

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
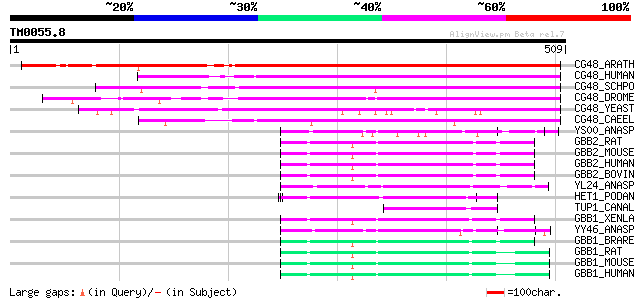
Query= TM0055.8
(509 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CG48_ARATH (Q9FMU5) Hypothetical WD-repeat protein At5g14050 592 e-168
CG48_HUMAN (Q9Y5J1) WD-repeat protein CGI-48 252 1e-66
CG48_SCHPO (P78750) Hypothetical WD-repeat protein C29A3.06 in c... 236 8e-62
CG48_DROME (Q9V7P1) Hypothetical WD-repeat protein l(2)k07824 175 2e-43
CG48_YEAST (P40362) Hypothetical 66.4 kDa Trp-Asp repeats contai... 171 4e-42
CG48_CAEEL (P42000) Hypothetical WD-repeat protein B0280.9 in ch... 135 2e-31
YS00_ANASP (Q8YTC2) Hypothetical WD-repeat protein alr2800 62 5e-09
GBB2_RAT (P54313) Guanine nucleotide-binding protein G(I)/G(S)/G... 56 2e-07
GBB2_MOUSE (P62880) Guanine nucleotide-binding protein G(I)/G(S)... 56 2e-07
GBB2_HUMAN (P62879) Guanine nucleotide-binding protein G(I)/G(S)... 56 2e-07
GBB2_BOVIN (P11017) Guanine nucleotide-binding protein G(I)/G(S)... 56 2e-07
YL24_ANASP (Q8YV57) Hypothetical WD-repeat protein all2124 55 6e-07
HET1_PODAN (Q00808) Vegetatible incompatibility protein HET-E-1 53 2e-06
TUP1_CANAL (P56093) Transcriptional repressor TUP1 51 6e-06
GBB1_XENLA (P79959) Guanine nucleotide-binding protein G(I)/G(S)... 51 6e-06
YY46_ANASP (Q8YRI1) Hypothetical WD-repeat protein alr3466 51 8e-06
GBB1_BRARE (Q6PH57) Guanine nucleotide-binding protein G(I)/G(S)... 50 1e-05
GBB1_RAT (P54311) Guanine nucleotide-binding protein G(I)/G(S)/G... 49 2e-05
GBB1_MOUSE (P62874) Guanine nucleotide-binding protein G(I)/G(S)... 49 2e-05
GBB1_HUMAN (P62873) Guanine nucleotide-binding protein G(I)/G(S)... 49 2e-05
>CG48_ARATH (Q9FMU5) Hypothetical WD-repeat protein At5g14050
Length = 546
Score = 592 bits (1525), Expect = e-168
Identities = 312/501 (62%), Positives = 391/501 (77%), Gaps = 23/501 (4%)
Query: 12 KIEEKEVVDREE-NSDVDTVKSKKRKRDRKREEHVVDMVEQVREMRKLESFLFGSLYSPL 70
K + ++V D EE SD D + K+RK ++++++ ++ E V EM+KLE+ +FGSLYSP+
Sbjct: 19 KKQYEDVEDEEEIGSDDDLTRGKRRKTEKEKQK--LEESELV-EMKKLENLIFGSLYSPV 75
Query: 71 ESGKEVDGEVEPSDLFFTDRSADSVLSVCDEDGEFSDGSGDGDDGL-----GRKPAWVDE 125
GKE E + S LF DRSA + ++DG+ + D ++G + AW DE
Sbjct: 76 TFGKEE--EEDGSALFHVDRSAVRQIPDYEDDGDDDEELSDEENGQVVAIRKGEAAWEDE 133
Query: 126 EEEKFTVNIAKVNRLRKLRKDEDESFISGSEYVARLRAHHVKLNRGTDWAQLDSRLKLDR 185
EE++ V+IA VNRLRKLRK+E+E ISGSEY+ARLRAHH KLN GTDWA+ DS++
Sbjct: 134 EEKQINVDIASVNRLRKLRKEENEGLISGSEYIARLRAHHAKLNPGTDWARPDSQI---- 189
Query: 186 SDYDGELTDDENEAVVRRGYENVDDILRTNEDLVANS-SSKLLPGHLEYSRLVDANIQDP 244
DGE +DD++ + G VDDILRTNEDLV S +KL G LEYS+LVDAN DP
Sbjct: 190 --VDGESSDDDD---TQDG--GVDDILRTNEDLVVKSRGNKLCAGRLEYSKLVDANAADP 242
Query: 245 SNGPVNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQ 304
SNGP+NSV FH+NAQLLLTAGLD++LRFFQIDGKRNTKIQSIFLEDCPIRKA+FLP+GSQ
Sbjct: 243 SNGPINSVHFHQNAQLLLTAGLDRRLRFFQIDGKRNTKIQSIFLEDCPIRKAAFLPNGSQ 302
Query: 305 VILSGRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVST 364
VI+SGRRKFFYSFDL K +KIGPLVGREEKSLE FE+S DS IAFVGNEGYILLVST
Sbjct: 303 VIVSGRRKFFYSFDLEKAKFDKIGPLVGREEKSLEYFEVSQDSNTIAFVGNEGYILLVST 362
Query: 365 KTKQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTA 424
KTK+L+G+LKM+G+ RSLAFSEDG+ LLS+GGDGQVY WDLRT C++KGVDEG T+
Sbjct: 363 KTKELIGTLKMNGSVRSLAFSEDGKHLLSSGGDGQVYVWDLRTMKCLYKGVDEGSTCGTS 422
Query: 425 LCTSPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQILAIC 484
LC+S +G LFA+G+D GIVN+Y + +F+GGKRKPIKT++NL +K+DFM+FNHD+QILAI
Sbjct: 423 LCSSLNGALFASGTDRGIVNIYKKSEFVGGKRKPIKTVDNLTSKIDFMKFNHDAQILAIV 482
Query: 485 SSMKKSSLKLIHIPSYTVFSN 505
S+M K+S+KL+H+PS TVFSN
Sbjct: 483 STMNKNSVKLVHVPSLTVFSN 503
>CG48_HUMAN (Q9Y5J1) WD-repeat protein CGI-48
Length = 532
Score = 252 bits (644), Expect = 1e-66
Identities = 149/392 (38%), Positives = 226/392 (57%), Gaps = 21/392 (5%)
Query: 118 RKPAWVDEEEEKFTVNIAKVNRLRK-LRKDEDESFISGSEYVARLRAHHVKLNRGTD-WA 175
+KP WVDEE+E + NR RK + K+ ES +S RL+ G WA
Sbjct: 114 KKPVWVDEEDEDEEMVDMMNNRFRKDMMKNASESKLSKDNLKKRLKEEFQHAMGGVPAWA 173
Query: 176 QLDSRLKLDRSDYDGELTDDENEAVVRRGYENVDDILRTNEDLVANSSSKLLPGHLEYSR 235
+ R +DDE+E E+ DD+L+ + ++ S+S L G L+
Sbjct: 174 ETTKRKTS---------SDDESE-------EDEDDLLQRTGNFISTSTS-LPRGILKMKN 216
Query: 236 LVDANIQDPSNGPVNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRK 295
AN + P+ ++SVQFH AQ+++ AGLD + FQ+DGK N KIQSI+LE PI K
Sbjct: 217 CQHANAERPTVARISSVQFHPGAQIVMVAGLDNAVSLFQVDGKTNPKIQSIYLERFPIFK 276
Query: 296 ASFLPDGSQVIL-SGRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVG 354
A F +G +V+ S K Y +D++ G L + + G +EK + FE+S D + G
Sbjct: 277 ACFSANGEEVLATSTHSKVLYVYDMLAGKLIPVHQVRGLKEKIVRSFEVSPDGSFLLING 336
Query: 355 NEGYILLVSTKTKQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKG 414
GY+ L++ KTK+L+GS+K++G + FS D +K+ ++ GDG+VY WD+ +R C+++
Sbjct: 337 IAGYLHLLAMKTKELIGSMKINGRVAASTFSSDSKKVYASSGDGEVYVWDVNSRKCLNRF 396
Query: 415 VDEGCINSTALCTSPSGTLFAAGSDSGIVNVYNREDFLGGKR-KPIKTIENLITKVDFMR 473
VDEG + ++ TS +G A GS+ G+VN+YN++ L KPIK I NL+T V +
Sbjct: 397 VDEGSLYGLSIATSRNGQYVACGSNCGVVNIYNQDSCLQETNPKPIKAIMNLVTGVTSLT 456
Query: 474 FNHDSQILAICSSMKKSSLKLIHIPSYTVFSN 505
FN ++ILAI S K +++L+H+PS TVFSN
Sbjct: 457 FNPTTEILAIASEKMKEAVRLVHLPSCTVFSN 488
>CG48_SCHPO (P78750) Hypothetical WD-repeat protein C29A3.06 in
chromosome II
Length = 556
Score = 236 bits (603), Expect = 8e-62
Identities = 136/433 (31%), Positives = 232/433 (53%), Gaps = 14/433 (3%)
Query: 79 EVEPSDLFFTDR-SADSVLSVCDEDGEFSDGSGDGDDGLGRK--PAWVDEEEEKFTVNIA 135
++E ++LF D SAD D D + D+ + W D ++E+ +++
Sbjct: 55 KLEDAELFMFDTGSADGAKDSVPLDIIAGDNTVKEDEEASNEIPSIWEDSDDERLMISLQ 114
Query: 136 KVNRLRKLRKDEDESFISGSEYVARLRAHHVKLNRGTDWAQLDSRLKLDRSDYDGELTDD 195
+RLRKLR+ EDE ++G +Y RLR ++ +WA+ K D ++ + E
Sbjct: 115 DHSRLRKLRQYEDEDMVNGLQYARRLRTQFERVYPVPEWAK-----KQDVTEEEDEFNAL 169
Query: 196 ENEAVVRRGYENVDDILRTNEDLVANSSSKLLPGHLEYSRLVDANIQDPSNGPVNSVQFH 255
++V+ + ++ + +++ + SS L PG + RL DAN Q PS+ + + H
Sbjct: 170 SEKSVIPKSLKS---LFKSSVSYINQSSKLLAPGTINIKRLKDANFQAPSHSGIRCMSIH 226
Query: 256 RNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILSGRRKFFY 315
LLLT G D+ LR +Q+DGK N + S+ L ++ A F PDG +VI +GRRK+ Y
Sbjct: 227 PYFPLLLTCGFDRTLRIYQLDGKVNPLVTSLHLRSSALQTALFHPDGKRVIAAGRRKYMY 286
Query: 316 SFDLVKGTLEKIGPLVGRE--EKSLEVFELSSDSQMIAFVGNEGYILLVSTKTKQLVGSL 373
+DL ++K+ + G+E + S+E F + + IA G G+I L+ T Q S
Sbjct: 287 IWDLESAQVQKVSRMYGQENFQPSMERFHVDPTGKYIALEGRSGHINLLHALTGQFATSF 346
Query: 374 KMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTALCTSPSGTL 433
K+ G + F+ DG ++L +V+H+++ R+ + + + +++T C PS
Sbjct: 347 KIEGVLSDVLFTSDGSEMLVLSYGAEVWHFNVEQRSVVRRWQVQDGVSTTHFCLDPSNKY 406
Query: 434 FAAGSDSGIVNVYN-REDFLGGKRKPIKTIENLITKVDFMRFNHDSQILAICSSMKKSSL 492
A GS SGIVN+Y+ + KP+ T++N+ ++ M F+ DSQ+LAI S KK +L
Sbjct: 407 LAIGSKSGIVNIYDLQTSNADAAPKPVTTLDNITFSINSMSFSQDSQVLAIASRGKKDTL 466
Query: 493 KLIHIPSYTVFSN 505
+L+H+PS++VF N
Sbjct: 467 RLVHVPSFSVFRN 479
>CG48_DROME (Q9V7P1) Hypothetical WD-repeat protein l(2)k07824
Length = 506
Score = 175 bits (444), Expect = 2e-43
Identities = 133/486 (27%), Positives = 221/486 (45%), Gaps = 55/486 (11%)
Query: 31 KSKKRKRDRKREEHVVDMVEQVREMR----KLESFLFGSLYSPLESGKEVDGEVEPSDLF 86
+ ++ K + + E + Q +E+ +E LFG L + + G+ P+D
Sbjct: 22 QQEQEKPAKIKRERYIPKASQAKELNYVEVPMEKVLFGDRQRLLTNLAKSVGQKLPND-- 79
Query: 87 FTDRSADSVLSVCDEDGEFSDGSGDGDDGLGRKPAWVDEEEEKFTVNIAK-----VNRLR 141
DED E + G G RK AW D ++E V K L
Sbjct: 80 -------------DED-EQEENPGQAKPGDKRKAAWSDSDDEDLQVGDVKKATKHTGPLN 125
Query: 142 KLRKDEDESFISGSEYVARLRAHHVKLNRGTDWAQLDSRLKLDRSDYDGELTDDENEAVV 201
LRKD+ Y L A + WA+ K +++ D +++ DE
Sbjct: 126 HLRKDKS--------YKEYLTARFQRTLNQPKWAE-----KKVKNEDDEDVSSDE----- 167
Query: 202 RRGYENVDDILRTNEDLVANSSSKLLPGH-LEYSRLVDANIQDPSNGPVNSVQFHRNAQL 260
++LRT + + + LP L + R+ D N + G S+QFH +
Sbjct: 168 --------ELLRTVGFIDRKARNSDLPQKTLNFKRVKDLNRATYAEGNATSIQFHPTSTA 219
Query: 261 LLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILSGRRKFFYSFDLV 320
L AG++ + +DG++N ++ ++ + P+ + P G++ + F+YS+DL+
Sbjct: 220 ALVAGMNGLATIYAVDGQKNERLHNMRFKKFPLACSRIAPCGTRAFFGSVKPFYYSYDLL 279
Query: 321 KGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTKQLVGSLKMSGTTR 380
+ K+ L G E + FE+S + I G G I L++ KT +L+ S K G +
Sbjct: 280 EAKESKL-KLPGAME-FMHRFEVSPCGKFIVTAGKFGAIHLLTAKTNELLHSFKQEGKVK 337
Query: 381 SLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTALCTSPSGTLFAAGSDS 440
+S D +++L G V +LR H +D+GCI+ ++ SP+ L A GS
Sbjct: 338 GFTWSSDSKRILVCGSTSNVSVLNLRQNLIEHIFMDDGCIHGESIQLSPNQRLLATGSQE 397
Query: 441 GIVNVYNREDFLGGKR-KPIKTIENLITKVDFMRFNHDSQILAICSSMKKSSLKLIHIPS 499
G+VN+Y+ E K +P K NL T + ++FNH S++LA+CSS ++ KL H PS
Sbjct: 398 GVVNIYDYESIFASKAPQPEKRFMNLRTAITDLQFNHSSELLAMCSSEAPNAFKLAHFPS 457
Query: 500 YTVFSN 505
TV+SN
Sbjct: 458 ATVYSN 463
>CG48_YEAST (P40362) Hypothetical 66.4 kDa Trp-Asp repeats
containing protein in SMC3-MRPL8 intergenic region
Length = 594
Score = 171 bits (433), Expect = 4e-42
Identities = 151/505 (29%), Positives = 236/505 (45%), Gaps = 73/505 (14%)
Query: 64 GSLYSPLESGKEVDGE---VEPSDLFFTDRSA--------DSVLSVCDEDGEFSDGSGDG 112
GS E + +GE V LFF D + + V DED SD +
Sbjct: 60 GSESDNSEEDEAQNGELDHVNNDQLFFVDDGGNEDSQDKNEDTMDVDDEDDSSSDDYSED 119
Query: 113 DDGLGRKPAWVDEEEEKFTVNIAKVNRLRKLRKDEDESFISGSEYVARLRAHHVKLNRGT 172
+ + AW+D ++EK V I N+ +KLR +ES I+G Y+ RLR+ K+
Sbjct: 120 SE----EAAWIDSDDEKIKVPILVTNKTKKLRTSYNESKINGVHYINRLRSQFEKIYPRP 175
Query: 173 DWAQLDSRLKLDRSDYDGELTDDENEAVVRRGYENVDDILRTNEDLVAN-SSSKLLPGH- 230
W +S +LD + D E + + V+ + IL T + S+SKLLP
Sbjct: 176 KWVDDESDSELDDEEDDEE---EGSNNVINGDINALTKILSTTYNYKDTLSNSKLLPPKK 232
Query: 231 LEYSRLVDANIQDPSNGPVNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLED 290
L+ RL DAN PS+ + S+ FH + LLLT G D+ LR + IDGK N + S+ L
Sbjct: 233 LDIVRLKDANASHPSHSAIQSLSFHPSKPLLLTGGYDKTLRIYHIDGKTNHLVTSLHLVG 292
Query: 291 CPIRKASFLPDGS-----QVILSGRRKFFYSFDL--------VKGTLEKIGPLVGRE--E 335
PI+ +F S + +GRR++ +S+DL +EK L G E +
Sbjct: 293 SPIQTCTFYTSLSNQNQQNIFTAGRRRYMHSWDLSLENLTHSQTAKIEKFSRLYGHESTQ 352
Query: 336 KSLEVFELS--SDSQ------MIAFVGNEGYI-LLVSTKTKQLVGSLKMSGTTRSLAFSE 386
+S E F+++ +SQ ++ GN G+I +L ST L+G K+ G F
Sbjct: 353 RSFENFKVAHLQNSQTNSVHGIVLLQGNNGWINILHSTSGLWLMG-CKIEGVITD--FCI 409
Query: 387 DGQK---------LLSAGGDGQVYHWDL-RTRTCIHKGVDEGCINSTAL-------CTSP 429
D Q L++ G+V+ +DL + I + D+G + T + T P
Sbjct: 410 DYQPISRGKFRTILIAVNAYGEVWEFDLNKNGHVIRRWKDQGGVGITKIQVGGGTTTTCP 469
Query: 430 S--------GTLFAAGSDSGIVNVYNREDFL-GGKRKPIKTIENLITKVDFMRFNHDSQI 480
+ A GS+SG VN+Y+R + + P+ ++ L T + ++F+ D QI
Sbjct: 470 ALQISKIKQNRWLAVGSESGFVNLYDRNNAMTSSTPTPVAALDQLTTTISNLQFSPDGQI 529
Query: 481 LAICSSMKKSSLKLIHIPSYTVFSN 505
L + S K +L+L+H+PS +VFSN
Sbjct: 530 LCMASRAVKDALRLVHLPSCSVFSN 554
>CG48_CAEEL (P42000) Hypothetical WD-repeat protein B0280.9 in
chromosome III
Length = 429
Score = 135 bits (341), Expect = 2e-31
Identities = 102/402 (25%), Positives = 185/402 (45%), Gaps = 41/402 (10%)
Query: 119 KPAWVDEEEEKFTVNIAKVNRLR-----KLRKDEDESFISGSEYVARLRAHHVKLNRGTD 173
KPAW DE+++ V + K ++ K DE+ + EYV RL+ K + GT
Sbjct: 11 KPAWHDEDDDNMVVAVPKKVKMTMRVELKRTTDEEHGELDSKEYVGRLQEAFRKRHGGTP 70
Query: 174 -WAQLDSRLKLDRSDYDGELTDDENEAVVRRGYENVDDILRTNEDLVANSSSKLLPGHLE 232
WA+ + G E +L+T +A + LP
Sbjct: 71 KWAKAAA------------------------GDEEEGSLLKTAAGYLAKDVN--LPKTTI 104
Query: 233 YSRLV-DANIQDPSNGPVNSVQFHRNAQLLLTAGLDQKLRFFQI--DGKRNTKIQSIFLE 289
++ L+ D NI + V+FH+ +L+ A ++ F++ + K++ +QS
Sbjct: 105 HTTLIKDFNIGHRYTRGITVVKFHKTRPVLIVADQGGNVQLFKVSREVKKDRFLQSANFS 164
Query: 290 DCPIRKASFLPDGSQVILSGRRK-FFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQ 348
PI G VI S RK + +++ + ++ P ++ + +F +S DSQ
Sbjct: 165 KFPIDSLEIADKGHSVICSSSRKEYLMQYNMETREVTQLKPPNTVPKQGIRLFAISHDSQ 224
Query: 349 MIAFVGNEGYILLVSTKTKQLVGSLKMSGTTRSLAF-SEDGQKLLSAGGDGQVYHWDLR- 406
+A G+ +I ++ + + + ++ + + F +++ GQ+ ++
Sbjct: 225 FLAIAGHNSHIYVLHATSMEHITTISLPANASEIKFFPSHSREIWIICETGQIVIANIGL 284
Query: 407 --TRTCIHKGVDEGCINSTALCTSPSGTLFAAGSDSGIVNVYNREDFLGGKR-KPIKTIE 463
T++ H D+G ++ T L S G FA GSD+GIVNVY+ D +P+ +
Sbjct: 285 PGTKSSQHTFTDDGAVHGTTLAISQHGDYFATGSDTGIVNVYSGNDCRNSTNPRPLFNVS 344
Query: 464 NLITKVDFMRFNHDSQILAICSSMKKSSLKLIHIPSYTVFSN 505
NL+T V + FN D+Q++AICS++K + L+L+H+ S T F N
Sbjct: 345 NLVTAVSSIAFNSDAQLMAICSNVKDNHLRLVHVASQTTFKN 386
>YS00_ANASP (Q8YTC2) Hypothetical WD-repeat protein alr2800
Length = 1258
Score = 61.6 bits (148), Expect = 5e-09
Identities = 59/244 (24%), Positives = 109/244 (44%), Gaps = 15/244 (6%)
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILS 308
V V F + ++L + G D+ ++ + + + I+++ + + +F PDG + +
Sbjct: 687 VRFVVFSPDGEILASCGADENVKLWSV--RDGVCIKTLTGHEHEVFSVAFHPDGETLASA 744
Query: 309 GRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTKQ 368
K +D+ GT + L G + + S D +A + I L +
Sbjct: 745 SGDKTIKLWDIQDGTC--LQTLTGHTDW-VRCVAFSPDGNTLASSAADHTIKLWDVSQGK 801
Query: 369 LVGSLKM-SGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINST-ALC 426
+ +LK +G RS+AFS DGQ L S GD + W+ T C+ + G NS ++
Sbjct: 802 CLRTLKSHTGWVRSVAFSADGQTLASGSGDRTIKIWNYHTGECLKTYI--GHTNSVYSIA 859
Query: 427 TSPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQILAICSS 486
SP + +GS + +++ + + IKT+ +V + F+ D Q LA C S
Sbjct: 860 YSPDSKILVSGSGDRTIKLWDCQTHI-----CIKTLHGHTNEVCSVAFSPDGQTLA-CVS 913
Query: 487 MKKS 490
+ +S
Sbjct: 914 LDQS 917
Score = 46.2 bits (108), Expect = 2e-04
Identities = 51/206 (24%), Positives = 87/206 (41%), Gaps = 19/206 (9%)
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILS 308
V +V FH +++ T D ++ + I + K S + I ++ PDG + +
Sbjct: 1023 VYAVVFHPQGKIIATGSADCTVKLWNISTGQCLKTLSEHSDK--ILGMAWSPDGQLLASA 1080
Query: 309 GRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTKQ 368
+ +D G +G L G + S + ++IA + + + + +
Sbjct: 1081 SADQSVRLWDCCTGRC--VGILRGHSNRVYSAI-FSPNGEIIATCSTDQTVKIWDWQQGK 1137
Query: 369 LVGSLKMSGTTR---SLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTAL 425
+ +L +G T +AFS DG+ L SA D V WD+ T C H CI T L
Sbjct: 1138 CLKTL--TGHTNWVFDIAFSPDGKILASASHDQTVRIWDVNTGKCHHI-----CIGHTHL 1190
Query: 426 CT----SPSGTLFAAGSDSGIVNVYN 447
+ SP G + A+GS V ++N
Sbjct: 1191 VSSVAFSPDGEVVASGSQDQTVRIWN 1216
Score = 45.8 bits (107), Expect = 3e-04
Identities = 70/295 (23%), Positives = 123/295 (40%), Gaps = 50/295 (16%)
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILS 308
V SV FH + + L +A D+ ++ + I T +Q++ +R +F PDG+ + S
Sbjct: 729 VFSVAFHPDGETLASASGDKTIKLWDIQD--GTCLQTLTGHTDWVRCVAFSPDGNTLASS 786
Query: 309 GRRKFFYSFDLVKG----TLEKIGPLV--------------GREEKSLEVFELSSDSQMI 350
+D+ +G TL+ V G +++++++ + +
Sbjct: 787 AADHTIKLWDVSQGKCLRTLKSHTGWVRSVAFSADGQTLASGSGDRTIKIWNYHTGECLK 846
Query: 351 AFVG--NEGYILLVSTKTKQLVGSL-----------------KMSGTTR---SLAFSEDG 388
++G N Y + S +K LV + G T S+AFS DG
Sbjct: 847 TYIGHTNSVYSIAYSPDSKILVSGSGDRTIKLWDCQTHICIKTLHGHTNEVCSVAFSPDG 906
Query: 389 QKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTALCTSPSGTLFAAGSDSGIVNVYNR 448
Q L D V W+ RT C+ K + + SP + A+GS+ V ++
Sbjct: 907 QTLACVSLDQSVRLWNCRTGQCL-KAWYGNTDWALPVAFSPDRQILASGSNDKTVKLW-- 963
Query: 449 EDFLGGKRKPIKTIENLITKVDFMRFNHDSQILAICSSMKKSSLKLIHIPSYTVF 503
D+ GK I ++E + + F+ DSQ LA S+ SS++L +I + F
Sbjct: 964 -DWQTGKY--ISSLEGHTDFIYGIAFSPDSQTLA--SASTDSSVRLWNISTGQCF 1013
Score = 34.3 bits (77), Expect = 0.78
Identities = 26/108 (24%), Positives = 49/108 (45%), Gaps = 2/108 (1%)
Query: 344 SSDSQMIAFVGNEGYILLVSTKTKQLVGSLK-MSGTTRSLAFSEDGQKLLSAGGDGQVYH 402
S + Q++A + ++ + K+ +L+ + S R + FS DG+ L S G D V
Sbjct: 651 SPEGQLLATCDTDCHVRVWEVKSGKLLLICRGHSNWVRFVVFSPDGEILASCGADENVKL 710
Query: 403 WDLRTRTCIHKGVDEGCINSTALCTSPSGTLFAAGSDSGIVNVYNRED 450
W +R CI K + ++ P G A+ S + +++ +D
Sbjct: 711 WSVRDGVCI-KTLTGHEHEVFSVAFHPDGETLASASGDKTIKLWDIQD 757
Score = 31.2 bits (69), Expect = 6.6
Identities = 42/186 (22%), Positives = 76/186 (40%), Gaps = 17/186 (9%)
Query: 257 NAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILSGRRKFFYS 316
+ QLL +A DQ +R + R I + + A F P+G + +
Sbjct: 1073 DGQLLASASADQSVRLWDCCTGRCVGI--LRGHSNRVYSAIFSPNGEIIATCSTDQTVKI 1130
Query: 317 FDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTKQL----VGS 372
+D +G K L G ++ S D +++A ++ + + T + +G
Sbjct: 1131 WDWQQGKCLKT--LTGHTNWVFDI-AFSPDGKILASASHDQTVRIWDVNTGKCHHICIGH 1187
Query: 373 LKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIH----KGVDEGCINSTALCTS 428
+ S+AFS DG+ + S D V W+++T C+ K + EG +N T +
Sbjct: 1188 THL---VSSVAFSPDGEVVASGSQDQTVRIWNVKTGECLQILRAKRLYEG-MNITGVTGL 1243
Query: 429 PSGTLF 434
T+F
Sbjct: 1244 TKATIF 1249
>GBB2_RAT (P54313) Guanine nucleotide-binding protein G(I)/G(S)/G(T)
beta subunit 2 (Transducin beta chain 2) (G protein beta
2 subunit)
Length = 340
Score = 56.2 bits (134), Expect = 2e-07
Identities = 55/236 (23%), Positives = 98/236 (41%), Gaps = 11/236 (4%)
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILS 308
+ ++ + +++LL++A D KL + D K+ +I L + ++ P G+ V
Sbjct: 58 IYAMHWGTDSRLLVSASQDGKLIIW--DSYTTNKVHAIPLRSSWVMTCAYAPSGNFVACG 115
Query: 309 GRRKF--FYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKT 366
G YS +G + L G L D+Q+I G+ L
Sbjct: 116 GLDNICSIYSLKTREGNVRVSRELPGHTGY-LSCCRFLDDNQIITSSGDTTCALWDIETG 174
Query: 367 KQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGV-DEGCINSTAL 425
+Q VG SG SL+ + DG+ +S D + WD+R C + E IN+ A
Sbjct: 175 QQTVGFAGHSGDVMSLSLAPDGRTFVSGACDASIKLWDVRDSMCRQTFIGHESDINAVAF 234
Query: 426 CTSPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQIL 481
P+G F GSD ++ D + + + +N+I + + F+ ++L
Sbjct: 235 --FPNGYAFTTGSDDATCRLF---DLRADQELLMYSHDNIICGITSVAFSRSGRLL 285
>GBB2_MOUSE (P62880) Guanine nucleotide-binding protein
G(I)/G(S)/G(T) beta subunit 2 (Transducin beta chain 2)
(G protein beta 2 subunit)
Length = 340
Score = 56.2 bits (134), Expect = 2e-07
Identities = 55/236 (23%), Positives = 98/236 (41%), Gaps = 11/236 (4%)
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILS 308
+ ++ + +++LL++A D KL + D K+ +I L + ++ P G+ V
Sbjct: 58 IYAMHWGTDSRLLVSASQDGKLIIW--DSYTTNKVHAIPLRSSWVMTCAYAPSGNFVACG 115
Query: 309 GRRKF--FYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKT 366
G YS +G + L G L D+Q+I G+ L
Sbjct: 116 GLDNICSIYSLKTREGNVRVSRELPGHTGY-LSCCRFLDDNQIITSSGDTTCALWDIETG 174
Query: 367 KQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGV-DEGCINSTAL 425
+Q VG SG SL+ + DG+ +S D + WD+R C + E IN+ A
Sbjct: 175 QQTVGFAGHSGDVMSLSLAPDGRTFVSGACDASIKLWDVRDSMCRQTFIGHESDINAVAF 234
Query: 426 CTSPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQIL 481
P+G F GSD ++ D + + + +N+I + + F+ ++L
Sbjct: 235 --FPNGYAFTTGSDDATCRLF---DLRADQELLMYSHDNIICGITSVAFSRSGRLL 285
>GBB2_HUMAN (P62879) Guanine nucleotide-binding protein
G(I)/G(S)/G(T) beta subunit 2 (Transducin beta chain 2)
(G protein beta 2 subunit)
Length = 340
Score = 56.2 bits (134), Expect = 2e-07
Identities = 55/236 (23%), Positives = 98/236 (41%), Gaps = 11/236 (4%)
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILS 308
+ ++ + +++LL++A D KL + D K+ +I L + ++ P G+ V
Sbjct: 58 IYAMHWGTDSRLLVSASQDGKLIIW--DSYTTNKVHAIPLRSSWVMTCAYAPSGNFVACG 115
Query: 309 GRRKF--FYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKT 366
G YS +G + L G L D+Q+I G+ L
Sbjct: 116 GLDNICSIYSLKTREGNVRVSRELPGHTGY-LSCCRFLDDNQIITSSGDTTCALWDIETG 174
Query: 367 KQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGV-DEGCINSTAL 425
+Q VG SG SL+ + DG+ +S D + WD+R C + E IN+ A
Sbjct: 175 QQTVGFAGHSGDVMSLSLAPDGRTFVSGACDASIKLWDVRDSMCRQTFIGHESDINAVAF 234
Query: 426 CTSPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQIL 481
P+G F GSD ++ D + + + +N+I + + F+ ++L
Sbjct: 235 --FPNGYAFTTGSDDATCRLF---DLRADQELLMYSHDNIICGITSVAFSRSGRLL 285
>GBB2_BOVIN (P11017) Guanine nucleotide-binding protein
G(I)/G(S)/G(T) beta subunit 2 (Transducin beta chain 2)
(G protein beta 2 subunit) (Fragment)
Length = 326
Score = 56.2 bits (134), Expect = 2e-07
Identities = 55/236 (23%), Positives = 98/236 (41%), Gaps = 11/236 (4%)
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILS 308
+ ++ + +++LL++A D KL + D K+ +I L + ++ P G+ V
Sbjct: 44 IYAMHWGTDSRLLVSASQDGKLIIW--DSYTTNKVHAIPLRSSWVMTCAYAPSGNFVACG 101
Query: 309 GRRKF--FYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKT 366
G YS +G + L G L D+Q+I G+ L
Sbjct: 102 GLDNICSIYSLKTREGNVRVSRELPGHTGY-LSCCRFLDDNQIITSSGDTTCALWDIETG 160
Query: 367 KQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGV-DEGCINSTAL 425
+Q VG SG SL+ + DG+ +S D + WD+R C + E IN+ A
Sbjct: 161 QQTVGFAGHSGDVMSLSLAPDGRTFVSGACDASIKLWDVRDSMCRQTFIGHESDINAVAF 220
Query: 426 CTSPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQIL 481
P+G F GSD ++ D + + + +N+I + + F+ ++L
Sbjct: 221 --FPNGYAFTTGSDDATCRLF---DLRADQELLMYSHDNIICGITSVAFSRSGRLL 271
>YL24_ANASP (Q8YV57) Hypothetical WD-repeat protein all2124
Length = 1683
Score = 54.7 bits (130), Expect = 6e-07
Identities = 60/248 (24%), Positives = 109/248 (43%), Gaps = 17/248 (6%)
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILS 308
V S+ R+ Q + + LD+ ++ + DG+ +++ + + SF PDG +
Sbjct: 1075 VISISISRDGQTIASGSLDKTIKLWSRDGR---LFRTLNGHEDAVYSVSFSPDGQTIASG 1131
Query: 309 GRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTKQ 368
G K + GTL K + G E+ V+ S D + +A ++ I L T + Q
Sbjct: 1132 GSDKTIKLWQTSDGTLLK--TITGHEQTVNNVY-FSPDGKNLASASSDHSIKLWDTTSGQ 1188
Query: 369 LVGSLK-MSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHK-GVDEGCINSTALC 426
L+ +L S ++ FS DGQ + + D V W + + + +NS L
Sbjct: 1189 LLMTLTGHSAGVITVRFSPDGQTIAAGSEDKTVKLWHRQDGKLLKTLNGHQDWVNS--LS 1246
Query: 427 TSPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQILAICSS 486
SP G A+ S + ++ D K +KT++ V + F+ D + AI S+
Sbjct: 1247 FSPDGKTLASASADKTIKLWRIAD-----GKLVKTLKGHNDSVWDVNFSSDGK--AIASA 1299
Query: 487 MKKSSLKL 494
+ +++KL
Sbjct: 1300 SRDNTIKL 1307
Score = 43.5 bits (101), Expect = 0.001
Identities = 55/240 (22%), Positives = 95/240 (38%), Gaps = 17/240 (7%)
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILS 308
V SV F + Q + + G D+ ++ +Q T +++I + + F PDG + +
Sbjct: 1116 VYSVSFSPDGQTIASGGSDKTIKLWQTSD--GTLLKTITGHEQTVNNVYFSPDGKNLASA 1173
Query: 309 GRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTKQ 368
+D G L + L G + V S D Q IA + + L + +
Sbjct: 1174 SSDHSIKLWDTTSGQL--LMTLTGHSAGVITV-RFSPDGQTIAAGSEDKTVKLWHRQDGK 1230
Query: 369 LVGSLK-MSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIH--KGVDEGCINSTAL 425
L+ +L SL+FS DG+ L SA D + W + + KG ++ + +
Sbjct: 1231 LLKTLNGHQDWVNSLSFSPDGKTLASASADKTIKLWRIADGKLVKTLKGHNDSVWD---V 1287
Query: 426 CTSPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQILAICS 485
S G A+ S + ++NR ++T V + F DS I+A S
Sbjct: 1288 NFSSDGKAIASASRDNTIKLWNRHGI------ELETFTGHSGGVYAVNFLPDSNIIASAS 1341
Score = 39.7 bits (91), Expect = 0.019
Identities = 51/250 (20%), Positives = 105/250 (41%), Gaps = 17/250 (6%)
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQI-DGKRNTKIQSIFLEDCPIRKASFLPDGSQVIL 307
VNS+ F + + L +A D+ ++ ++I DGK ++++ + + +F DG +
Sbjct: 1242 VNSLSFSPDGKTLASASADKTIKLWRIADGKL---VKTLKGHNDSVWDVNFSSDGKAIAS 1298
Query: 308 SGRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTK 367
+ R ++ LE G + DS +IA + I L
Sbjct: 1299 ASRDNTIKLWNRHGIELETFTGHSG----GVYAVNFLPDSNIIASASLDNTIRLWQRPLI 1354
Query: 368 QLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTALCT 427
+ L + +++F DG + +AG DG + W + + + I +
Sbjct: 1355 SPLEVLAGNSGVYAVSFLHDGSIIATAGADGNIQLWHSQDGSLLKTLPGNKAIYGISF-- 1412
Query: 428 SPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQILAICSSM 487
+P G L A+ + V ++ D K +KT+ +V+ + F+ D + LA S+
Sbjct: 1413 TPQGDLIASANADKTVKIWRVRD-----GKALKTLIGHDNEVNKVNFSPDGKTLA--SAS 1465
Query: 488 KKSSLKLIHI 497
+ +++KL ++
Sbjct: 1466 RDNTVKLWNV 1475
Score = 35.4 bits (80), Expect = 0.35
Identities = 39/159 (24%), Positives = 64/159 (39%), Gaps = 8/159 (5%)
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQI-DGKRNTKIQSIFLEDCPIRKASFLPDGSQVIL 307
VN V F + + L +A D ++ + + DGK ++ E + SF PDG +
Sbjct: 1449 VNKVNFSPDGKTLASASRDNTVKLWNVSDGKFKKTLKGHTDE---VFWVSFSPDGKIIAS 1505
Query: 308 SGRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTK 367
+ K +D G L K P S+ + D M+A + + L +
Sbjct: 1506 ASADKTIRLWDSFSGNLIKSLPAHNDLVYSVN---FNPDGSMLASTSADKTVKLWRSHDG 1562
Query: 368 QLVGSLK-MSGTTRSLAFSEDGQKLLSAGGDGQVYHWDL 405
L+ + S S +FS DG+ + SA D V W +
Sbjct: 1563 HLLHTFSGHSNVVYSSSFSPDGRYIASASEDKTVKIWQI 1601
Score = 34.3 bits (77), Expect = 0.78
Identities = 49/213 (23%), Positives = 87/213 (40%), Gaps = 22/213 (10%)
Query: 246 NGPVNSVQFHRNAQLLLTAGLDQKLRFFQI-DGKRNTKIQSIFLEDCPIRKA----SFLP 300
N V +V F + ++ TAG D ++ + DG L+ P KA SF P
Sbjct: 1363 NSGVYAVSFLHDGSIIATAGADGNIQLWHSQDGS--------LLKTLPGNKAIYGISFTP 1414
Query: 301 DGSQVILSGRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYIL 360
G + + K + + G K L+G + + +V S D + +A + +
Sbjct: 1415 QGDLIASANADKTVKIWRVRDGKALKT--LIGHDNEVNKV-NFSPDGKTLASASRDNTVK 1471
Query: 361 LVSTKTKQLVGSLKMSGTTRSL---AFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDE 417
L + + +LK G T + +FS DG+ + SA D + WD + I K +
Sbjct: 1472 LWNVSDGKFKKTLK--GHTDEVFWVSFSPDGKIIASASADKTIRLWDSFSGNLI-KSLPA 1528
Query: 418 GCINSTALCTSPSGTLFAAGSDSGIVNVYNRED 450
++ +P G++ A+ S V ++ D
Sbjct: 1529 HNDLVYSVNFNPDGSMLASTSADKTVKLWRSHD 1561
>HET1_PODAN (Q00808) Vegetatible incompatibility protein HET-E-1
Length = 1356
Score = 52.8 bits (125), Expect = 2e-06
Identities = 53/203 (26%), Positives = 85/203 (41%), Gaps = 9/203 (4%)
Query: 247 GPVNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVI 306
G V SV F + Q + + D+ ++ + D T Q++ ++ F PDG +V
Sbjct: 1010 GSVWSVAFSPDGQRVASGSDDKTIKIW--DTASGTCTQTLEGHGGWVQSVVFSPDGQRVA 1067
Query: 307 LSGRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKT 366
+D V GT + L G + V S D Q +A +G I + +
Sbjct: 1068 SGSDDHTIKIWDAVSGTCTQT--LEGHGDSVWSV-AFSPDGQRVASGSIDGTIKIWDAAS 1124
Query: 367 KQLVGSLK-MSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDE-GCINSTA 424
+L+ G S+AFS DGQ++ S DG + WD + TC G + S A
Sbjct: 1125 GTCTQTLEGHGGWVHSVAFSPDGQRVASGSIDGTIKIWDAASGTCTQTLEGHGGWVQSVA 1184
Query: 425 LCTSPSGTLFAAGSDSGIVNVYN 447
SP G A+GS + +++
Sbjct: 1185 F--SPDGQRVASGSSDKTIKIWD 1205
Score = 50.8 bits (120), Expect = 8e-06
Identities = 48/200 (24%), Positives = 86/200 (43%), Gaps = 7/200 (3%)
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILS 308
V SV F + Q + + D+ ++ + D T Q++ + +F PD +V
Sbjct: 844 VLSVAFSADGQRVASGSDDKTIKIW--DTASGTGTQTLEGHGGSVWSVAFSPDRERVASG 901
Query: 309 GRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTKQ 368
K +D GT + G +S+ S D Q +A ++ I + +
Sbjct: 902 SDDKTIKIWDAASGTCTQTLEGHGGRVQSVA---FSPDGQRVASGSDDHTIKIWDAASGT 958
Query: 369 LVGSLKMSGTT-RSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTALCT 427
+L+ G++ S+AFS DGQ++ S GD + WD + TC G + ++
Sbjct: 959 CTQTLEGHGSSVLSVAFSPDGQRVASGSGDKTIKIWDTASGTCTQTLEGHGG-SVWSVAF 1017
Query: 428 SPSGTLFAAGSDSGIVNVYN 447
SP G A+GSD + +++
Sbjct: 1018 SPDGQRVASGSDDKTIKIWD 1037
Score = 45.4 bits (106), Expect = 3e-04
Identities = 44/179 (24%), Positives = 74/179 (40%), Gaps = 10/179 (5%)
Query: 251 SVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILSGR 310
SV F + Q + + +D ++ + D T Q++ + +F PDG +V
Sbjct: 1098 SVAFSPDGQRVASGSIDGTIKIW--DAASGTCTQTLEGHGGWVHSVAFSPDGQRVASGSI 1155
Query: 311 RKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTKQLV 370
+D GT + G +S+ S D Q +A ++ I + T +
Sbjct: 1156 DGTIKIWDAASGTCTQTLEGHGGWVQSVA---FSPDGQRVASGSSDKTIKIWDTASGTCT 1212
Query: 371 GSLK-MSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTALCTS 428
+L+ G +S+AFS DGQ++ S D + WD + TC + STA C S
Sbjct: 1213 QTLEGHGGWVQSVAFSPDGQRVASGSSDNTIKIWDTASGTC----TQTLNVGSTATCLS 1267
>TUP1_CANAL (P56093) Transcriptional repressor TUP1
Length = 514
Score = 51.2 bits (121), Expect = 6e-06
Identities = 35/106 (33%), Positives = 53/106 (49%), Gaps = 5/106 (4%)
Query: 344 SSDSQMIAFVGNEGYILLVSTKTKQLVGSLK-MSGTTRSLAFSEDGQKLLSAGGDGQVYH 402
S D +++A + I + TK+++ L+ SL F DG +L+S GD V
Sbjct: 266 SPDGKLLATGAEDKLIRIWDLSTKRIIKILRGHEQDIYSLDFFPDGDRLVSGSGDRSVRI 325
Query: 403 WDLRTRTC-IHKGVDEGCINSTALCTSPSGTLFAAGSDSGIVNVYN 447
WDLRT C + +++G T + SP G L AAGS V V++
Sbjct: 326 WDLRTSQCSLTLSIEDGV---TTVAVSPDGKLIAAGSLDRTVRVWD 368
Score = 37.7 bits (86), Expect = 0.071
Identities = 47/203 (23%), Positives = 83/203 (40%), Gaps = 24/203 (11%)
Query: 290 DCPIRKASFLPDGSQVILSGRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQM 349
D IR F PDG + K +DL + ++I ++ E+ + + D
Sbjct: 257 DLYIRSVCFSPDGKLLATGAEDKLIRIWDL---STKRIIKILRGHEQDIYSLDFFPDGDR 313
Query: 350 IAFVGNEGYILLVSTKTKQLVGSLKMSGTTRSLAFSEDGQKLLSAGG-DGQVYHWDLRTR 408
+ + + + +T Q +L + ++A S DG KL++AG D V WD T
Sbjct: 314 LVSGSGDRSVRIWDLRTSQCSLTLSIEDGVTTVAVSPDG-KLIAAGSLDRTVRVWDSTTG 372
Query: 409 TCIHKGVDEGCINST-------ALCTSPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKT 461
+ + +D G N ++ S +G A+GS V +++ E GK T
Sbjct: 373 FLVER-LDSGNENGNGHEDSVYSVAFSNNGEQIASGSLDRTVKLWHLE----GKSDKKST 427
Query: 462 IENLITKVDFMRFNHDSQILAIC 484
E +T + H +L++C
Sbjct: 428 CE--VTYI-----GHKDFVLSVC 443
>GBB1_XENLA (P79959) Guanine nucleotide-binding protein
G(I)/G(S)/G(T) beta subunit 1 (Transducin beta chain 1)
(XGbeta1)
Length = 340
Score = 51.2 bits (121), Expect = 6e-06
Identities = 52/236 (22%), Positives = 95/236 (40%), Gaps = 11/236 (4%)
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILS 308
+ ++ + +++LL++A D KL + D K+ +I L + ++ P G+ V
Sbjct: 58 IYAMHWGTDSRLLVSASQDGKLIIW--DSYTTNKVHAIPLRSSWVMTCAYAPSGNYVACG 115
Query: 309 GRRKF--FYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKT 366
G Y+ +G + L G L D+Q+I G+ L
Sbjct: 116 GLDNICPIYNLKTREGNVRVSRELAGHTGY-LSCCRFLDDNQIITSSGDTTCALWDIETG 174
Query: 367 KQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVD-EGCINSTAL 425
+Q +G SL+ + D + +S D WD+R C E IN A+
Sbjct: 175 QQTTTFTGHTGDVMSLSLAPDSRCFVSGACDASAKLWDVREGMCRQTFTGHESDIN--AI 232
Query: 426 CTSPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQIL 481
C P+G FA GSD ++ D + + + +N+I + + F+ ++L
Sbjct: 233 CFFPNGNAFATGSDDATCRLF---DLRADQELMVYSHDNIICGITSVAFSKSGRLL 285
>YY46_ANASP (Q8YRI1) Hypothetical WD-repeat protein alr3466
Length = 1526
Score = 50.8 bits (120), Expect = 8e-06
Identities = 47/201 (23%), Positives = 83/201 (40%), Gaps = 51/201 (25%)
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCP-IRKASFLPDGSQVIL 307
VNSV F+ + L + DQ +R ++I+ ++K F + F PDGS +
Sbjct: 1203 VNSVVFNPDGSTLASGSSDQTVRLWEIN---SSKCLCTFQGHTSWVNSVVFNPDGSMLAS 1259
Query: 308 SGRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTK 367
G +K++ ++++SS + F G+ ++
Sbjct: 1260 ------------------------GSSDKTVRLWDISSSKCLHTFQGHTNWV-------- 1287
Query: 368 QLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTALCT 427
S+AF+ DG L S GD V W++ + C+H +G + + T
Sbjct: 1288 ------------NSVAFNPDGSMLASGSGDQTVRLWEISSSKCLH--TFQGHTSWVSSVT 1333
Query: 428 -SPSGTLFAAGSDSGIVNVYN 447
SP GT+ A+GSD V +++
Sbjct: 1334 FSPDGTMLASGSDDQTVRLWS 1354
Score = 50.4 bits (119), Expect = 1e-05
Identities = 52/235 (22%), Positives = 95/235 (40%), Gaps = 54/235 (22%)
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILS 308
VNSV F+ + +L + DQ +R ++I + + + + +F PDG+ +
Sbjct: 1287 VNSVAFNPDGSMLASGSGDQTVRLWEISSSKC--LHTFQGHTSWVSSVTFSPDGTMLAS- 1343
Query: 309 GRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTKQ 368
G +++++ ++ +SS + F+G+ ++
Sbjct: 1344 -----------------------GSDDQTVRLWSISSGECLYTFLGHTNWV--------- 1371
Query: 369 LVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINST-ALCT 427
S+ FS DG L S GD V W + + C++ +G N ++
Sbjct: 1372 -----------GSVIFSPDGAILASGSGDQTVRLWSISSGKCLY--TLQGHNNWVGSIVF 1418
Query: 428 SPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQILA 482
SP GTL A+GSD V ++N + + T+ I V + F+ D ILA
Sbjct: 1419 SPDGTLLASGSDDQTVRLWNI-----SSGECLYTLHGHINSVRSVAFSSDGLILA 1468
Score = 49.7 bits (117), Expect = 2e-05
Identities = 61/257 (23%), Positives = 108/257 (41%), Gaps = 22/257 (8%)
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILS 308
V SV F + +L + DQ +R + I + ++ +R F PDG+ +
Sbjct: 1035 VRSVVFSSDGAMLASGSDDQTVRLWDISS--GNCLYTLQGHTSCVRSVVFSPDGAMLASG 1092
Query: 309 GRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTKQ 368
G + +D+ G + L G + S + +A ++ + L +K+
Sbjct: 1093 GDDQIVRLWDISSGNC--LYTLQGYTSW-VRFLVFSPNGVTLANGSSDQIVRLWDISSKK 1149
Query: 369 LVGSLK-MSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIH--KGVDEGCINSTAL 425
+ +L+ + ++AFS DG L S GD V WD+ + C++ +G +NS
Sbjct: 1150 CLYTLQGHTNWVNAVAFSPDGATLASGSGDQTVRLWDISSSKCLYILQG-HTSWVNSVVF 1208
Query: 426 CTSPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQILAICS 485
+P G+ A+GS V ++ K + T + + V+ + FN D +LA S
Sbjct: 1209 --NPDGSTLASGSSDQTVRLWEI-----NSSKCLCTFQGHTSWVNSVVFNPDGSMLASGS 1261
Query: 486 SMKK------SSLKLIH 496
S K SS K +H
Sbjct: 1262 SDKTVRLWDISSSKCLH 1278
Score = 43.1 bits (100), Expect = 0.002
Identities = 46/204 (22%), Positives = 83/204 (40%), Gaps = 51/204 (25%)
Query: 246 NGPVNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQV 305
N VNSV F ++ ++L + DQ +R + I + K + +R F P+ S +
Sbjct: 906 NSWVNSVGFSQDGKMLASGSDDQTVRLWDISSGQCLK--TFKGHTSRVRSVVFSPN-SLM 962
Query: 306 ILSGRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTK 365
+ SG ++++ ++++SS + F G+ G++
Sbjct: 963 LASGS-----------------------SDQTVRLWDISSGECLYIFQGHTGWVY----- 994
Query: 366 TKQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTC--IHKGVDEGCINST 423
S+AF+ DG L + GD V WD+ + C I +G C+ S
Sbjct: 995 ---------------SVAFNLDGSMLATGSGDQTVRLWDISSSQCFYIFQG-HTSCVRSV 1038
Query: 424 ALCTSPSGTLFAAGSDSGIVNVYN 447
S G + A+GSD V +++
Sbjct: 1039 VF--SSDGAMLASGSDDQTVRLWD 1060
Score = 35.8 bits (81), Expect = 0.27
Identities = 26/105 (24%), Positives = 47/105 (44%), Gaps = 2/105 (1%)
Query: 344 SSDSQMIAFVGNEGYILLVSTKT-KQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYH 402
S D ++ A + G + T K+L+ + S+ FS+DG+ L S D V
Sbjct: 873 SPDGKLFATGDSGGIVRFWEAATGKELLTCKGHNSWVNSVGFSQDGKMLASGSDDQTVRL 932
Query: 403 WDLRTRTCIHKGVDEGCINSTALCTSPSGTLFAAGSDSGIVNVYN 447
WD+ + C+ K ++ SP+ + A+GS V +++
Sbjct: 933 WDISSGQCL-KTFKGHTSRVRSVVFSPNSLMLASGSSDQTVRLWD 976
Score = 35.0 bits (79), Expect = 0.46
Identities = 35/117 (29%), Positives = 56/117 (46%), Gaps = 14/117 (11%)
Query: 374 KMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRT----RTCIHKGVDEGCINSTALCTSP 429
K+ G+ ++AFS DG+ + G V W+ T TC KG +NS S
Sbjct: 862 KILGSVLTVAFSPDGKLFATGDSGGIVRFWEAATGKELLTC--KG-HNSWVNSVGF--SQ 916
Query: 430 SGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQILAICSS 486
G + A+GSD V ++ D G + +KT + ++V + F+ +S +LA SS
Sbjct: 917 DGKMLASGSDDQTVRLW---DISSG--QCLKTFKGHTSRVRSVVFSPNSLMLASGSS 968
>GBB1_BRARE (Q6PH57) Guanine nucleotide-binding protein
G(I)/G(S)/G(T) beta subunit 1 (Transducin beta chain 1)
Length = 340
Score = 50.4 bits (119), Expect = 1e-05
Identities = 51/236 (21%), Positives = 95/236 (39%), Gaps = 11/236 (4%)
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILS 308
+ ++ + +++LL++A D KL + D K+ +I L + ++ P G+ V
Sbjct: 58 IYAMHWGTDSRLLVSASQDGKLIIW--DSYTTNKVHAIPLRSSWVMTCAYAPSGNYVACG 115
Query: 309 GRRKF--FYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKT 366
G Y+ +G + L G L D+Q++ G+ L
Sbjct: 116 GLDNICSIYNLKTREGNVRVSRELAGHTGY-LSCCRFLDDNQIVTSSGDTTCALWDIETG 174
Query: 367 KQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVD-EGCINSTAL 425
+Q +G SL+ + D + +S D WD+R C E IN A+
Sbjct: 175 QQTTTFAGHTGDVMSLSLAPDTRLFVSGACDASAKLWDVREGMCRQTFTGHESDIN--AI 232
Query: 426 CTSPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQIL 481
C P+G FA GSD ++ D + + + +N+I + + F+ ++L
Sbjct: 233 CFFPNGNAFATGSDDATCRLF---DLRADQELMVYSHDNIICGITSVAFSKSGRLL 285
>GBB1_RAT (P54311) Guanine nucleotide-binding protein G(I)/G(S)/G(T)
beta subunit 1 (Transducin beta chain 1)
Length = 340
Score = 49.3 bits (116), Expect = 2e-05
Identities = 57/251 (22%), Positives = 97/251 (37%), Gaps = 26/251 (10%)
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILS 308
+ ++ + +++LL++A D KL + D K+ +I L + ++ P G+ V
Sbjct: 58 IYAMHWGTDSRLLVSASQDGKLIIW--DSYTTNKVHAIPLRSSWVMTCAYAPSGNYVACG 115
Query: 309 GRRKF--FYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKT 366
G Y+ +G + L G L D+Q++ G+ L
Sbjct: 116 GLDNICSIYNLKTREGNVRVSRELAGHTGY-LSCCRFLDDNQIVTSSGDTTCALWDIETG 174
Query: 367 KQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVD-EGCINSTAL 425
+Q +G SL+ + D + +S D WD+R C E IN A+
Sbjct: 175 QQTTTFTGHTGDVMSLSLAPDTRLFVSGACDASAKLWDVREGMCRQTFTGHESDIN--AI 232
Query: 426 CTSPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQILAICS 485
C P+G FA GSD ++ +L + M ++HD+ I I S
Sbjct: 233 CFFPNGNAFATGSDDATCRLF-----------------DLRADQELMTYSHDNIICGITS 275
Query: 486 -SMKKSSLKLI 495
S KS L+
Sbjct: 276 VSFSKSGRLLL 286
>GBB1_MOUSE (P62874) Guanine nucleotide-binding protein
G(I)/G(S)/G(T) beta subunit 1 (Transducin beta chain 1)
Length = 340
Score = 49.3 bits (116), Expect = 2e-05
Identities = 57/251 (22%), Positives = 97/251 (37%), Gaps = 26/251 (10%)
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILS 308
+ ++ + +++LL++A D KL + D K+ +I L + ++ P G+ V
Sbjct: 58 IYAMHWGTDSRLLVSASQDGKLIIW--DSYTTNKVHAIPLRSSWVMTCAYAPSGNYVACG 115
Query: 309 GRRKF--FYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKT 366
G Y+ +G + L G L D+Q++ G+ L
Sbjct: 116 GLDNICSIYNLKTREGNVRVSRELAGHTGY-LSCCRFLDDNQIVTSSGDTTCALWDIETG 174
Query: 367 KQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVD-EGCINSTAL 425
+Q +G SL+ + D + +S D WD+R C E IN A+
Sbjct: 175 QQTTTFTGHTGDVMSLSLAPDTRLFVSGACDASAKLWDVREGMCRQTFTGHESDIN--AI 232
Query: 426 CTSPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQILAICS 485
C P+G FA GSD ++ +L + M ++HD+ I I S
Sbjct: 233 CFFPNGNAFATGSDDATCRLF-----------------DLRADQELMTYSHDNIICGITS 275
Query: 486 -SMKKSSLKLI 495
S KS L+
Sbjct: 276 VSFSKSGRLLL 286
>GBB1_HUMAN (P62873) Guanine nucleotide-binding protein
G(I)/G(S)/G(T) beta subunit 1 (Transducin beta chain 1)
Length = 340
Score = 49.3 bits (116), Expect = 2e-05
Identities = 57/251 (22%), Positives = 97/251 (37%), Gaps = 26/251 (10%)
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILS 308
+ ++ + +++LL++A D KL + D K+ +I L + ++ P G+ V
Sbjct: 58 IYAMHWGTDSRLLVSASQDGKLIIW--DSYTTNKVHAIPLRSSWVMTCAYAPSGNYVACG 115
Query: 309 GRRKF--FYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKT 366
G Y+ +G + L G L D+Q++ G+ L
Sbjct: 116 GLDNICSIYNLKTREGNVRVSRELAGHTGY-LSCCRFLDDNQIVTSSGDTTCALWDIETG 174
Query: 367 KQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVD-EGCINSTAL 425
+Q +G SL+ + D + +S D WD+R C E IN A+
Sbjct: 175 QQTTTFTGHTGDVMSLSLAPDTRLFVSGACDASAKLWDVREGMCRQTFTGHESDIN--AI 232
Query: 426 CTSPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQILAICS 485
C P+G FA GSD ++ +L + M ++HD+ I I S
Sbjct: 233 CFFPNGNAFATGSDDATCRLF-----------------DLRADQELMTYSHDNIICGITS 275
Query: 486 -SMKKSSLKLI 495
S KS L+
Sbjct: 276 VSFSKSGRLLL 286
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.134 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 59,083,333
Number of Sequences: 164201
Number of extensions: 2658723
Number of successful extensions: 8255
Number of sequences better than 10.0: 282
Number of HSP's better than 10.0 without gapping: 57
Number of HSP's successfully gapped in prelim test: 226
Number of HSP's that attempted gapping in prelim test: 7595
Number of HSP's gapped (non-prelim): 693
length of query: 509
length of database: 59,974,054
effective HSP length: 115
effective length of query: 394
effective length of database: 41,090,939
effective search space: 16189829966
effective search space used: 16189829966
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0055.8


