Y
Sbjct: 924 LTVGEVYKLLLRY 936
>PARG_RAT (Q9QYM2) Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143)
Length = 972
Score = 192 bits (489), Expect = 1e-48
Identities = 137/398 (34%), Positives = 195/398 (48%), Gaps = 62/398 (15%)
Query: 155 VFLSQELIAALLVCSFFCLFPVNERYGKHLQSINFDELFGSLYDYYSQKQESKIQCIIHY 214
V +SQE IA+LL +FFC FP K S D F L++ S ++ K++ + Y
Sbjct: 619 VTMSQEQIASLLANAFFCTFPRRNAKMKSEYSSYPDINFNRLFEGRSSRKPEKLKTLFCY 678
Query: 215 FQRITSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSSGLIEDQLS 274
F+R+T P G+V+F R+ LED +P W PL R V G IE
Sbjct: 679 FRRVTEKKPTGLVTFTRQ--SLED------FPE---WERCDKPLTRLHVTYEGTIEGNGR 727
Query: 275 EAVEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGVERFSS 334
++VDFAN ++GGG G VQEEIRF+I+PELIV LF + NE + I G E++S
Sbjct: 728 GMLQVDFANRFVGGGVTGAGLVQEEIRFLINPELIVSRLFTEVLDHNECLIITGTEQYSE 787
Query: 335 YTGYASSFRFSGNYVDERDVDTLGRRKTRIVAIDAL-CSPGMRQYRPKFLLREINKAFCG 393
YTGYA ++R++ ++ D + D R T IVAIDAL + Q+ P+ + RE+NKA+CG
Sbjct: 788 YTGYAETYRWARSHEDGSEKDDWQRCCTEIVAIDALHFRRYLDQFVPEKVRRELNKAYCG 847
Query: 394 FLYGSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSNHEIRDSQNDYHRMEQCNNI 453
FL G P N
Sbjct: 848 FL-------------RPGVPPE-----------------------------------NLS 859
Query: 454 GVATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTF-RLEALHNIDKVAHWILSHR 512
VATGNWGCGAFGGD +K +IQ LAA+ A R + Y+TF E + +I + ++ +
Sbjct: 860 AVATGNWGCGAFGGDARLKALIQLLAAAAAERD-VVYFTFGDSELMRDIYSMHTFLTERK 918
Query: 513 WTVGDLWNMLTEYSMNRSKGETNVGFLQWVLPSMYDDA 550
VG ++ +L Y + ++ G + P +Y A
Sbjct: 919 LNVGKVYRLLLRYYREECRDCSSPGPDTKLYPFIYHAA 956
>PARG_CAEEL (Q867X0) Poly(ADP-ribose) glycohydrolase pme-3 (EC
3.2.1.143) (Poly ADP-ribose metabolism enzyme-3)
Length = 781
Score = 119 bits (298), Expect = 2e-26
Identities = 98/328 (29%), Positives = 143/328 (42%), Gaps = 69/328 (21%)
Query: 207 KIQCIIHYFQRITSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSS 266
K++ + YF +++ + P G VSF ++ ++ D + + + + S L E
Sbjct: 471 KLKFLFTYFDKMSMDPPDGAVSF--RLTKMDKDTFNEEWKDKKLRS-----LPEVEFFDE 523
Query: 267 GLIEDQLSEAVEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEI 326
LIED + +VDFANE+LGGG L G VQEEIRF++ PE++V ML M EAI I
Sbjct: 524 MLIEDT-ALCTQVDFANEHLGGGVLNHGSVQEEIRFLMCPEMMVGMLLCEKMKQLEAISI 582
Query: 327 VGVERFSSYTGYASSFRFS------GNYVDERDVDTLGRRKTRIVAIDALCSPGMR---- 376
VG FSSYTGY + +++ D GR + +AIDA+ G +
Sbjct: 583 VGAYVFSSYTGYGHTLKWAELQPNHSRQNTNEFRDRFGRLRVETIAIDAILFKGSKLDCQ 642
Query: 377 --QYRPKFLLREINKAFCGFLYGSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSN 434
Q ++RE+ KA GF+ S+G
Sbjct: 643 TEQLNKANIIREMKKASIGFM--------------------------------SQG---- 666
Query: 435 HEIRDSQNDYHRMEQCNNIGVATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTFR 494
+ NI + TG WGCGAF GD +K IIQ +AA A RP
Sbjct: 667 -------------PKFTNIPIVTGWWGCGAFNGDKPLKFIIQVIAAGVADRPLHFCSFGE 713
Query: 495 LEALHNIDKVAHWILSHRWTVGDLWNML 522
E K+ + T+G L++M+
Sbjct: 714 PELAAKCKKIIERMKQKDVTLGMLFSMI 741
>POL2_GCMV (P13026) RNA2 polyprotein (P2) [Contains: P2A protein
(2A); Movement protein (2B-MP); Coat protein (2C-CP)]
Length = 1324
Score = 39.7 bits (91), Expect = 0.021
Identities = 39/125 (31%), Positives = 53/125 (42%), Gaps = 32/125 (25%)
Query: 391 FCGFLYGSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSN-HEIRDSQN-----DY 444
FCG Y KI PS D TS +E + C + H +RDS+ D+
Sbjct: 874 FCGVAIWYIFDAYGKI------PS---DVTTSLELEIARSLCPHVHVLRDSKTSVWTIDF 924
Query: 445 HRMEQCNNIGVATGNWGCGA---FGGDPEVKTIIQWLAASQALRPFIAYYTFRLEALHNI 501
H++ CG F G K + +AAS A P+ A T+RLEAL
Sbjct: 925 HKI--------------CGQSLNFSGRGFSKPTLWVIAASTAQLPWSAQVTYRLEALAQG 970
Query: 502 DKVAH 506
D++AH
Sbjct: 971 DEIAH 975
>P300_HUMAN (Q09472) E1A-associated protein p300 (EC 2.3.1.48)
Length = 2414
Score = 33.9 bits (76), Expect = 1.1
Identities = 21/53 (39%), Positives = 28/53 (52%), Gaps = 8/53 (15%)
Query: 61 PATNCSAPFPT--SEPPFPSLPNLYPLPLQTDTPSSSTIEESRKWFQEVVPAL 111
PAT AP PT + PP P L+P P QT TP ++ + Q+V P+L
Sbjct: 855 PATTIPAPVPTPPAMPPGPQSQALHPPPRQTPTPPTTQLP------QQVQPSL 901
>COAD_BACHD (Q9K9Q6) Phosphopantetheine adenylyltransferase (EC
2.7.7.3) (Pantetheine-phosphate adenylyltransferase)
(PPAT) (Dephospho-CoA pyrophosphorylase)
Length = 165
Score = 33.9 bits (76), Expect = 1.1
Identities = 32/145 (22%), Positives = 64/145 (44%), Gaps = 11/145 (7%)
Query: 147 LDSQEPGIVFLSQELIAALLVCSFFCLFPVNERYG------KHLQSINFDELFGSLYDYY 200
LD + G + ++A L + LF V ER +H+ ++ D G L DY
Sbjct: 19 LDIIQRGANVFDEVIVAVLHNRNKVPLFSVEERLELLKKATEHIPNVTIDSFNGLLIDYV 78
Query: 201 SQKQESKIQCIIHYFQRITS-NMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSII-PL 258
QKQ + II + ++ S +K+ P + + ++ S+SI+ +
Sbjct: 79 KQKQ---AKAIIRGLRAVSDFEYEMQAASINKKLGPDVETFFMMTSNQYSYLSSSIVKEV 135
Query: 259 CRFEVHSSGLIEDQLSEAVEVDFAN 283
++E S ++ ++EA++ F++
Sbjct: 136 AKYEADVSDIVPPVVAEALKAKFSS 160
>WSC4_YEAST (P38739) Cell wall integrity and stress response
component 4 precursor
Length = 605
Score = 32.7 bits (73), Expect = 2.6
Identities = 29/97 (29%), Positives = 40/97 (40%), Gaps = 5/97 (5%)
Query: 3 SEKCQSNICTVTCFLSSHFTSLLRENGKQTRLEIDSSISTPRD---AFLLSVLAFPSRRI 59
S K ++I T T SS T+ + T + + SS ST + L+S S
Sbjct: 184 STKLSTSIPTSTT--SSTSTTTSTSSSTSTTVSVTSSTSTTTSTTSSTLISTSTSSSSSS 241
Query: 60 TPATNCSAPFPTSEPPFPSLPNLYPLPLQTDTPSSST 96
TP T SAP TS S P + P+SS+
Sbjct: 242 TPTTTSSAPISTSTTSSTSTSTSTTSPTSSSAPTSSS 278
>GP10_DICDI (Q06885) Glycoprotein gp100 precursor (P29F8)
Length = 544
Score = 32.7 bits (73), Expect = 2.6
Identities = 18/47 (38%), Positives = 23/47 (48%), Gaps = 4/47 (8%)
Query: 55 PSRRITPATNCSAP----FPTSEPPFPSLPNLYPLPLQTDTPSSSTI 97
PS+ I P + + P PTS+P P P P T TP+S TI
Sbjct: 136 PSQTIPPTVSPTVPPQTTSPTSKPTSTPTPTSTPTPTSTPTPTSQTI 182
>ADA1_METKA (Q8TXF7) Acetyl-CoA decarbonylase/synthase complex alpha
subunit 1 (EC 1.2.99.2) (ACDS complex alpha subunit 1)
(ACDS complex carbon monoxide dehydrogenase 1) (ACDS
CODH 1)
Length = 760
Score = 32.3 bits (72), Expect = 3.3
Identities = 15/58 (25%), Positives = 30/58 (50%), Gaps = 5/58 (8%)
Query: 500 NIDKVAHWILSHRWTVGDLWNMLTEYSMNRSKGETNVGFLQWVLPSMYDDAGMDFSNI 557
N ++A +IL+ G LW +++ ++ S GF +W +P +Y AG+ + +
Sbjct: 585 NFKEIADYILNRIGACGVLWGTMSQKALAIS-----TGFTRWGIPIVYGPAGLKYQTL 637
>YP79_CAEEL (Q09439) Hypothetical protein B0228.9 in chromosome II
Length = 338
Score = 31.6 bits (70), Expect = 5.7
Identities = 22/74 (29%), Positives = 31/74 (41%), Gaps = 16/74 (21%)
Query: 363 RIVAIDALCSPGMRQYRP----KFLLREINKAFCGFLYGSKHQLYQKILQEKGCPSTLFD 418
+I + C+ + QY +L E+NK FCG + G +YQ L LFD
Sbjct: 260 KIYWVTKRCTVDVTQYPAVLDGSVILGEVNKRFCGMIDGENWDVYQSFLD-------LFD 312
Query: 419 AATSTPMETSEGKC 432
P +T G C
Sbjct: 313 -----PNDTKWGSC 321
>YL54_CAEEL (P34434) Hypothetical protein F44E2.4 in chromosome III
Length = 1283
Score = 31.6 bits (70), Expect = 5.7
Identities = 34/113 (30%), Positives = 46/113 (40%), Gaps = 23/113 (20%)
Query: 2 ISEKCQSNICTVTCFLSSHFTSLLRENGKQTRLEIDSSISTPRD--AFLLSVLAFPSRRI 59
+ KC+++ T L + TSL E G PRD A L +++ P R
Sbjct: 576 VKNKCEADALTDLLSLKTKMTSLGEEEG------------CPRDPPANLDEIISRPVARP 623
Query: 60 TPATNCSAPFPTSEP-PFPSLPNLYPLPLQTDTPSSSTIEESRKWFQEVVPAL 111
TP T PT++P P PS P SS E +K F+E V L
Sbjct: 624 TPVT-MPPRAPTAKPLPIPSAPT-------PPVASSKCSIEDQKKFEECVKPL 668
>TEA1_HUMAN (P28347) Transcriptional enhancer factor TEF-1 (TEA
domain family member 1) (TEAD-1) (Protein GT-IIC)
(Transcription factor 13) (NTEF-1)
Length = 426
Score = 31.6 bits (70), Expect = 5.7
Identities = 26/96 (27%), Positives = 40/96 (41%), Gaps = 7/96 (7%)
Query: 17 LSSHFTSLLRENGKQTRLEIDSSISTPRDAFLLSVLAFPSRRITPAT---NCSAPFPTSE 73
+SSH L R + ++ T +D L + A S +I AT N
Sbjct: 90 VSSHIQVLARRKSRDFHSKLKDQ--TAKDKALQHMAAMSSAQIVSATAIHNKLGLPGIPR 147
Query: 74 PPFPSLPNLYPLPLQTDTPSSSTIEESRKWFQEVVP 109
P FP P +P +QT P SS ++ + + Q+ P
Sbjct: 148 PTFPGAPGFWPGMIQTGQPGSS--QDVKPFVQQAYP 181
>SON_HUMAN (P18583) SON protein (SON3) (Negative regulatory
element-binding protein) (NRE-binding protein) (DBP-5)
(Bax antagonist selected in saccharomyces 1) (BASS1)
Length = 2426
Score = 31.6 bits (70), Expect = 5.7
Identities = 23/81 (28%), Positives = 35/81 (42%), Gaps = 7/81 (8%)
Query: 27 ENGKQTRLEIDSSISTPRDAFL-------LSVLAFPSRRITPATNCSAPFPTSEPPFPSL 79
E+ +T+ D +I D+FL L P+R P+ +P EPP S+
Sbjct: 135 ESESKTKSHDDGNIDLESDSFLKFDSEPSAVALELPTRAFGPSETNESPAVVLEPPVVSM 194
Query: 80 PNLYPLPLQTDTPSSSTIEES 100
P L+T P++ T E S
Sbjct: 195 EVSEPHILETLKPATKTAELS 215
>MUTS_PSEPK (Q88ME7) DNA mismatch repair protein mutS
Length = 857
Score = 31.6 bits (70), Expect = 5.7
Identities = 22/79 (27%), Positives = 39/79 (48%), Gaps = 6/79 (7%)
Query: 256 IPLCRFEVHSSGLIEDQLSEAVEVDFAN---EYLGGGALRRGCVQEEIRFMISPELIVDM 312
IP+C HS +E L++ V++ + E +G A +G V+ ++ +I+P + D
Sbjct: 63 IPMCGIPFHS---LEGYLAKLVKLGESVVICEQIGDPATSKGPVERQVVRIITPGTVSDE 119
Query: 313 LFLPSMADNEAIEIVGVER 331
L DN ++G ER
Sbjct: 120 ALLDERRDNLIAALLGDER 138
>AK11_HUMAN (Q9UKA4) A-kinase anchor protein 11 (Protein kinase A
anchoring protein 11) (PRKA11) (A kinase anchor protein
220 kDa) (AKAP 220) (hAKAP220)
Length = 1901
Score = 31.6 bits (70), Expect = 5.7
Identities = 18/58 (31%), Positives = 29/58 (49%), Gaps = 3/58 (5%)
Query: 468 DPEVKTIIQWLAASQALRPFIAYYTFRLEALHNIDKVAHWILSHRWTVGDLWNMLTEY 525
DP+++ I+QWL AS+A +A F A ++ + W VGDL + +Y
Sbjct: 1823 DPQLRIILQWLIASEA---EVAELYFHDSANKEFMLLSKQLQEKGWKVGDLLQAVLQY 1877
>TLE4_HUMAN (Q04727) Transducin-like enhancer protein 4
Length = 766
Score = 31.2 bits (69), Expect = 7.4
Identities = 21/68 (30%), Positives = 32/68 (46%), Gaps = 5/68 (7%)
Query: 22 TSLLRENGKQTRLEIDSSISTPRDAFLLSVLAFPSRRITPATNCSAPFPTSEPPFP---S 78
T LL+++ + I SS STP L+ + TP + + P P ++ P P S
Sbjct: 275 TRLLKKDAPISPASIASSSSTPSSKS--KELSLNEKSTTPVSKSNTPTPRTDAPTPGSNS 332
Query: 79 LPNLYPLP 86
P L P+P
Sbjct: 333 TPGLRPVP 340
>MUTS_PSESM (Q87XW6) DNA mismatch repair protein mutS
Length = 855
Score = 31.2 bits (69), Expect = 7.4
Identities = 21/76 (27%), Positives = 34/76 (44%)
Query: 256 IPLCRFEVHSSGLIEDQLSEAVEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFL 315
IP+C H++ +L + E E +G A +G V ++ +I+P I D L
Sbjct: 63 IPMCGIPYHAAEGYLAKLVKLGESVVICEQIGDPATSKGPVDRQVVRIITPGTISDEALL 122
Query: 316 PSMADNEAIEIVGVER 331
DN ++G ER
Sbjct: 123 DERRDNLIAAVLGDER 138
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.138 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 68,858,664
Number of Sequences: 164201
Number of extensions: 3029374
Number of successful extensions: 7389
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 7355
Number of HSP's gapped (non-prelim): 39
length of query: 558
length of database: 59,974,054
effective HSP length: 115
effective length of query: 443
effective length of database: 41,090,939
effective search space: 18203285977
effective search space used: 18203285977
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0057b.8


