

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0052b.4
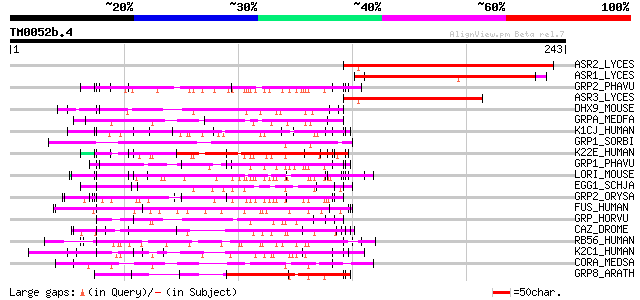
(243 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ASR2_LYCES (P37219) Abscisic stress ripening protein 2 139 6e-33
ASR1_LYCES (Q08655) Abscisic stress ripening protein 1 136 5e-32
GRP2_PHAVU (P10496) Glycine-rich cell wall structural protein 1.... 87 3e-17
ASR3_LYCES (P37220) Abscisic stress ripening protein 3 87 3e-17
DHX9_MOUSE (O70133) ATP-dependent RNA helicase A (Nuclear DNA he... 84 3e-16
GRPA_MEDFA (Q09134) Abscisic acid and environmental stress induc... 82 2e-15
K1CJ_HUMAN (P13645) Keratin, type I cytoskeletal 10 (Cytokeratin... 80 3e-15
GRP1_SORBI (Q99069) Glycine-rich RNA-binding protein 1 (Fragment) 78 2e-14
K22E_HUMAN (P35908) Keratin, type II cytoskeletal 2 epidermal (C... 77 4e-14
GRP1_PHAVU (P10495) Glycine-rich cell wall structural protein 1.... 77 5e-14
LORI_MOUSE (P18165) Loricrin 76 8e-14
EGG1_SCHJA (P19470) Eggshell protein 1 precursor 76 8e-14
GRP2_ORYSA (P29834) Glycine-rich cell wall structural protein 2 ... 75 1e-13
FUS_HUMAN (P35637) RNA-binding protein FUS (Oncogene FUS) (Oncog... 75 1e-13
GRP_HORVU (P17816) Glycine-rich cell wall structural protein pre... 74 2e-13
CAZ_DROME (Q27294) RNA-binding protein cabeza (Sarcoma-associate... 74 3e-13
RB56_HUMAN (Q92804) TATA-binding protein associated factor 2N (R... 74 4e-13
K2C1_HUMAN (P04264) Keratin, type II cytoskeletal 1 (Cytokeratin... 73 5e-13
CORA_MEDSA (Q07202) Cold and drought-regulated protein CORA 73 5e-13
GRP8_ARATH (Q03251) Glycine-rich RNA-binding protein 8 (CCR1 pro... 73 7e-13
>ASR2_LYCES (P37219) Abscisic stress ripening protein 2
Length = 114
Score = 139 bits (350), Expect = 6e-33
Identities = 63/95 (66%), Positives = 78/95 (81%), Gaps = 3/95 (3%)
Query: 147 NRRDE---VDYKEEEKHHKKLEHLGELGTTAAGAYALHEKHKSEKDPENAHRHKLEEEVA 203
N+ DE VDY++E KHH LE +GELG AAGA ALHEKHK++KDPE+AH+HK+EEE+
Sbjct: 17 NKEDEGGPVDYEKEVKHHSHLEKIGELGAVAAGALALHEKHKAKKDPEHAHKHKIEEEIM 76
Query: 204 AAAAVGAGGFAFHEHHEKKEAKEEEEESHGKKHHH 238
A AAVGAGGFAFHEHH+KK+AK+E++E G HHH
Sbjct: 77 AVAAVGAGGFAFHEHHQKKDAKKEKKEVEGGHHHH 111
>ASR1_LYCES (Q08655) Abscisic stress ripening protein 1
Length = 115
Score = 136 bits (342), Expect = 5e-32
Identities = 60/79 (75%), Positives = 75/79 (93%)
Query: 152 VDYKEEEKHHKKLEHLGELGTTAAGAYALHEKHKSEKDPENAHRHKLEEEVAAAAAVGAG 211
VDY++E KHHK LE +G+LGT AAGAYALHEKH+++KDPE+AH+HK+EEE+AAAAAVGAG
Sbjct: 23 VDYEKEIKHHKHLEQIGKLGTVAAGAYALHEKHEAKKDPEHAHKHKIEEEIAAAAAVGAG 82
Query: 212 GFAFHEHHEKKEAKEEEEE 230
GFAFHEHHEKK+AK+EE++
Sbjct: 83 GFAFHEHHEKKDAKKEEKK 101
Score = 42.7 bits (99), Expect = 8e-04
Identities = 29/84 (34%), Positives = 39/84 (45%), Gaps = 21/84 (25%)
Query: 156 EEEKHHKKLEHLGELGTTAAGAYALHEKHKSEKDPENAHR----HKLEEEVAAAAAVGAG 211
EEEKHH HL H K K+E+ P + + HK E++ V AG
Sbjct: 2 EEEKHHH--HHL------------FHHKDKAEEGPVDYEKEIKHHKHLEQIGKLGTVAAG 47
Query: 212 GFAFHEHHEKKEAKEEEEESHGKK 235
+A HE HE AK++ E +H K
Sbjct: 48 AYALHEKHE---AKKDPEHAHKHK 68
>GRP2_PHAVU (P10496) Glycine-rich cell wall structural protein 1.8
precursor (GRP 1.8)
Length = 465
Score = 87.4 bits (215), Expect = 3e-17
Identities = 51/113 (45%), Positives = 59/113 (52%), Gaps = 7/113 (6%)
Query: 38 SGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSG--YNKTAYSTDEPS 95
+GG++ GY +GGGY TG Y + GGGGY +G + Y E
Sbjct: 355 TGGEHGGGYGGGQGGGAGGGYGTGGEHGGGYGGGQ--GGGGGYGAGGDHGAAGYGGGEGG 412
Query: 96 GGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGG-GGGYSTGGGGYGNDDSGN 147
GGG G GG G G + GGGYG G GGGG Y GG GGGY GGGG G GN
Sbjct: 413 GGGSG-GGYGDGGAHGGGYGGGAGGGGGYGAGGAHGGGYG-GGGGIGGGHGGN 463
Score = 84.3 bits (207), Expect = 2e-16
Identities = 49/117 (41%), Positives = 56/117 (46%), Gaps = 15/117 (12%)
Query: 40 GDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSG-YNKTAYSTDEPSGGG 98
G++ GY GGGY TG Y + GGGY +G + Y + GGG
Sbjct: 335 GEHGGGYGGGQGGGDGGGYGTGGEHGGGYGGGQGGGAGGGYGTGGEHGGGYGGGQGGGGG 394
Query: 99 YGTGG----------AGGGYSTGGGYGTGGGGGGEYSTG-GGGGGYSTG---GGGYG 141
YG GG GGG +GGGYG GG GG Y G GGGGGY G GGGYG
Sbjct: 395 YGAGGDHGAAGYGGGEGGGGGSGGGYGDGGAHGGGYGGGAGGGGGYGAGGAHGGGYG 451
Score = 79.0 bits (193), Expect = 1e-14
Identities = 50/118 (42%), Positives = 53/118 (44%), Gaps = 10/118 (8%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGG-GGYDSGYNKTAYSTDEPSGG 97
GG GY GGG G Y+ + GG GG G + Y T GG
Sbjct: 302 GGGAGGGYGAGGEHGGGGGGGQGGGAGGGYAAVGEHGGGYGGGQGGGDGGGYGTGGEHGG 361
Query: 98 GYGT---GGAGGGYST----GGGYGTGGGGGGEYSTGG--GGGGYSTGGGGYGNDDSG 146
GYG GGAGGGY T GGGYG G GGGG Y GG G GY G GG G G
Sbjct: 362 GYGGGQGGGAGGGYGTGGEHGGGYGGGQGGGGGYGAGGDHGAAGYGGGEGGGGGSGGG 419
Score = 77.8 bits (190), Expect = 2e-14
Identities = 50/113 (44%), Positives = 57/113 (50%), Gaps = 11/113 (9%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYN-KTSSYSTDEPNSGGGGYDSG-YNKTAYSTDEPSG 96
GGD +GY GGG G + Y + GGGY +G + Y SG
Sbjct: 89 GGDQGAGYGGGGGSGGGGGVAYGGGGERGGYGGGQGGGAGGGYGAGGEHGIGYGGGGGSG 148
Query: 97 ----GGYGTGGA-GGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTG---GGGYG 141
GGY GGA GGGY TGGG G GGGGGG++ GG GGG G GGGYG
Sbjct: 149 AGGGGGYNAGGAQGGGYGTGGGAGGGGGGGGDHG-GGYGGGQGAGGGAGGGYG 200
Score = 77.4 bits (189), Expect = 3e-14
Identities = 50/115 (43%), Positives = 55/115 (47%), Gaps = 18/115 (15%)
Query: 38 SGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGG 97
+GG++ G +GGGY G E GGGG G Y+ GG
Sbjct: 289 AGGEHGGGAGGGQGGGAGGGYGAG---------GEHGGGGGGGQGGGAGGGYAAVGEHGG 339
Query: 98 GYGTGGAGGGYSTGGGYGTGGGGGGEY---STGGGGGGYSTG---GGGYGNDDSG 146
GYG GG GGG GGGYGTGG GG Y GG GGGY TG GGGYG G
Sbjct: 340 GYG-GGQGGG--DGGGYGTGGEHGGGYGGGQGGGAGGGYGTGGEHGGGYGGGQGG 391
Score = 75.1 bits (183), Expect = 1e-13
Identities = 46/118 (38%), Positives = 49/118 (40%), Gaps = 22/118 (18%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG GY +GGGY G Y GGGG Y+ GGG
Sbjct: 112 GGGERGGYGGGQGGGAGGGYGAGGEHGIGY-------GGGGGSGAGGGGGYNAGGAQGGG 164
Query: 99 YGTGGA---------------GGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYG 141
YGTGG GGG GGG G G GGGGE+ GGGGG GGGYG
Sbjct: 165 YGTGGGAGGGGGGGGDHGGGYGGGQGAGGGAGGGYGGGGEHGGGGGGGQGGGAGGGYG 222
Score = 75.1 bits (183), Expect = 1e-13
Identities = 47/117 (40%), Positives = 56/117 (47%), Gaps = 13/117 (11%)
Query: 45 GYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGG--YDSGYNKTAYSTDEP--SGGGYG 100
GY S GGG G ++ + Y + GGGG Y G + Y + +GGGYG
Sbjct: 73 GYGGGSGGGQGGGVGYGGDQGAGYGGGGGSGGGGGVAYGGGGERGGYGGGQGGGAGGGYG 132
Query: 101 TGGAGG-GYSTGGGYGTGGGGG--------GEYSTGGGGGGYSTGGGGYGNDDSGNR 148
GG G GY GGG G GGGGG G Y TGGG GG GGG +G G +
Sbjct: 133 AGGEHGIGYGGGGGSGAGGGGGYNAGGAQGGGYGTGGGAGGGGGGGGDHGGGYGGGQ 189
Score = 72.4 bits (176), Expect = 9e-13
Identities = 44/125 (35%), Positives = 53/125 (42%), Gaps = 16/125 (12%)
Query: 38 SGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGG 97
+GG++ G +GGGY G + GGGY +G + GG
Sbjct: 245 AGGEHGGGAGGGQGGGAGGGYGAGGEHGGGAGGGQGGGAGGGYGAGGEHGGGAGGGQGGG 304
Query: 98 GYGTGGAGGGYSTGGGYGTGGGGGGEYST-------------GGGGGGYSTG---GGGYG 141
G GAGG + GGG G GGG GG Y+ GG GGGY TG GGGYG
Sbjct: 305 AGGGYGAGGEHGGGGGGGQGGGAGGGYAAVGEHGGGYGGGQGGGDGGGYGTGGEHGGGYG 364
Query: 142 NDDSG 146
G
Sbjct: 365 GGQGG 369
Score = 71.2 bits (173), Expect = 2e-12
Identities = 47/114 (41%), Positives = 51/114 (44%), Gaps = 16/114 (14%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GGD+ GY + +GGG GY E GGGG G Y GGG
Sbjct: 178 GGDHGGGY--GGGQGAGGGAGGGYG-----GGGEHGGGGGGGQGGGAGGGYGAGGEHGGG 230
Query: 99 YGTG---GAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTG---GGGYGNDDSG 146
G G GAGGGY GG +G G GGG GG GGGY G GGG G G
Sbjct: 231 AGGGQGGGAGGGYGAGGEHGGGAGGG---QGGGAGGGYGAGGEHGGGAGGGQGG 281
Score = 70.9 bits (172), Expect = 3e-12
Identities = 49/117 (41%), Positives = 54/117 (45%), Gaps = 20/117 (17%)
Query: 38 SGGDYDSGYKKS--SYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPS 95
+GG++ GY S GGGY G + Y T GGGG +
Sbjct: 133 AGGEHGIGYGGGGGSGAGGGGGYNAGGAQGGGYGTGGGAGGGGGGGGDH----------- 181
Query: 96 GGGYGTGGAGGGYSTGGGYGTGG--GGGGEYSTGGG-GGGYSTG---GGGYGNDDSG 146
GGGYG GG G G GGGYG GG GGGG GGG GGGY G GGG G G
Sbjct: 182 GGGYG-GGQGAGGGAGGGYGGGGEHGGGGGGGQGGGAGGGYGAGGEHGGGAGGGQGG 237
Score = 69.7 bits (169), Expect = 6e-12
Identities = 46/122 (37%), Positives = 50/122 (40%), Gaps = 10/122 (8%)
Query: 32 YSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGG-GGYDSGYNKTAYS 90
Y + +GG GY GGG G Y + GG GG G Y
Sbjct: 185 YGGGQGAGGGAGGGYGGGGEHGGGGGGGQGGGAGGGYGAGGEHGGGAGGGQGGGAGGGYG 244
Query: 91 TDEPSGGGYGTG---GAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTG---GGGYGNDD 144
GGG G G GAGGGY GG +G G GGG GG GGGY G GGG G
Sbjct: 245 AGGEHGGGAGGGQGGGAGGGYGAGGEHGGGAGGG---QGGGAGGGYGAGGEHGGGAGGGQ 301
Query: 145 SG 146
G
Sbjct: 302 GG 303
Score = 68.2 bits (165), Expect = 2e-11
Identities = 44/113 (38%), Positives = 51/113 (44%), Gaps = 15/113 (13%)
Query: 38 SGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGG 97
+GG++ G +GGGY G + GGGY +G +GG
Sbjct: 223 AGGEHGGGAGGGQGGGAGGGYGAGGEHGGGAGGGQGGGAGGGYGAGGEHGG-----GAGG 277
Query: 98 GYGTGGAGGGYSTGG------GYGTGGGGGGEYSTG---GGGGGYSTGGGGYG 141
G G GGAGGGY GG G G GGG GG Y G GGGGG GGG G
Sbjct: 278 GQG-GGAGGGYGAGGEHGGGAGGGQGGGAGGGYGAGGEHGGGGGGGQGGGAGG 329
Score = 62.0 bits (149), Expect = 1e-09
Identities = 47/128 (36%), Positives = 53/128 (40%), Gaps = 15/128 (11%)
Query: 32 YSTDEPSGGDYDSGYKKSSYE------TSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYN 85
Y +GG Y G SY + GGG G + Y GGG G +
Sbjct: 34 YGLGHGTGGGY--GGAAGSYGGGGGGGSGGGGGYAGEHGVVGYGGGSGGGQGGGVGYGGD 91
Query: 86 KTA-YSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGG------GGGGYSTGGG 138
+ A Y SGGG G GGG G G G GGG GG Y GG GGGG S GG
Sbjct: 92 QGAGYGGGGGSGGGGGVAYGGGGERGGYGGGQGGGAGGGYGAGGEHGIGYGGGGGSGAGG 151
Query: 139 GYGNDDSG 146
G G + G
Sbjct: 152 GGGYNAGG 159
Score = 56.6 bits (135), Expect = 5e-08
Identities = 36/96 (37%), Positives = 42/96 (43%), Gaps = 8/96 (8%)
Query: 53 TSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGG 112
T GY G+ Y + GGGG Y+ G +G G GGG +GG
Sbjct: 29 TLDAGYGLGHGTGGGYGGAAGSYGGGGGGGSGGGGGYA------GEHGVVGYGGG--SGG 80
Query: 113 GYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGNR 148
G G G G GG+ G GGGG S GGGG G R
Sbjct: 81 GQGGGVGYGGDQGAGYGGGGGSGGGGGVAYGGGGER 116
Score = 49.7 bits (117), Expect = 6e-06
Identities = 29/58 (50%), Positives = 31/58 (53%), Gaps = 3/58 (5%)
Query: 98 GYGTG-GAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGNRRDEVDY 154
GYG G G GGGY GG G+ GGGGG S GGGG G GYG G + V Y
Sbjct: 33 GYGLGHGTGGGY--GGAAGSYGGGGGGGSGGGGGYAGEHGVVGYGGGSGGGQGGGVGY 88
>ASR3_LYCES (P37220) Abscisic stress ripening protein 3
Length = 78
Score = 87.4 bits (215), Expect = 3e-17
Identities = 39/64 (60%), Positives = 52/64 (80%), Gaps = 3/64 (4%)
Query: 147 NRRDE---VDYKEEEKHHKKLEHLGELGTTAAGAYALHEKHKSEKDPENAHRHKLEEEVA 203
NR +E VD+K++ KHH L+ +GELG AAG YALHEKHK++KDPENAH+HK+++E+A
Sbjct: 15 NREEEGGPVDHKKKVKHHSHLQKIGELGAVAAGRYALHEKHKAKKDPENAHKHKIKQEIA 74
Query: 204 AAAA 207
A AA
Sbjct: 75 AVAA 78
>DHX9_MOUSE (O70133) ATP-dependent RNA helicase A (Nuclear DNA
helicase II) (NDH II) (DEAH-box protein 9) (mHEL-5)
Length = 1380
Score = 84.0 bits (206), Expect = 3e-16
Identities = 56/132 (42%), Positives = 65/132 (48%), Gaps = 23/132 (17%)
Query: 22 DTETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYD 81
D +GY + Y GG Y GY + GGG+ +G + NSGGGG+
Sbjct: 1169 DNGSGYRR-GYGGGGYGGGGYGGGYGSGGF---GGGFGSGGGFGGGF-----NSGGGGFG 1219
Query: 82 SGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYG----TGGGGGGEYSTGG---GGGGYS 134
SG SGGG G GG GGG+S GGG G GGGGGG +GG GGGGY
Sbjct: 1220 SGGGGFG------SGGG-GFGGGGGGFSGGGGGGFGGGRGGGGGGFGGSGGFGNGGGGYG 1272
Query: 135 TGGGGYGNDDSG 146
GGGGYG G
Sbjct: 1273 VGGGGYGGGGGG 1284
Score = 75.1 bits (183), Expect = 1e-13
Identities = 50/115 (43%), Positives = 59/115 (50%), Gaps = 17/115 (14%)
Query: 40 GDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGY 99
GD K + Y+ +G GY GY GGGGY GY + SGGG+
Sbjct: 1157 GDGPRPPKMARYD-NGSGYRRGYGGGGY--------GGGGYGGGYGSGGFGGGFGSGGGF 1207
Query: 100 GTG--GAGGGY-STGGGYGTG----GGGGGEYSTGGGGG-GYSTGGGGYGNDDSG 146
G G GGG+ S GGG+G+G GGGGG +S GGGGG G GGGG G SG
Sbjct: 1208 GGGFNSGGGGFGSGGGGFGSGGGGFGGGGGGFSGGGGGGFGGGRGGGGGGFGGSG 1262
Score = 72.8 bits (177), Expect = 7e-13
Identities = 46/121 (38%), Positives = 57/121 (47%), Gaps = 11/121 (9%)
Query: 26 GYNKTSYSTDEPSGGDYDSGYKKSS--YETSGGGYETGYNKTSSYSTDEPNSGGGGYDSG 83
GY + SGG + G+ + + GGG+ +G GGGG+ G
Sbjct: 1191 GYGSGGFGGGFGSGGGFGGGFNSGGGGFGSGGGGFGSGGGGFGGGGGGFSGGGGGGFGGG 1250
Query: 84 YNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGND 143
+ GG+G G GGGY GGG G GGGGGG Y GGG GGY GGGGYG
Sbjct: 1251 --RGGGGGGFGGSGGFGNG--GGGYGVGGG-GYGGGGGGGY--GGGSGGY--GGGGYGGG 1301
Query: 144 D 144
+
Sbjct: 1302 E 1302
Score = 59.7 bits (143), Expect = 6e-09
Identities = 41/108 (37%), Positives = 47/108 (42%), Gaps = 12/108 (11%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGG------YDSGYNKTAYSTD 92
GG GY S GGGY G + S ++ N GGGG GY
Sbjct: 1279 GGGGGGGYGGGSGGYGGGGYGGGEGYSISPNSYRGNYGGGGGGYRGGSQGGYRNNFGGDY 1338
Query: 93 EPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGY 140
S G Y G+GGGY GG+ G GGG + G GGG GGGGY
Sbjct: 1339 RGSSGDYR--GSGGGYRGSGGFQRRGYGGGYFGQGRGGG----GGGGY 1380
Score = 38.5 bits (88), Expect = 0.015
Identities = 30/94 (31%), Positives = 36/94 (37%), Gaps = 23/94 (24%)
Query: 31 SYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYS 90
SY + GG G + Y + GG G S+ + GGGY
Sbjct: 1310 SYRGNYGGGGGGYRGGSQGGYRNNFGGDYRG-------SSGDYRGSGGGY---------- 1352
Query: 91 TDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEY 124
GG+ G GGGY G G GGGGGG Y
Sbjct: 1353 ---RGSGGFQRRGYGGGYF---GQGRGGGGGGGY 1380
>GRPA_MEDFA (Q09134) Abscisic acid and environmental stress
inducible protein
Length = 159
Score = 81.6 bits (200), Expect = 2e-15
Identities = 53/124 (42%), Positives = 62/124 (49%), Gaps = 30/124 (24%)
Query: 34 TDEPSGGDYDSGYKKSSYETSGGGYETG---YNKTSSYSTDEPNSGGGGYDSGYNKTAYS 90
T+E + Y GY +GGGY G YN Y N GGGGY++G
Sbjct: 29 TNEVNDAKYGGGYNHGGGGYNGGGYNHGGGGYNNGGGY-----NHGGGGYNNG------- 76
Query: 91 TDEPSGGGYGTGGAGGGYSTGGG-YGTGGGG----GGEYSTGGGG---GGYSTGGGGYGN 142
GGGY GG GGY+ GGG Y GGGG GG Y+ GGGG GGY+ GGGGY +
Sbjct: 77 -----GGGYNHGG--GGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGGYNH 129
Query: 143 DDSG 146
G
Sbjct: 130 GGGG 133
Score = 76.6 bits (187), Expect = 5e-14
Identities = 53/124 (42%), Positives = 61/124 (48%), Gaps = 31/124 (25%)
Query: 29 KTSYSTDEPSGGDYD---SGYKKSSYETSGGGYETG--YNKTSSYSTDEPNSGGGGYDSG 83
KT+ D GG Y+ GY Y GGGY G YN + N+GGGGY+ G
Sbjct: 28 KTNEVNDAKYGGGYNHGGGGYNGGGYNHGGGGYNNGGGYN----HGGGGYNNGGGGYNHG 83
Query: 84 YNKTAYSTDEPSGGGYGTGGAG-----GGYSTGGG-YGTGGGG--GGEYSTGGGGGGYST 135
GGGY GG G GGY+ GGG Y GGGG GG Y+ GGGGY+
Sbjct: 84 ------------GGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYN--HGGGGYNH 129
Query: 136 GGGG 139
GGGG
Sbjct: 130 GGGG 133
Score = 63.2 bits (152), Expect = 6e-10
Identities = 37/69 (53%), Positives = 40/69 (57%), Gaps = 11/69 (15%)
Query: 86 KTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGG---GGEYSTGG-----GGGGYSTGG 137
KT D GGGY GG GGY+ GGGY GGGG GG Y+ GG GGGGY+ GG
Sbjct: 28 KTNEVNDAKYGGGYNHGG--GGYN-GGGYNHGGGGYNNGGGYNHGGGGYNNGGGGYNHGG 84
Query: 138 GGYGNDDSG 146
GGY N G
Sbjct: 85 GGYNNGGGG 93
Score = 38.5 bits (88), Expect = 0.015
Identities = 19/37 (51%), Positives = 20/37 (53%), Gaps = 7/37 (18%)
Query: 111 GGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGN 147
GGGY GGGG GGGY+ GGGGY N N
Sbjct: 38 GGGYNHGGGGYN-------GGGYNHGGGGYNNGGGYN 67
>K1CJ_HUMAN (P13645) Keratin, type I cytoskeletal 10 (Cytokeratin
10) (K10) (CK 10)
Length = 593
Score = 80.5 bits (197), Expect = 3e-15
Identities = 47/113 (41%), Positives = 57/113 (49%), Gaps = 4/113 (3%)
Query: 39 GGDYDSGYK--KSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSG 96
GG + GY S +SGGGY G+ +S +SGGG GY S+ G
Sbjct: 466 GGSFGGGYGGGSSGGGSSGGGYGGGHGGSSGGGYGGGSSGGGSSGGGYG--GGSSSGGHG 523
Query: 97 GGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGNRR 149
GG +GG GG S G G G+ GGGGG Y G GGG S+GGG G SG +
Sbjct: 524 GGSSSGGHGGSSSGGYGGGSSGGGGGGYGGGSSGGGSSSGGGYGGGSSSGGHK 576
Score = 67.8 bits (164), Expect = 2e-11
Identities = 42/108 (38%), Positives = 52/108 (47%), Gaps = 7/108 (6%)
Query: 38 SGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSG-GGGYDSGYNKTAYS------ 90
SGG Y G+ SS GGG G + Y + G GGG SG + + S
Sbjct: 483 SGGGYGGGHGGSSGGGYGGGSSGGGSSGGGYGGGSSSGGHGGGSSSGGHGGSSSGGYGGG 542
Query: 91 TDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGG 138
+ GGGYG G +GGG S+GGGYG G GG S+ G G S+ G
Sbjct: 543 SSGGGGGGYGGGSSGGGSSSGGGYGGGSSSGGHKSSSSGSVGESSSKG 590
Score = 65.5 bits (158), Expect = 1e-10
Identities = 39/78 (50%), Positives = 45/78 (57%), Gaps = 4/78 (5%)
Query: 72 EPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGY--STGGGYGTGGGGGGEYSTGGG 129
E +SGGGG G Y SGGG GG GGG+ S+GGGYG G GGG S GG
Sbjct: 456 EGSSGGGGRGGGSFGGGYGGGS-SGGGSSGGGYGGGHGGSSGGGYGGGSSGGGS-SGGGY 513
Query: 130 GGGYSTGGGGYGNDDSGN 147
GGG S+GG G G+ G+
Sbjct: 514 GGGSSSGGHGGGSSSGGH 531
Score = 62.8 bits (151), Expect = 7e-10
Identities = 45/122 (36%), Positives = 57/122 (45%), Gaps = 13/122 (10%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEP-------NSGG----GGYDSGYNKT 87
GG S SS + GGG+ +G S+S +SGG GG+ G
Sbjct: 28 GGGVSSLRISSSKGSLGGGFSSGGFSGGSFSRGSSGGGCFGGSSGGYGGLGGFGGGSFHG 87
Query: 88 AYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGN 147
+Y + GG YG G GGG GG +G G GGG + GG GGG+ G GG G SGN
Sbjct: 88 SYGSSS-FGGSYG-GSFGGGNFGGGSFGGGSFGGGGFGGGGFGGGFGGGFGGDGGLLSGN 145
Query: 148 RR 149
+
Sbjct: 146 EK 147
Score = 53.1 bits (126), Expect = 6e-07
Identities = 35/95 (36%), Positives = 47/95 (48%), Gaps = 6/95 (6%)
Query: 55 GGGYETGYNKTSSYSTDEPNSGGGGYDSG-YNKTAYSTDEPSGGGYGTGGAGGGYSTGGG 113
GGG G +S + S GGG+ SG ++ ++S GG +G G+ GGY GG
Sbjct: 22 GGGCGGGGGVSSLRISSSKGSLGGGFSSGGFSGGSFSRGSSGGGCFG--GSSGGYGGLGG 79
Query: 114 YGTGGGGGGEYSTGGGGGGY--STGGGGYGNDDSG 146
+G GG G Y + GG Y S GGG +G G
Sbjct: 80 FG-GGSFHGSYGSSSFGGSYGGSFGGGNFGGGSFG 113
Score = 48.1 bits (113), Expect = 2e-05
Identities = 33/89 (37%), Positives = 40/89 (44%), Gaps = 6/89 (6%)
Query: 38 SGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGG 97
SGG Y G SS GG G+ +SS +SGGGG GY + SGG
Sbjct: 509 SGGGYGGG--SSSGGHGGGSSSGGHGGSSSGGYGGGSSGGGG--GGYGGGSSGGGSSSGG 564
Query: 98 GYGTGGAGGGY--STGGGYGTGGGGGGEY 124
GYG G + GG+ S+ G G G Y
Sbjct: 565 GYGGGSSSGGHKSSSSGSVGESSSKGPRY 593
Score = 47.4 bits (111), Expect = 3e-05
Identities = 26/63 (41%), Positives = 32/63 (50%), Gaps = 11/63 (17%)
Query: 90 STDEPSGGGYGTGGAGGGYST----------GGGYGTGGGGGGEYSTGGGGGG-YSTGGG 138
S+ GGG G G GGG S+ GGG+ +GG GG +S G GGG + G
Sbjct: 13 SSRSGGGGGGGGCGGGGGVSSLRISSSKGSLGGGFSSGGFSGGSFSRGSSGGGCFGGSSG 72
Query: 139 GYG 141
GYG
Sbjct: 73 GYG 75
Score = 47.4 bits (111), Expect = 3e-05
Identities = 31/81 (38%), Positives = 39/81 (47%), Gaps = 5/81 (6%)
Query: 62 YNKTSSYSTDEPNSGGGGYDSGYNKTAYSTD-EPSGGGYGTGGAGGGYSTGGGYGTGGGG 120
Y+ + YS+ GGGG G S S G G G + GG+S GG + G G
Sbjct: 5 YSSSKHYSSSRSGGGGGGGGCGGGGGVSSLRISSSKGSLGGGFSSGGFS-GGSFSRGSSG 63
Query: 121 GGEYSTGGGGGGYSTGGGGYG 141
GG + GG GGY G GG+G
Sbjct: 64 GGCF--GGSSGGYG-GLGGFG 81
Score = 45.8 bits (107), Expect = 9e-05
Identities = 30/96 (31%), Positives = 44/96 (45%), Gaps = 8/96 (8%)
Query: 26 GYNKTSYSTDEPSGGDY--DSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSG 83
G++ S+S GG + SG GG + Y +S + + GGG + G
Sbjct: 51 GFSGGSFSRGSSGGGCFGGSSGGYGGLGGFGGGSFHGSYGSSSFGGSYGGSFGGGNFGGG 110
Query: 84 YNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGG 119
++ GGG+G GG GGG+ GGG+G GG
Sbjct: 111 ----SFGGGSFGGGGFGGGGFGGGF--GGGFGGDGG 140
Score = 40.4 bits (93), Expect = 0.004
Identities = 23/44 (52%), Positives = 24/44 (54%), Gaps = 3/44 (6%)
Query: 105 GGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTG--GGGYGNDDSG 146
G G S GGG G G GGG Y G GGG S G GGG+G G
Sbjct: 455 GEGSSGGGGRGGGSFGGG-YGGGSSGGGSSGGGYGGGHGGSSGG 497
Score = 38.1 bits (87), Expect = 0.019
Identities = 24/56 (42%), Positives = 29/56 (50%), Gaps = 11/56 (19%)
Query: 99 YGTGGAGGGYSTGGGYGTGGGGGG-----EYSTGGGGGGYSTG---GGGYGNDDSG 146
Y + +GGG GGG G GGGGG S G GGG+S+G GG + SG
Sbjct: 11 YSSSRSGGG---GGGGGCGGGGGVSSLRISSSKGSLGGGFSSGGFSGGSFSRGSSG 63
>GRP1_SORBI (Q99069) Glycine-rich RNA-binding protein 1 (Fragment)
Length = 142
Score = 78.2 bits (191), Expect = 2e-14
Identities = 55/138 (39%), Positives = 64/138 (45%), Gaps = 21/138 (15%)
Query: 18 DRPTDTETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGG 77
DR T G+ ++ST+E +S+ E G G N T + + GG
Sbjct: 21 DRETQRSRGFGFVTFSTEEAM---------RSAIEGMNGKELDGRNITVNEAQSRGGRGG 71
Query: 78 GGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGG----GGGEYSTGGGG-GG 132
GG Y GGGYG GGG GGG G GGG GGG Y GGGG GG
Sbjct: 72 GGGGG------YGGGRGGGGGYGRRDGGGGGYGGGGGGYGGGRGGYGGGGYGGGGGGYGG 125
Query: 133 YSTGGGGYGNDDSGNRRD 150
S GGGGYGN D GN R+
Sbjct: 126 GSRGGGGYGNSD-GNWRN 142
>K22E_HUMAN (P35908) Keratin, type II cytoskeletal 2 epidermal
(Cytokeratin 2e) (K2e) (CK 2e)
Length = 645
Score = 77.0 bits (188), Expect = 4e-14
Identities = 45/99 (45%), Positives = 57/99 (57%), Gaps = 10/99 (10%)
Query: 53 TSGGGYETG---YNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAG-GGY 108
+SGGGY +G Y S SGGGG SG Y + SGG YG+GG GG
Sbjct: 533 SSGGGYSSGSSSYGSGGRQSGSRGGSGGGGSISG---GGYGSGGGSGGRYGSGGGSKGGS 589
Query: 109 STGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGN 147
+GGGYG+ GGG++S+GGG G S+ GGGYG+ G+
Sbjct: 590 ISGGGYGS---GGGKHSSGGGSRGGSSSGGGYGSGGGGS 625
Score = 72.0 bits (175), Expect = 1e-12
Identities = 35/69 (50%), Positives = 43/69 (61%), Gaps = 3/69 (4%)
Query: 74 NSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGY 133
+S GGGY SG ++Y + G G G GGG +GGGYG+GGG GG Y +GGG G
Sbjct: 532 SSSGGGYSSG--SSSYGSGGRQSGSRG-GSGGGGSISGGGYGSGGGSGGRYGSGGGSKGG 588
Query: 134 STGGGGYGN 142
S GGGYG+
Sbjct: 589 SISGGGYGS 597
Score = 60.8 bits (146), Expect = 3e-09
Identities = 45/107 (42%), Positives = 50/107 (46%), Gaps = 12/107 (11%)
Query: 38 SGGDYDSGYKKSSYETSG--GGYETGYNKTSSYSTDEPNSGGGG---YDSGYNKTAYSTD 92
SGG Y SG SSY + G G G S S SGGG Y SG S
Sbjct: 534 SGGGYSSG--SSSYGSGGRQSGSRGGSGGGGSISGGGYGSGGGSGGRYGSGGGSKGGSI- 590
Query: 93 EPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGG 139
SGGGYG+G GG +S+GGG G GG Y +GGGG G G
Sbjct: 591 --SGGGYGSG--GGKHSSGGGSRGGSSSGGGYGSGGGGSSSVKGSSG 633
Score = 60.5 bits (145), Expect = 4e-09
Identities = 43/120 (35%), Positives = 54/120 (44%), Gaps = 21/120 (17%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGG------------YDSGYNK 86
GG G+ S SGG + TSS+S + GGGG G K
Sbjct: 15 GGGGFRGFSSGSAVVSGGSRRS----TSSFSCLSRHGGGGGGFGGGGFGSRSLVGLGGTK 70
Query: 87 TAYSTDEPSGGGYGT----GGAGGGYSTGGGYGTGGG-GGGEYSTGGGGGGYSTGGGGYG 141
+ + GGG+G GG GGG+ G G+G G G GGG +GGG GG GGG +G
Sbjct: 71 SISISVAGGGGGFGAAGGFGGRGGGFGGGSGFGGGSGFGGGSGFSGGGFGGGGFGGGRFG 130
Score = 54.3 bits (129), Expect = 3e-07
Identities = 36/94 (38%), Positives = 44/94 (46%), Gaps = 7/94 (7%)
Query: 55 GGGYET----GYNKTSSYSTDEPNSGGG-GYDSGYNKTAYSTDEPSGGGYGTGGAGGGYS 109
GGG+ + G T S S GGG G G+ SG G G+G GG
Sbjct: 55 GGGFGSRSLVGLGGTKSISISVAGGGGGFGAAGGFGGRGGGFGGGSGFGGGSGFGGGSGF 114
Query: 110 TGGGYGTGGGGGGEYSTGGGGGGYS--TGGGGYG 141
+GGG+G GG GGG + GG GG G GG+G
Sbjct: 115 SGGGFGGGGFGGGRFGGFGGPGGVGGLGGPGGFG 148
Score = 48.5 bits (114), Expect = 1e-05
Identities = 30/56 (53%), Positives = 34/56 (60%), Gaps = 5/56 (8%)
Query: 96 GGGYGTGGA-GGGYSTGGG-YGTGGGGGGEYSTGGGGGGYSTGGGGYGN-DDSGNR 148
GG G G + GGGYS+G YG+GG G S GG GGG S GGGYG+ SG R
Sbjct: 525 GGSGGRGSSSGGGYSSGSSSYGSGGRQSG--SRGGSGGGGSISGGGYGSGGGSGGR 578
Score = 48.1 bits (113), Expect = 2e-05
Identities = 33/98 (33%), Positives = 38/98 (38%), Gaps = 33/98 (33%)
Query: 32 YSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYST 91
Y + SGG Y SG SGGGY SGGG + SG
Sbjct: 569 YGSGGGSGGRYGSGGGSKGGSISGGGYG---------------SGGGKHSSG-------- 605
Query: 92 DEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGG 129
GG+ GG S+GGGYG+GGGG G
Sbjct: 606 ----------GGSRGGSSSGGGYGSGGGGSSSVKGSSG 633
Score = 42.7 bits (99), Expect = 8e-04
Identities = 37/96 (38%), Positives = 43/96 (44%), Gaps = 12/96 (12%)
Query: 45 GYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGG---GYDSGYNKTAYSTDEPSGGGYGT 101
G K S +GGG G+ + GGG G SG+ + SGGG+G
Sbjct: 68 GTKSISISVAGGG--GGFGAAGGFGGRGGGFGGGSGFGGGSGFGGGSGF----SGGGFGG 121
Query: 102 GGAGGGYSTG-GGYGTGGGGGGEYSTGGGGGGYSTG 136
GG GGG G GG G GG GG G G GGY G
Sbjct: 122 GGFGGGRFGGFGGPGGVGGLGG--PGGFGPGGYPGG 155
Score = 36.2 bits (82), Expect = 0.073
Identities = 22/52 (42%), Positives = 28/52 (53%), Gaps = 11/52 (21%)
Query: 100 GTGGAGGG---YSTGGGYGTGGGGGGEYS------TGGGGGGYSTGGGGYGN 142
G GG GGG +S+G +GG S GGGGGG+ GGGG+G+
Sbjct: 11 GRGGGGGGFRGFSSGSAVVSGGSRRSTSSFSCLSRHGGGGGGF--GGGGFGS 60
Score = 35.8 bits (81), Expect = 0.096
Identities = 32/90 (35%), Positives = 37/90 (40%), Gaps = 13/90 (14%)
Query: 38 SGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGG 97
SGG Y SG SGGG + G Y SGGG + SG S+ SGG
Sbjct: 565 SGGGYGSGGGSGGRYGSGGGSKGGSISGGGYG-----SGGGKHSSGGGSRGGSS---SGG 616
Query: 98 GYG--TGGAGGGYSTGGGYGTGGGGGGEYS 125
GYG GG+ S G G G +S
Sbjct: 617 GYGSGGGGSS---SVKGSSGEAFGSSVTFS 643
Score = 35.0 bits (79), Expect = 0.16
Identities = 24/78 (30%), Positives = 30/78 (37%), Gaps = 15/78 (19%)
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGG------------- 122
G GG G+ + + SGG + + S GG G G GGGG
Sbjct: 11 GRGGGGGGFRGFSSGSAVVSGGSRRSTSSFSCLSRHGGGGGGFGGGGFGSRSLVGLGGTK 70
Query: 123 --EYSTGGGGGGYSTGGG 138
S GGGGG+ GG
Sbjct: 71 SISISVAGGGGGFGAAGG 88
>GRP1_PHAVU (P10495) Glycine-rich cell wall structural protein 1.0
precursor (GRP 1.0)
Length = 252
Score = 76.6 bits (187), Expect = 5e-14
Identities = 46/111 (41%), Positives = 51/111 (45%), Gaps = 7/111 (6%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG Y G K E GGG G Y + GGGG +G + Y E SG G
Sbjct: 104 GGGYGGGAGKGGGEGYGGGGANG----GGYGGGGGSGGGGGGGAGGAGSGYGGGEGSGAG 159
Query: 99 YGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTG---GGGYGNDDSG 146
G GGA GG G G G GGG GG + G GGG G GGGYG +G
Sbjct: 160 GGYGGANGGGGGGNGGGGGGGSGGAHGGGAAGGGEGAGQGAGGGYGGGAAG 210
Score = 71.2 bits (173), Expect = 2e-12
Identities = 46/112 (41%), Positives = 50/112 (44%), Gaps = 4/112 (3%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG GY GGG G S Y E + GGGY G GGG
Sbjct: 122 GGANGGGYGGGGGSGGGGGGGAG-GAGSGYGGGEGSGAGGGY--GGANGGGGGGNGGGGG 178
Query: 99 YGTGGAGGGYSTGGGYGTGGGGGGEYSTGG-GGGGYSTGGGGYGNDDSGNRR 149
G+GGA GG + GGG G G G GG Y G GGGG +GGGG G G R
Sbjct: 179 GGSGGAHGGGAAGGGEGAGQGAGGGYGGGAAGGGGRGSGGGGGGGYGGGGAR 230
Score = 70.5 bits (171), Expect = 4e-12
Identities = 46/115 (40%), Positives = 54/115 (46%), Gaps = 10/115 (8%)
Query: 36 EPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNK---TAYSTD 92
E +GG +G + Y +GGG+ G + GGGY G K Y
Sbjct: 69 EGAGGGEGAG---AGYGAAGGGHGGGGGNGGGGGG---GADGGGYGGGAGKGGGEGYGGG 122
Query: 93 EPSGGGYGTGGAGGGYSTGGGYGTGGG-GGGEYSTGGGGGGYSTGGGGYGNDDSG 146
+GGGYG GG GG GG G G G GGGE S GGG G + GGGG GN G
Sbjct: 123 GANGGGYGGGGGSGGGGGGGAGGAGSGYGGGEGSGAGGGYGGANGGGGGGNGGGG 177
Score = 68.9 bits (167), Expect = 1e-11
Identities = 47/109 (43%), Positives = 52/109 (47%), Gaps = 12/109 (11%)
Query: 40 GDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGY 99
G SGY +GGGY G N N GGGG SG + G G
Sbjct: 144 GGAGSGYGGGEGSGAGGGYG-GANGGGGGG----NGGGGGGGSGGAHGGGAAGGGEGAGQ 198
Query: 100 GTGGA-GGGYSTGGGYGTGGGGGGEYSTGG------GGGGYSTGGGGYG 141
G GG GGG + GGG G+GGGGGG Y GG GGGG S GGG+G
Sbjct: 199 GAGGGYGGGAAGGGGRGSGGGGGGGYGGGGARGSGYGGGGGSGEGGGHG 247
Score = 67.8 bits (164), Expect = 2e-11
Identities = 48/117 (41%), Positives = 54/117 (46%), Gaps = 24/117 (20%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG G ++ E GGGY G GG G +GY GGG
Sbjct: 48 GGGSGGGGGGAAVELGGGGYGEGAG------------GGEGAGAGYGAAGGG----HGGG 91
Query: 99 YGTGGAGGGYSTGGGYGTGGG-GGGE-YSTGG------GGGGYSTGGGGYGNDDSGN 147
G GG GGG + GGGYG G G GGGE Y GG GGGG S GGGG G +G+
Sbjct: 92 GGNGGGGGGGADGGGYGGGAGKGGGEGYGGGGANGGGYGGGGGSGGGGGGGAGGAGS 148
Score = 63.9 bits (154), Expect = 3e-10
Identities = 35/66 (53%), Positives = 35/66 (53%), Gaps = 4/66 (6%)
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYST 135
GGGG G A E GGGYG G AGGG G GYG GGG G GGGGG
Sbjct: 47 GGGGSGGGGGGAAV---ELGGGGYGEG-AGGGEGAGAGYGAAGGGHGGGGGNGGGGGGGA 102
Query: 136 GGGGYG 141
GGGYG
Sbjct: 103 DGGGYG 108
Score = 56.6 bits (135), Expect = 5e-08
Identities = 32/60 (53%), Positives = 35/60 (58%), Gaps = 9/60 (15%)
Query: 96 GGGYGTGGAGGGYST---GGGYGTGGGGG-----GEYSTGGG-GGGYSTGGGGYGNDDSG 146
GGG G+GG GGG + GGGYG G GGG G + GGG GGG GGGG G D G
Sbjct: 46 GGGGGSGGGGGGAAVELGGGGYGEGAGGGEGAGAGYGAAGGGHGGGGGNGGGGGGGADGG 105
Score = 44.3 bits (103), Expect = 3e-04
Identities = 25/51 (49%), Positives = 28/51 (54%), Gaps = 7/51 (13%)
Query: 98 GYGTGGAGGGYSTGGGYG-----TGGGGGGEYSTG--GGGGGYSTGGGGYG 141
G GG GGG +GGG G GGGG GE + G G G GY GGG+G
Sbjct: 39 GTVVGGYGGGGGSGGGGGGAAVELGGGGYGEGAGGGEGAGAGYGAAGGGHG 89
Score = 40.4 bits (93), Expect = 0.004
Identities = 30/87 (34%), Positives = 33/87 (37%), Gaps = 23/87 (26%)
Query: 38 SGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGG 97
SGG + G +GGG G Y GG G G GG
Sbjct: 181 SGGAHGGG-------AAGGGEGAGQGAGGGYGGGAAGGGGRGSGGG-----------GGG 222
Query: 98 GYGTGGA-----GGGYSTGGGYGTGGG 119
GYG GGA GGG +G G G GGG
Sbjct: 223 GYGGGGARGSGYGGGGGSGEGGGHGGG 249
Score = 40.0 bits (92), Expect = 0.005
Identities = 30/87 (34%), Positives = 31/87 (35%), Gaps = 36/87 (41%)
Query: 38 SGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGG 97
+GG Y G SGGG GGGGY GG
Sbjct: 200 AGGGYGGGAAGGGGRGSGGG------------------GGGGY---------------GG 226
Query: 98 GYGTGGAGGGYSTGGGYGTGGGGGGEY 124
G G G GY GGG G GGG GG Y
Sbjct: 227 G---GARGSGYGGGGGSGEGGGHGGGY 250
Score = 36.6 bits (83), Expect = 0.056
Identities = 20/36 (55%), Positives = 20/36 (55%), Gaps = 6/36 (16%)
Query: 112 GGYGTGGGGGGEYSTGGGGG-GYSTGGGGYGNDDSG 146
GGYG GGG S GGGGG GGGGYG G
Sbjct: 43 GGYGGGGG-----SGGGGGGAAVELGGGGYGEGAGG 73
>LORI_MOUSE (P18165) Loricrin
Length = 486
Score = 75.9 bits (185), Expect = 8e-14
Identities = 48/110 (43%), Positives = 50/110 (44%), Gaps = 7/110 (6%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG Y G +SGGG G SY GGGG G + GGG
Sbjct: 32 GGYYSGGGSGCGGGSSGGGSSCGGGGGGSYGGGSSCGGGGGSGGGVKYSGGGGGSSCGGG 91
Query: 99 YGTGGAGGGYSTGGGYGTGGGG---GGEYSTGGGGGGYSTGGGGYGNDDS 145
Y G GGG S GGGY GGGG GG YS GGGGG S GGG Y S
Sbjct: 92 YSGG--GGGSSCGGGYSGGGGGSSCGGGYS--GGGGGSSCGGGSYSGGGS 137
Score = 73.2 bits (178), Expect = 5e-13
Identities = 52/130 (40%), Positives = 55/130 (42%), Gaps = 22/130 (16%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKT------------SSYSTDEPNSGGGGYDSGYNK 86
GG Y G S + GGGY G + SS GGGG Y
Sbjct: 218 GGGYSGGGGSSGGSSCGGGYSGGGGSSCGGGGGYSGGGGSSCGGGSSGGGGGGSSQQYQC 277
Query: 87 TAY----STDEPSGGGYGTGGA---GGGYSTGGGYGTGGG--GGGEYSTGGGGGGYSTGG 137
+Y S GG Y GG GGGYS GGG GGG GGG G GGGGYS GG
Sbjct: 278 QSYGGGSSGGSSCGGRYSGGGGSSCGGGYSGGGGSSCGGGSSGGGSSCGGSGGGGYSGGG 337
Query: 138 GG-YGNDDSG 146
GG G SG
Sbjct: 338 GGSCGGGSSG 347
Score = 72.8 bits (177), Expect = 7e-13
Identities = 53/148 (35%), Positives = 59/148 (39%), Gaps = 28/148 (18%)
Query: 27 YNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKT-----SSYSTDEPNSGGGGYD 81
Y SY G Y + GGGY G + S + SGGGGY
Sbjct: 275 YQCQSYGGGSSGGSSCGGRYSGGGGSSCGGGYSGGGGSSCGGGSSGGGSSCGGSGGGGYS 334
Query: 82 SGYNKTAYSTDEPSGGGY-----------------GTGGAGGGYSTGGGY-GTGGGG--- 120
G + GGGY G G +GGG S GGG G GGGG
Sbjct: 335 GGGGGSCGGGSSGGGGGYYSSQQTSQTSCAPQQSYGGGSSGGGGSCGGGSSGGGGGGGCY 394
Query: 121 --GGEYSTGGGGGGYSTGGGGYGNDDSG 146
GG S+GG GGGYS GGGG G SG
Sbjct: 395 SSGGGGSSGGCGGGYSGGGGGCGGGSSG 422
Score = 71.6 bits (174), Expect = 2e-12
Identities = 46/117 (39%), Positives = 51/117 (43%), Gaps = 8/117 (6%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG Y G SGGG + S + GGGG G + GGG
Sbjct: 58 GGSYGGGSSCGGGGGSGGGVKYSGGGGGSSCGGGYSGGGGGSSCGGGYSGGGGGSSCGGG 117
Query: 99 YGTGGAG-----GGYSTGG---GYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGN 147
Y GG G G YS GG G G G GGG +YS GGGGGG S GGG G G+
Sbjct: 118 YSGGGGGSSCGGGSYSGGGSSCGGGGGSGGGVKYSGGGGGGGSSCGGGSSGGGGGGS 174
Score = 68.9 bits (167), Expect = 1e-11
Identities = 42/107 (39%), Positives = 52/107 (48%), Gaps = 1/107 (0%)
Query: 53 TSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGG 112
+SGGG G + S GGG SG YS GG +GG+GGG G
Sbjct: 373 SSGGGGSCGGGSSGGGGGGGCYSSGGGGSSGGCGGGYSGGGGGCGGGSSGGSGGGCGGGS 432
Query: 113 GYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGNRRDEVDYKEEEK 159
G+GGG GG YS GGGGG S GGG G G + V ++ ++K
Sbjct: 433 SGGSGGGCGGGYSGGGGGGS-SCGGGSSGGGSGGGKGVPVCHQTQQK 478
Score = 67.8 bits (164), Expect = 2e-11
Identities = 57/141 (40%), Positives = 61/141 (42%), Gaps = 41/141 (29%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTS----SYSTDEPNSGGGGYDSGYNKTAYSTDEP 94
GG Y G SS GGGY G +S SYS + GGGG G K YS
Sbjct: 102 GGGYSGGGGGSS---CGGGYSGGGGGSSCGGGSYSGGGSSCGGGGGSGGGVK--YSG--- 153
Query: 95 SGGGYGT-------GGAGGGYSTGGGYG------------------TGGGGGGEYSTGGG 129
GGG G+ GG GGG S GGG G GG GGG+YS GGG
Sbjct: 154 GGGGGGSSCGGGSSGGGGGGSSCGGGSGGGGSYCGGSSGGGSSGGCGGGSGGGKYSGGGG 213
Query: 130 ----GGGYSTGGGGYGNDDSG 146
GGGYS GGG G G
Sbjct: 214 GSSCGGGYSGGGGSSGGSSCG 234
Score = 67.4 bits (163), Expect = 3e-11
Identities = 45/113 (39%), Positives = 47/113 (40%), Gaps = 8/113 (7%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTS---SYSTDEPNS--GGGGYDSGYNKTAYSTDE 93
GG Y G SS GGGY G +S YS S GGG Y G +
Sbjct: 89 GGGYSGGGGGSS---CGGGYSGGGGGSSCGGGYSGGGGGSSCGGGSYSGGGSSCGGGGGS 145
Query: 94 PSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSG 146
G Y GG GGG S GGG GGGGG G GGGG GG G G
Sbjct: 146 GGGVKYSGGGGGGGSSCGGGSSGGGGGGSSCGGGSGGGGSYCGGSSGGGSSGG 198
Score = 66.6 bits (161), Expect = 5e-11
Identities = 51/114 (44%), Positives = 56/114 (48%), Gaps = 15/114 (13%)
Query: 39 GGDYDSGYKKSSYETS---GGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPS 95
GG Y G S S GGGY + +TS S S GGG G
Sbjct: 330 GGGYSGGGGGSCGGGSSGGGGGYYSS-QQTSQTSCAPQQSYGGGSSGGGGSCG------- 381
Query: 96 GGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGG-GGYSTG--GGGYGNDDSG 146
GG G GG GG YS+GGG G+ GG GG YS GGGG GG S+G GGG G SG
Sbjct: 382 GGSSGGGGGGGCYSSGGG-GSSGGCGGGYSGGGGGCGGGSSGGSGGGCGGGSSG 434
Score = 65.1 bits (157), Expect = 1e-10
Identities = 50/123 (40%), Positives = 55/123 (44%), Gaps = 18/123 (14%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG Y G SS GGGY G + S SGGGG G YS S G
Sbjct: 205 GGKYSGGGGGSS---CGGGYSGGGGSSGGSSCGGGYSGGGGSSCG-GGGGYSGGGGSSCG 260
Query: 99 YGTGGAGGGYST--------GGGYGTGGGGGGEYSTGGG---GGGYSTGGG---GYGNDD 144
G+ G GGG S+ GGG G GG YS GGG GGGYS GGG G G+
Sbjct: 261 GGSSGGGGGGSSQQYQCQSYGGGSSGGSSCGGRYSGGGGSSCGGGYSGGGGSSCGGGSSG 320
Query: 145 SGN 147
G+
Sbjct: 321 GGS 323
Score = 63.9 bits (154), Expect = 3e-10
Identities = 52/138 (37%), Positives = 58/138 (41%), Gaps = 24/138 (17%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG G K Y GGG + SS +S GGG G + S+ S GG
Sbjct: 142 GGGSGGGVK---YSGGGGGGGSSCGGGSSGGGGGGSSCGGGSGGGGSYCGGSSGGGSSGG 198
Query: 99 YGTG-------GAGGGYSTGGGYGTGGGG------GGEYSTGGG-----GGGYSTGGG-- 138
G G G GGG S GGGY GGG GG YS GGG GGGYS GGG
Sbjct: 199 CGGGSGGGKYSGGGGGSSCGGGYSGGGGSSGGSSCGGGYSGGGGSSCGGGGGYSGGGGSS 258
Query: 139 -GYGNDDSGNRRDEVDYK 155
G G+ G Y+
Sbjct: 259 CGGGSSGGGGGGSSQQYQ 276
Score = 63.9 bits (154), Expect = 3e-10
Identities = 43/98 (43%), Positives = 46/98 (46%), Gaps = 11/98 (11%)
Query: 61 GYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSG---GGYGTGGAGGGYSTGGGYGTG 117
G KTS GGGGY SG G GG G G GGG S GGG G+G
Sbjct: 15 GCGKTSGGGGGGGGGGGGGYYSGGGSGCGGGSSGGGSSCGGGGGGSYGGGSSCGGGGGSG 74
Query: 118 GG-----GGGEYSTGGG---GGGYSTGGGGYGNDDSGN 147
GG GGG S GGG GGG S+ GGGY G+
Sbjct: 75 GGVKYSGGGGGSSCGGGYSGGGGGSSCGGGYSGGGGGS 112
Score = 62.8 bits (151), Expect = 7e-10
Identities = 43/112 (38%), Positives = 51/112 (45%), Gaps = 5/112 (4%)
Query: 28 NKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKT 87
++TS + + GG G +SGGG G SS GGGY G
Sbjct: 359 SQTSCAPQQSYGGGSSGGGGSCGGGSSGGG--GGGGCYSSGGGGSSGGCGGGYSGGGGGC 416
Query: 88 AYSTDEPSGGGYGTGGAGG-GYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGG 138
+ SGGG G G +GG G GGGY GGGGG S GGG G +GGG
Sbjct: 417 GGGSSGGSGGGCGGGSSGGSGGGCGGGYSGGGGGGS--SCGGGSSGGGSGGG 466
Score = 62.4 bits (150), Expect = 1e-09
Identities = 45/118 (38%), Positives = 48/118 (40%), Gaps = 26/118 (22%)
Query: 38 SGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGG 97
S G G Y GGG G YS +SGG GY SGG
Sbjct: 195 SSGGCGGGSGGGKYSGGGGGSSCG----GGYSGGGGSSGGSSCGGGY----------SGG 240
Query: 98 GYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGG-------YSTGGGGYGNDDSGNR 148
G + G GGGYS GGG GGG S+GGGGGG S GGG G G R
Sbjct: 241 GGSSCGGGGGYSGGGGSSCGGG-----SSGGGGGGSSQQYQCQSYGGGSSGGSSCGGR 293
Score = 59.3 bits (142), Expect = 8e-09
Identities = 41/89 (46%), Positives = 45/89 (50%), Gaps = 8/89 (8%)
Query: 55 GGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGY 114
GGG GY +SGGG G +Y GGG G+GG G YS GGG
Sbjct: 27 GGGGGGGYYSGGGSGCGGGSSGGGSSCGGGGGGSYGGGSSCGGGGGSGG-GVKYSGGGG- 84
Query: 115 GTGGGGGGEYSTGGG----GGGYSTGGGG 139
G+ GGG YS GGG GGGYS GGGG
Sbjct: 85 GSSCGGG--YSGGGGGSSCGGGYSGGGGG 111
Score = 45.8 bits (107), Expect = 9e-05
Identities = 36/86 (41%), Positives = 40/86 (45%), Gaps = 11/86 (12%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG Y SG SS GGGY G S+ G GG SG + GGG
Sbjct: 391 GGCYSSGGGGSS-GGCGGGYSGGGGGCGGGSSGGSGGGCGGGSSG------GSGGGCGGG 443
Query: 99 YGTGGAGGGYSTGGGY---GTGGGGG 121
Y +GG GGG S GGG G+GGG G
Sbjct: 444 Y-SGGGGGGSSCGGGSSGGGSGGGKG 468
>EGG1_SCHJA (P19470) Eggshell protein 1 precursor
Length = 212
Score = 75.9 bits (185), Expect = 8e-14
Identities = 54/143 (37%), Positives = 60/143 (41%), Gaps = 24/143 (16%)
Query: 32 YSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGG------YDSGYN 85
Y+ P DYDSGY GGGY + Y GGGG Y GY
Sbjct: 15 YTIAYPPPSDYDSGYGGGGGGGGGGGYGGWCGGSDCYGGGNGGGGGGGGGNGGEYGGGYG 74
Query: 86 KT---AYSTDEPSGGGYGT---GGAGGGYSTGGGYGTGGGGGGEYSTGGGGGG------- 132
+Y GGGYG GG GG GGG G G GGGG + GG GGG
Sbjct: 75 DVYGGSYGGGSYGGGGYGDVYGGGCGGPDCYGGGNGGGNGGGGGCNGGGCGGGPDFYGKG 134
Query: 133 --YSTGGGGYGND---DSGNRRD 150
S GG YGND DS R++
Sbjct: 135 YEDSYGGDSYGNDYYGDSNGRKN 157
Score = 62.8 bits (151), Expect = 7e-10
Identities = 39/86 (45%), Positives = 42/86 (48%), Gaps = 6/86 (6%)
Query: 57 GYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGT 116
GY Y S Y + GGGG GY +D GG G GG GGG GG Y
Sbjct: 14 GYTIAYPPPSDYDSGYGGGGGGGGGGGYGGWCGGSDCYGGGNGGGGGGGGG--NGGEY-- 69
Query: 117 GGGGGGEYSTGGGGGGYSTGGGGYGN 142
GGG G Y GG GG S GGGGYG+
Sbjct: 70 GGGYGDVY--GGSYGGGSYGGGGYGD 93
Score = 56.2 bits (134), Expect = 7e-08
Identities = 38/99 (38%), Positives = 45/99 (45%), Gaps = 9/99 (9%)
Query: 54 SGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTD---EPSG--GGYGTGGAGGGY 108
+GGG G P+ G GY+ Y +Y D + +G G+G GG GG
Sbjct: 109 NGGGNGGGGGCNGGGCGGGPDFYGKGYEDSYGGDSYGNDYYGDSNGRKNGHGKGGKGG-- 166
Query: 109 STGGGYGTGGG-GGGEYSTGGGGGGYSTGGGGYGNDDSG 146
GGG G GGG GGG G GGG GGG GN G
Sbjct: 167 -NGGGGGKGGGKGGGNGKGNGKGGGGKNGGGKGGNGGKG 204
Score = 46.2 bits (108), Expect = 7e-05
Identities = 36/99 (36%), Positives = 43/99 (43%), Gaps = 9/99 (9%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG + G + G GYE Y SY D G +G+ K + GGG
Sbjct: 116 GGGCNGGGCGGGPDFYGKGYEDSYGG-DSYGNDYYGDSNGR-KNGHGKGGKGGN---GGG 170
Query: 99 YGTGGAGGGYSTGGGYGTGGG---GGGEYSTGGGGGGYS 134
G GG GG G G G GGG GGG+ GG GG Y+
Sbjct: 171 GGKGGGKGG-GNGKGNGKGGGGKNGGGKGGNGGKGGSYA 208
Score = 35.0 bits (79), Expect = 0.16
Identities = 29/91 (31%), Positives = 30/91 (32%), Gaps = 16/91 (17%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
G Y+ Y SY G G N GGGG G GGG
Sbjct: 132 GKGYEDSYGGDSYGNDYYGDSNGRKNGHGKGGKGGNGGGGGKGGG-----------KGGG 180
Query: 99 YGTG-GAGGGYSTGGGY----GTGGGGGGEY 124
G G G GGG GGG G GG Y
Sbjct: 181 NGKGNGKGGGGKNGGGKGGNGGKGGSYAPSY 211
>GRP2_ORYSA (P29834) Glycine-rich cell wall structural protein 2
precursor
Length = 183
Score = 75.5 bits (184), Expect = 1e-13
Identities = 51/125 (40%), Positives = 60/125 (47%), Gaps = 17/125 (13%)
Query: 36 EPSGGDYD-SGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEP 94
E GG Y SGY S GGG +G Y GGGG + G + + Y + +
Sbjct: 40 EGGGGGYGGSGYGSGSGYGEGGG--SGGAAGGGYGRGGGGGGGGG-EGGGSGSGYGSGQG 96
Query: 95 SG--------GGYGTGGAGGGYSTGG--GYGTGGGGGGEYSTGGG---GGGYSTGGGGYG 141
SG GGYG+GG GGG GG GYG G G G Y +G G GGGY +GGGG G
Sbjct: 97 SGYGAGVGGAGGYGSGGGGGGGQGGGAGGYGQGSGYGSGYGSGAGGAHGGGYGSGGGGGG 156
Query: 142 NDDSG 146
G
Sbjct: 157 GGGQG 161
Score = 72.4 bits (176), Expect = 9e-13
Identities = 46/113 (40%), Positives = 50/113 (43%), Gaps = 19/113 (16%)
Query: 38 SGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGG 97
+GG Y G GGG +GY G GGY SG GG
Sbjct: 67 AGGGYGRGGGGGGGGGEGGGSGSGYGSGQGSGYGAGVGGAGGYGSG-----------GGG 115
Query: 98 GYGTGGAGGGYSTGGGYGTGGG-------GGGEYSTGGGGGGYSTGGG-GYGN 142
G G GG GGY G GYG+G G GGG S GGGGGG GGG GYG+
Sbjct: 116 GGGQGGGAGGYGQGSGYGSGYGSGAGGAHGGGYGSGGGGGGGGGQGGGSGYGS 168
Score = 71.2 bits (173), Expect = 2e-12
Identities = 47/109 (43%), Positives = 52/109 (47%), Gaps = 16/109 (14%)
Query: 37 PSGGDYDSGYKKSSYETSGGGYE-TGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPS 95
P GG G E GGGY +GY S Y E GG GY +
Sbjct: 31 PGGGGGGGG------EGGGGGYGGSGYGSGSGYG--EGGGSGGAAGGGYGRGGGG----G 78
Query: 96 GGGYGTGGAGGGYST--GGGYGTGGGGGGEYSTGGGGGGYSTGG-GGYG 141
GGG GG+G GY + G GYG G GG G Y +GGGGGG GG GGYG
Sbjct: 79 GGGGEGGGSGSGYGSGQGSGYGAGVGGAGGYGSGGGGGGGQGGGAGGYG 127
Score = 69.3 bits (168), Expect = 8e-12
Identities = 45/108 (41%), Positives = 53/108 (48%), Gaps = 14/108 (12%)
Query: 39 GGDYDSGYKKSSYETS-GGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGG 97
GG + G S Y + G GY G Y + GGGG G Y G
Sbjct: 79 GGGGEGGGSGSGYGSGQGSGYGAGVGGAGGYGS----GGGGGGGQGGGAGGYG----QGS 130
Query: 98 GYGTG-GAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTG---GGGYG 141
GYG+G G+G G + GGGYG+GGGGGG GGG G Y +G G GYG
Sbjct: 131 GYGSGYGSGAGGAHGGGYGSGGGGGGGGGQGGGSG-YGSGSGYGSGYG 177
Score = 68.9 bits (167), Expect = 1e-11
Identities = 49/124 (39%), Positives = 57/124 (45%), Gaps = 14/124 (11%)
Query: 25 TGYNKTS-YSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSG 83
+GY S Y SGG GY + GGG G S Y + + G GY +G
Sbjct: 49 SGYGSGSGYGEGGGSGGAAGGGYGRGGGGGGGGG--EGGGSGSGYGSGQ----GSGYGAG 102
Query: 84 YNKTA-YSTDEPSGGGYGTGGAGG-----GYSTGGGYGTGGGGGGEYSTGGGGGGYSTGG 137
Y + GGG G GGAGG GY +G G G GG GG Y +GGGGGG G
Sbjct: 103 VGGAGGYGSGGGGGGGQG-GGAGGYGQGSGYGSGYGSGAGGAHGGGYGSGGGGGGGGGQG 161
Query: 138 GGYG 141
GG G
Sbjct: 162 GGSG 165
Score = 63.9 bits (154), Expect = 3e-10
Identities = 37/85 (43%), Positives = 42/85 (48%), Gaps = 14/85 (16%)
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGA-GGGYSTGGGYG----TGGGGGGEYSTG--- 127
GG G GY + Y + G G G+GGA GGGY GGG G GGG G Y +G
Sbjct: 38 GGEGGGGGYGGSGYGSGSGYGEGGGSGGAAGGGYGRGGGGGGGGGEGGGSGSGYGSGQGS 97
Query: 128 ------GGGGGYSTGGGGYGNDDSG 146
GG GGY +GGGG G G
Sbjct: 98 GYGAGVGGAGGYGSGGGGGGGQGGG 122
Score = 60.8 bits (146), Expect = 3e-09
Identities = 32/70 (45%), Positives = 34/70 (47%), Gaps = 11/70 (15%)
Query: 73 PNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGG 132
P GGGG G GGGYG G G G G G G+GG GG Y GGGGGG
Sbjct: 31 PGGGGGGGGEG-----------GGGGYGGSGYGSGSGYGEGGGSGGAAGGGYGRGGGGGG 79
Query: 133 YSTGGGGYGN 142
GGG G+
Sbjct: 80 GGGEGGGSGS 89
Score = 55.5 bits (132), Expect = 1e-07
Identities = 30/57 (52%), Positives = 31/57 (53%), Gaps = 6/57 (10%)
Query: 96 GGGYGTGGAGGGYSTGGGY-GTGGGGGGEYSTGGG-----GGGYSTGGGGYGNDDSG 146
G G G GG GGG GGGY G+G G G Y GGG GGGY GGGG G G
Sbjct: 28 GYGPGGGGGGGGEGGGGGYGGSGYGSGSGYGEGGGSGGAAGGGYGRGGGGGGGGGEG 84
Score = 54.7 bits (130), Expect = 2e-07
Identities = 39/107 (36%), Positives = 48/107 (44%), Gaps = 19/107 (17%)
Query: 24 ETGYNKTSYSTDEPSG--------GDYDSGYKKSSYETSG-GGYETGYNKTSSYSTDEPN 74
E G + + Y + + SG G Y SG + G GGY G S Y +
Sbjct: 83 EGGGSGSGYGSGQGSGYGAGVGGAGGYGSGGGGGGGQGGGAGGYGQGSGYGSGYGSGAGG 142
Query: 75 SGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGG 121
+ GGGY SG GGG G GG G GY +G GYG+G GGG
Sbjct: 143 AHGGGYGSGGG---------GGGGGGQGG-GSGYGSGSGYGSGYGGG 179
>FUS_HUMAN (P35637) RNA-binding protein FUS (Oncogene FUS) (Oncogene
TLS) (Translocated in liposarcoma protein) (POMp75) (75
kDa DNA-pairing protein)
Length = 526
Score = 75.5 bits (184), Expect = 1e-13
Identities = 50/146 (34%), Positives = 67/146 (45%), Gaps = 19/146 (13%)
Query: 20 PTDTETGYNKTSYSTD--EPSGGDYDSGYKKSSYETSGGGYET-----GYNKTSSYSTDE 72
P+ T Y +S S+ +P G Y + S G ++ GY + + Y++
Sbjct: 106 PSSTSGSYGSSSQSSSYGQPQSGSYSQQPSYGGQQQSYGQQQSYNPPQGYGQQNQYNSSS 165
Query: 73 PNSGGGGYDSGY--NKTAYSTDEPSGGGYGT-----GGAGGGYSTG--GGYGTGGGGGGE 123
GGGG Y ++++ S+ SGGGYG GG GGY GG G GG GGG
Sbjct: 166 GGGGGGGGGGNYGQDQSSMSSGGGSGGGYGNQDQSGGGGSGGYGQQDRGGRGRGGSGGGG 225
Query: 124 YSTGGGGGGYSTGGGGYGNDDSGNRR 149
GGGGGGY+ GGY G R
Sbjct: 226 ---GGGGGGYNRSSGGYEPRGRGGGR 248
Score = 58.5 bits (140), Expect = 1e-08
Identities = 55/178 (30%), Positives = 70/178 (38%), Gaps = 56/178 (31%)
Query: 21 TDTETGYNKTSYSTDEPSGGDYDSGYKKSSYET---SGGGYETGYNKTSSYSTDE----- 72
TDT +GY ++SYS+ G ++GY S S GGY + + SSY
Sbjct: 45 TDT-SGYGQSSYSS---YGQSQNTGYGTQSTPQGYGSTGGYGSSQSSQSSYGQQSSYPGY 100
Query: 73 -----PNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGY------------------- 108
P+S G Y S ++Y +P G Y + GG
Sbjct: 101 GQQPAPSSTSGSYGSSSQSSSYG--QPQSGSYSQQPSYGGQQQSYGQQQSYNPPQGYGQQ 158
Query: 109 ----STGGGYGTGGGGG------GEYSTGGG-GGGYST-------GGGGYGNDDSGNR 148
S+ GG G GGGGG S+GGG GGGY G GGYG D G R
Sbjct: 159 NQYNSSSGGGGGGGGGGNYGQDQSSMSSGGGSGGGYGNQDQSGGGGSGGYGQQDRGGR 216
Score = 51.2 bits (121), Expect = 2e-06
Identities = 41/114 (35%), Positives = 52/114 (44%), Gaps = 13/114 (11%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG+Y G +SS +SGGG GY + D+ GG G ++ GGG
Sbjct: 174 GGNY--GQDQSSM-SSGGGSGGGYG-----NQDQSGGGGSGGYGQQDRGGRGRGGSGGGG 225
Query: 99 YGTGGAGGGYS-TGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGND-DSGNRRD 150
G GGGY+ + GGY G GGG GG GG G +G D G+R D
Sbjct: 226 G---GGGGGYNRSSGGYEPRGRGGGRGGRGGMGGSDRGGFNKFGGPRDQGSRHD 276
Score = 48.5 bits (114), Expect = 1e-05
Identities = 36/98 (36%), Positives = 47/98 (47%), Gaps = 16/98 (16%)
Query: 59 ETGYNK-TSSYSTDEPNSG--------GGGYDSGYNKTAYST---DEPSGGGYGTGGAGG 106
ETG K ++ S D+P S G + K +++T D GGG G GG G
Sbjct: 329 ETGKLKGEATVSFDDPPSAKAAIDWFDGKEFSGNPIKVSFATRRADFNRGGGNGRGGRGR 388
Query: 107 GYSTG----GGYGTGGGGGGEYSTGGGGGGYSTGGGGY 140
G G GG G+GGGG G + +GGGGGG G +
Sbjct: 389 GGPMGRGGYGGGGSGGGGRGGFPSGGGGGGGQQRAGDW 426
Score = 39.3 bits (90), Expect = 0.009
Identities = 32/127 (25%), Positives = 47/127 (36%), Gaps = 4/127 (3%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYD--SGYNKTAYSTDEPSG 96
GG G + + + GGG G + + P + + N+ + G
Sbjct: 398 GGGGSGGGGRGGFPSGGGG-GGGQQRAGDWKCPNPTCENMNFSWRNECNQCKAPKPDGPG 456
Query: 97 GGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGNRRDEVDYKE 156
GG G GG Y G GG G Y GG G GG G G D G ++D +
Sbjct: 457 GGPGGSHMGGNYGDDRRGGRGGYDRGGYRGRGGDRGGFRGGRG-GGDRGGFGPGKMDSRG 515
Query: 157 EEKHHKK 163
E + ++
Sbjct: 516 EHRQDRR 522
Score = 38.1 bits (87), Expect = 0.019
Identities = 19/39 (48%), Positives = 22/39 (55%)
Query: 108 YSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSG 146
++ GGG G GG G G GG GG +GGGG G SG
Sbjct: 375 FNRGGGNGRGGRGRGGPMGRGGYGGGGSGGGGRGGFPSG 413
Score = 35.4 bits (80), Expect = 0.13
Identities = 33/98 (33%), Positives = 44/98 (44%), Gaps = 21/98 (21%)
Query: 45 GYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYST-DEPSGGGYGTGG 103
GY + S + G +GY S STD SGY +++YS+ + GYGT
Sbjct: 24 GYSQQSSQPYGQQSYSGY----SQSTDT---------SGYGQSSYSSYGQSQNTGYGT-- 68
Query: 104 AGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYG 141
ST GYG+ GG G S+ G S+ GYG
Sbjct: 69 ----QSTPQGYGSTGGYGSSQSSQSSYGQQSS-YPGYG 101
>GRP_HORVU (P17816) Glycine-rich cell wall structural protein
precursor
Length = 200
Score = 74.3 bits (181), Expect = 2e-13
Identities = 50/116 (43%), Positives = 53/116 (45%), Gaps = 19/116 (16%)
Query: 39 GGDYDSGYKKSSYETSGGGYETG---YNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPS 95
GG Y G Y GGGY G Y GGGGY G + +
Sbjct: 59 GGGYGGG---GGYGGGGGGYPGGGGGYGGGGGGYPGHGGEGGGGYGGGGGYPGHGGE--G 113
Query: 96 GGGYGTGGA--------GGGYSTGGGY-GTGGGGGGEYSTGGGGGGYSTGGGGYGN 142
GGGYG GG GGGY GGGY G GG GGG Y GGGGGGY GGG G+
Sbjct: 114 GGGYGGGGGYHGHGGEGGGGYGGGGGYHGHGGEGGGGY--GGGGGGYPGHGGGGGH 167
Score = 65.9 bits (159), Expect = 9e-11
Identities = 42/95 (44%), Positives = 46/95 (48%), Gaps = 12/95 (12%)
Query: 55 GGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGY 114
GGG G+ + GGGGY G P GGG G GG GGGY GG
Sbjct: 45 GGGGHHGHGRGGH--------GGGGYGGGGGYGGGGGGYPGGGG-GYGGGGGGYPGHGGE 95
Query: 115 GTGG-GGGGEY--STGGGGGGYSTGGGGYGNDDSG 146
G GG GGGG Y G GGGGY GGG +G+ G
Sbjct: 96 GGGGYGGGGGYPGHGGEGGGGYGGGGGYHGHGGEG 130
Score = 60.1 bits (144), Expect = 5e-09
Identities = 45/109 (41%), Positives = 48/109 (43%), Gaps = 19/109 (17%)
Query: 39 GGDYDSGYKKSSYETSGGGYET-------GYNKTSSYSTDEPNSGGGGYDSGYNKTAYST 91
GG Y G Y GGGY GY Y GGGGY G +
Sbjct: 72 GGGYPGG--GGGYGGGGGGYPGHGGEGGGGYGGGGGYP-GHGGEGGGGYGGGGGYHGHGG 128
Query: 92 DEPSGGGYGTGGA--GGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGG 138
+ GGGYG GG G G GGGYG GGGG Y GGGGG+ GGG
Sbjct: 129 E--GGGGYGGGGGYHGHGGEGGGGYG---GGGGGYPGHGGGGGH--GGG 170
Score = 48.5 bits (114), Expect = 1e-05
Identities = 37/96 (38%), Positives = 42/96 (43%), Gaps = 12/96 (12%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG Y G + GGG GY Y GGGGY G + + GGG
Sbjct: 97 GGGYGGGGGYPGHGGEGGG---GYGGGGGYH-GHGGEGGGGYGGGGGYHGHGGE--GGGG 150
Query: 99 YGTGGAGGGYSTGGGYGTGGG-GGGEYSTGGGGGGY 133
YG G GGGY G+G GGG GGG G G G+
Sbjct: 151 YG--GGGGGYP---GHGGGGGHGGGRCKWGCCGHGF 181
>CAZ_DROME (Q27294) RNA-binding protein cabeza (Sarcoma-associated
RNA-binding fly homolog) (P19)
Length = 399
Score = 73.9 bits (180), Expect = 3e-13
Identities = 49/116 (42%), Positives = 57/116 (48%), Gaps = 19/116 (16%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSG--GGGYDSGYNKTAYSTDEPSG 96
GG GY +++ Y+ NKT +Y+ PN G GGGYDSG S SG
Sbjct: 7 GGGSGQGY--NNFAVPPPNYQQMPNKTGNYNEPPPNYGKQGGGYDSG------SGHRGSG 58
Query: 97 GGYGTGGAGGGYSTGGG--YGTGGG-------GGGEYSTGGGGGGYSTGGGGYGND 143
G GG GG ++ GG YG GG G G YS GGGGGG GGG GND
Sbjct: 59 GSGNGGGGGGSWNDRGGNSYGNGGASKDSYNKGHGGYSGGGGGGGGGGGGGSGGND 114
Score = 68.9 bits (167), Expect = 1e-11
Identities = 43/115 (37%), Positives = 53/115 (45%), Gaps = 26/115 (22%)
Query: 43 DSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTG 102
D +K +S + + N+ + D+ S GGG GGGYG G
Sbjct: 276 DGDWKCNSCNNTNFAWRNECNRCKTPKGDDEGSSGGG---------------GGGGYGGG 320
Query: 103 GAGGGYSTGGGYGTGGGG------GGEYSTGGGGGGYSTGGGGYG--NDDSGNRR 149
G GGGY G G+GGGG GG GGGGGG GGGGY ND++G R
Sbjct: 321 GGGGGYDRGNDRGSGGGGYHNRDRGGNSQGGGGGGG---GGGGYSRFNDNNGGGR 372
Score = 67.4 bits (163), Expect = 3e-11
Identities = 37/76 (48%), Positives = 38/76 (49%), Gaps = 17/76 (22%)
Query: 74 NSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGG- 132
N GGGG GGG G GG GG GGG G GGGGGG + GGGGGG
Sbjct: 211 NKGGGG---------------GGGGGGRGGFGGRRGGGGGGGGGGGGGGRFDRGGGGGGG 255
Query: 133 -YSTGGGGYGNDDSGN 147
Y GGGG G GN
Sbjct: 256 RYDRGGGGGGGGGGGN 271
Score = 62.4 bits (150), Expect = 1e-09
Identities = 37/99 (37%), Positives = 45/99 (45%), Gaps = 15/99 (15%)
Query: 53 TSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGG 112
+SGGG GY GGGGYD G ++ + GGGY GG GG
Sbjct: 308 SSGGGGGGGYGG---------GGGGGGYDRGNDRGS------GGGGYHNRDRGGNSQGGG 352
Query: 113 GYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGNRRDE 151
G G GGGG ++ GGG GGGG D G R++
Sbjct: 353 GGGGGGGGYSRFNDNNGGGRGGRGGGGGNRRDGGPMRND 391
Score = 56.2 bits (134), Expect = 7e-08
Identities = 42/106 (39%), Positives = 48/106 (44%), Gaps = 24/106 (22%)
Query: 29 KTSYSTDE-PSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKT 87
KT DE SGG GY GGGY+ G ++ SGGGGY +N+
Sbjct: 299 KTPKGDDEGSSGGGGGGGYGGGG---GGGGYDRGNDR---------GSGGGGY---HNRD 343
Query: 88 AYSTDEPSGGGYGTGGAGGGYS-----TGGGYGTGGGGGGEYSTGG 128
+ GGG GG GGGYS GGG G GGGGG GG
Sbjct: 344 RGGNSQGGGGG---GGGGGGYSRFNDNNGGGRGGRGGGGGNRRDGG 386
Score = 52.4 bits (124), Expect = 1e-06
Identities = 41/120 (34%), Positives = 50/120 (41%), Gaps = 29/120 (24%)
Query: 28 NKT-SYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNK 86
NKT +Y+ P+ G GY S GG G GGGG
Sbjct: 29 NKTGNYNEPPPNYGKQGGGYDSGSGHRGSGGSGNG-------------GGGGG------- 68
Query: 87 TAYSTDEPSGGGYGTGGAG-GGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDS 145
S ++ G YG GGA Y+ G G +GGGGGG GGGGGG S G +D+
Sbjct: 69 ---SWNDRGGNSYGNGGASKDSYNKGHGGYSGGGGGG----GGGGGGGSGGNDMITQEDT 121
Score = 37.0 bits (84), Expect = 0.043
Identities = 18/34 (52%), Positives = 18/34 (52%)
Query: 117 GGGGGGEYSTGGGGGGYSTGGGGYGNDDSGNRRD 150
GGGGGG GG GG GGGG G G R D
Sbjct: 214 GGGGGGGGGRGGFGGRRGGGGGGGGGGGGGGRFD 247
Score = 35.4 bits (80), Expect = 0.13
Identities = 29/77 (37%), Positives = 32/77 (40%), Gaps = 30/77 (38%)
Query: 97 GGYGTGGAGGGYS---------------TGG------GYGTGGGGGGEYST-----GGGG 130
GGYG GG+G GY+ TG YG GGG Y + G GG
Sbjct: 4 GGYG-GGSGQGYNNFAVPPPNYQQMPNKTGNYNEPPPNYGKQGGG---YDSGSGHRGSGG 59
Query: 131 GGYSTGGGGYGNDDSGN 147
G GGGG ND GN
Sbjct: 60 SGNGGGGGGSWNDRGGN 76
Score = 34.7 bits (78), Expect = 0.21
Identities = 17/36 (47%), Positives = 18/36 (49%)
Query: 114 YGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGNRR 149
+ GGGGGG GG GG GGGG G G R
Sbjct: 210 WNKGGGGGGGGGGRGGFGGRRGGGGGGGGGGGGGGR 245
>RB56_HUMAN (Q92804) TATA-binding protein associated factor 2N
(RNA-binding protein 56) (TAFII68) (TAF(II)68)
Length = 592
Score = 73.6 bits (179), Expect = 4e-13
Identities = 63/152 (41%), Positives = 68/152 (44%), Gaps = 29/152 (19%)
Query: 16 NEDRPTDTETGYNKTSYSTDEPSGGDYDS-GYK-KSSYETSGG--GYETGYNKTSS---Y 68
NE RP D+ PSGGD+ GY + Y GG G GY S Y
Sbjct: 380 NEPRPEDSR------------PSGGDFRGRGYGGERGYRGRGGRGGDRGGYGGDRSGGGY 427
Query: 69 STDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYS--TGGGYG--TGGGGGGEY 124
D S GGGY + Y D SGGGYG G GGGY GGGYG GGG GG+
Sbjct: 428 GGDR--SSGGGYSGDRSGGGYGGDR-SGGGYG-GDRGGGYGGDRGGGYGGDRGGGYGGDR 483
Query: 125 STGGG--GGGYSTGGGGYGNDDSGNRRDEVDY 154
GG GGGY GGYG D G D Y
Sbjct: 484 GGYGGDRGGGYGGDRGGYGGDRGGYGGDRGGY 515
Score = 69.3 bits (168), Expect = 8e-12
Identities = 48/123 (39%), Positives = 52/123 (42%), Gaps = 10/123 (8%)
Query: 32 YSTDEPSGGDYDSGYKKSSY--ETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAY 89
Y D SGG Y Y + SGGGY G ++ Y D GG GY
Sbjct: 427 YGGDRSSGGGYSGDRSGGGYGGDRSGGGY--GGDRGGGYGGDRGGGYGGDRGGGYGGDRG 484
Query: 90 STDEPSGGGY-----GTGGAGGGY-STGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGND 143
GGGY G GG GGY GGYG GG G + GG GG GG GYG D
Sbjct: 485 GYGGDRGGGYGGDRGGYGGDRGGYGGDRGGYGGDRGGYGGDRSRGGYGGDRGGGSGYGGD 544
Query: 144 DSG 146
SG
Sbjct: 545 RSG 547
Score = 61.2 bits (147), Expect = 2e-09
Identities = 50/137 (36%), Positives = 61/137 (44%), Gaps = 26/137 (18%)
Query: 39 GGDYDSGY---KKSSYETSGGGYETGYNKTSSYSTDEPNSGG--GGYDSGYNKTAYSTDE 93
GGD GY + Y GGY G ++ Y D GG GGY G ++ Y D
Sbjct: 464 GGDRGGGYGGDRGGGYGGDRGGY--GGDRGGGYGGDRGGYGGDRGGY--GGDRGGYGGDR 519
Query: 94 PSGGGYG----TGGAGGGYSTGGGYG--TGGGGGGEYSTGG----GGGGYSTGGGGYGND 143
GGYG GG GG G GYG GG GG+ S GG GGGY GGYG
Sbjct: 520 ---GGYGGDRSRGGYGGDRGGGSGYGGDRSGGYGGDRSGGGYGGDRGGGYGGDRGGYGGK 576
Query: 144 DSGNRRDEVDYKEEEKH 160
G DY+ ++++
Sbjct: 577 MGGRN----DYRNDQRN 589
Score = 45.8 bits (107), Expect = 9e-05
Identities = 40/125 (32%), Positives = 52/125 (41%), Gaps = 12/125 (9%)
Query: 33 STDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYST- 91
S D+P G DS ++S Y+ G Y+ N + + N Y S ++ T
Sbjct: 104 SYDQPDYGQQDSYDQQSGYDQHQGSYDEQSNYDQQHDSYSQNQQS--YHSQRENYSHHTQ 161
Query: 92 -DEPSGGGYGTGGAGGGYSTGGGYGTGG---GGGGEY--STGGGGGGYSTGGGGYGNDDS 145
D YG G G S GGG G GG G G S+GG GG+ G G+ D
Sbjct: 162 DDRRDVSRYGEDNRGYGGSQGGGRGRGGYDKDGRGPMTGSSGGDRGGFKNFG---GHRDY 218
Query: 146 GNRRD 150
G R D
Sbjct: 219 GPRTD 223
Score = 33.1 bits (74), Expect = 0.62
Identities = 33/122 (27%), Positives = 43/122 (35%), Gaps = 8/122 (6%)
Query: 38 SGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGG 97
SG SG ++ SY T G GY + S + + Y G N + YS+ S
Sbjct: 4 SGSYGQSGGEQQSYSTYGNPGSQGYGQASQSYSGYGQTTDSSY--GQNYSGYSSYGQSQS 61
Query: 98 GYGTGGAG------GGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGNRRDE 151
GY G YS G E S GG S YG DS +++
Sbjct: 62 GYSQSYGGYENQKQSSYSQQPYNNQGQQQNMESSGSQGGRAPSYDQPDYGQQDSYDQQSG 121
Query: 152 VD 153
D
Sbjct: 122 YD 123
>K2C1_HUMAN (P04264) Keratin, type II cytoskeletal 1 (Cytokeratin 1)
(K1) (CK 1) (67 kDa cytokeratin) (Hair alpha protein)
Length = 643
Score = 73.2 bits (178), Expect = 5e-13
Identities = 45/108 (41%), Positives = 53/108 (48%), Gaps = 19/108 (17%)
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG Y SG SSY + GG Y +G GGGG G + S+ GG
Sbjct: 521 GGGYGSG--GSSYGSGGGSYGSG--------------GGGGGGRGSYGSGGSSYGSGGGS 564
Query: 99 YGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSG 146
YG+GG GGG+ G YG+G GG GGGGG S+GG G G SG
Sbjct: 565 YGSGGGGGGH---GSYGSGSSSGGYRGGSGGGGGGSSGGRGSGGGSSG 609
Score = 70.5 bits (171), Expect = 4e-12
Identities = 46/98 (46%), Positives = 52/98 (52%), Gaps = 21/98 (21%)
Query: 55 GGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGG-GYSTGGG 113
GGGY +G SSY SGGG Y SG GGG G+ G+GG Y +GGG
Sbjct: 521 GGGYGSG---GSSYG-----SGGGSYGSGGG---------GGGGRGSYGSGGSSYGSGGG 563
Query: 114 -YGTGGGGG--GEYSTGGGGGGYSTGGGGYGNDDSGNR 148
YG+GGGGG G Y +G GGY G GG G SG R
Sbjct: 564 SYGSGGGGGGHGSYGSGSSSGGYRGGSGGGGGGSSGGR 601
Score = 68.6 bits (166), Expect = 1e-11
Identities = 51/124 (41%), Positives = 57/124 (45%), Gaps = 27/124 (21%)
Query: 30 TSYSTDEPSGGDYDSGYKKSSYETSGGGYETG--------YNKTSSYSTDEPNSGGGGYD 81
TS ST GG G + SS GG + G N S S + GGG
Sbjct: 31 TSSSTRRSGGG----GGRFSSCGGGGGSFGAGGGFGSRSLVNLGGSKSISISVARGGGRG 86
Query: 82 SGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEY----STGGGGGGYSTGG 137
SG+ GGGYG GG GGG GGG+G GG GGG + S GGG GG GG
Sbjct: 87 SGF-----------GGGYGGGGFGGGGFGGGGFGGGGIGGGGFGGFGSGGGGFGGGGFGG 135
Query: 138 GGYG 141
GGYG
Sbjct: 136 GGYG 139
Score = 66.6 bits (161), Expect = 5e-11
Identities = 42/105 (40%), Positives = 51/105 (48%), Gaps = 20/105 (19%)
Query: 59 ETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGG 118
E N + S ST GGG G GGGYG+GG+ G S GG YG+GG
Sbjct: 495 ECAPNVSVSVSTSHTTISGGGSRGG-----------GGGGYGSGGSSYG-SGGGSYGSGG 542
Query: 119 GGGG---EYSTGG-----GGGGYSTGGGGYGNDDSGNRRDEVDYK 155
GGGG Y +GG GGG Y +GGGG G+ G+ Y+
Sbjct: 543 GGGGGRGSYGSGGSSYGSGGGSYGSGGGGGGHGSYGSGSSSGGYR 587
Score = 63.9 bits (154), Expect = 3e-10
Identities = 44/135 (32%), Positives = 62/135 (45%), Gaps = 15/135 (11%)
Query: 9 GLFHHHKNEDRPTDTETGYNKTSYSTDEPSGGDYDSG--YKKSSYETSGGGYETGYNKTS 66
G+ ++ + + +G +S+ GG + +G + S GG +K+
Sbjct: 22 GIINYQRRTTSSSTRRSGGGGGRFSSCGGGGGSFGAGGGFGSRSLVNLGG------SKSI 75
Query: 67 SYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYST 126
S S G G+ GY + GGG+G GG GGG GG+G+GGGG G
Sbjct: 76 SISVARGGGRGSGFGGGYGGGGFGGGGFGGGGFGGGGIGGG--GFGGFGSGGGGFG--GG 131
Query: 127 GGGGGGYSTGGGGYG 141
G GGGGY GGGYG
Sbjct: 132 GFGGGGY---GGGYG 143
Score = 52.4 bits (124), Expect = 1e-06
Identities = 37/109 (33%), Positives = 47/109 (42%), Gaps = 12/109 (11%)
Query: 26 GYNKTSYSTDEPSGGDYDSGYK----KSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYD 81
G +SY + GG Y SG + SY + G Y +G SY + G G Y
Sbjct: 525 GSGGSSYGS---GGGSYGSGGGGGGGRGSYGSGGSSYGSG---GGSYGSGGGGGGHGSYG 578
Query: 82 SGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGY--GTGGGGGGEYSTGG 128
SG + Y GGG +GG G G + GG G G GG S+GG
Sbjct: 579 SGSSSGGYRGGSGGGGGGSSGGRGSGGGSSGGSIGGRGSSSGGVKSSGG 627
Score = 51.6 bits (122), Expect = 2e-06
Identities = 39/122 (31%), Positives = 49/122 (39%), Gaps = 44/122 (36%)
Query: 68 YSTDEPNSGGGGYDSG------YNKTAYSTDEPS-----------GGGYGTGGAGGGYST 110
+S+ GGG+ SG Y + S+ GGG G+ GAGGG+ +
Sbjct: 4 FSSRSGYRSGGGFSSGSAGIINYQRRTTSSSTRRSGGGGGRFSSCGGGGGSFGAGGGFGS 63
Query: 111 --------------------------GGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDD 144
GGGYG GG GGG + GG GGG GGGG+G
Sbjct: 64 RSLVNLGGSKSISISVARGGGRGSGFGGGYGGGGFGGGGFGGGGFGGG-GIGGGGFGGFG 122
Query: 145 SG 146
SG
Sbjct: 123 SG 124
>CORA_MEDSA (Q07202) Cold and drought-regulated protein CORA
Length = 204
Score = 73.2 bits (178), Expect = 5e-13
Identities = 58/146 (39%), Positives = 69/146 (46%), Gaps = 38/146 (26%)
Query: 21 TDTETGYNKTSYS-TDEPSGGDYDSGYKKSSYETSGGGYETG-YNKTSSYSTDEPNSGGG 78
T+T T K T+E + Y GY GGGY G YN Y N GGG
Sbjct: 28 TETSTDAKKEVVEKTNEVNDAKYGGGYNH------GGGYNGGGYNHGGGY-----NHGGG 76
Query: 79 GYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGG-----GGGY 133
GY +G GGGY GG GGY+ GGG+G GGGG Y+ GGG GGGY
Sbjct: 77 GYHNG------------GGGYNHGG--GGYNGGGGHGGHGGGG--YNGGGGHGGHGGGGY 120
Query: 134 STGGGGYGNDDSGNRRDEVDYKEEEK 159
+ GGGG+G + V + EEK
Sbjct: 121 N-GGGGHGGHGGA---ESVAVQTEEK 142
Score = 64.7 bits (156), Expect = 2e-10
Identities = 50/154 (32%), Positives = 60/154 (38%), Gaps = 47/154 (30%)
Query: 29 KTSYSTDEPSGGDYD--SGYKKSSYETSGGGYETG----YNKTSSYSTDEPNSGGGGYDS 82
KT+ D GG Y+ GY Y GGGY G +N Y N GGGGY+
Sbjct: 41 KTNEVNDAKYGGGYNHGGGYNGGGYN-HGGGYNHGGGGYHNGGGGY-----NHGGGGYNG 94
Query: 83 GYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGG----------------------- 119
G + +GGG G GGGY+ GGG+G GG
Sbjct: 95 GGGHGGHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGGAESVAVQTEEKTNEVNDAKYGGG 154
Query: 120 ------------GGGEYSTGGGGGGYSTGGGGYG 141
GGG Y+ GGGG G G GG+G
Sbjct: 155 SNYNDGRGGYNHGGGGYNHGGGGHGGHGGHGGHG 188
>GRP8_ARATH (Q03251) Glycine-rich RNA-binding protein 8 (CCR1
protein)
Length = 169
Score = 72.8 bits (177), Expect = 7e-13
Identities = 34/54 (62%), Positives = 37/54 (67%), Gaps = 1/54 (1%)
Query: 96 GGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGNRR 149
GGG G GG+GGGY +GGG G GGGGG YS GGGGGGY GGYG+ G R
Sbjct: 89 GGGGGRGGSGGGYRSGGGGGYSGGGGGGYS-GGGGGGYERRSGGYGSGGGGGGR 141
Score = 70.5 bits (171), Expect = 4e-12
Identities = 43/97 (44%), Positives = 45/97 (46%), Gaps = 29/97 (29%)
Query: 54 SGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGG 113
SGGGY +G GGGGY G GGGY GG GG GG
Sbjct: 97 SGGGYRSG--------------GGGGYSGG-----------GGGGYSGGGGGGYERRSGG 131
Query: 114 YGTGGGGGGEYSTGGG---GGGYSTG-GGGYGNDDSG 146
YG+GGGGGG GGG GGGY G GG YG G
Sbjct: 132 YGSGGGGGGRGYGGGGRREGGGYGGGDGGSYGGGGGG 168
Score = 67.0 bits (162), Expect = 4e-11
Identities = 42/86 (48%), Positives = 45/86 (51%), Gaps = 24/86 (27%)
Query: 75 SGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGG-------GEYSTG 127
SGGGG G SGGGY +GG GGGYS GGG G GGGG G Y +G
Sbjct: 87 SGGGGGGRG----------GSGGGYRSGG-GGGYSGGGGGGYSGGGGGGYERRSGGYGSG 135
Query: 128 GGGGGYSTG------GGGYGNDDSGN 147
GGGGG G GGGYG D G+
Sbjct: 136 GGGGGRGYGGGGRREGGGYGGGDGGS 161
Score = 46.2 bits (108), Expect = 7e-05
Identities = 34/85 (40%), Positives = 35/85 (41%), Gaps = 13/85 (15%)
Query: 38 SGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGG 97
SGG Y SG GGGY G + SGGGG GY GG
Sbjct: 97 SGGGYRSGGGGGYSGGGGGGYSGGGGGGYERRSGGYGSGGGGGGRGYG----GGGRREGG 152
Query: 98 GYGTGGAGGGYSTGGGYGTGGGGGG 122
GYG GG GG Y GGGGGG
Sbjct: 153 GYG-GGDGGSY--------GGGGGG 168
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.303 0.131 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 40,949,016
Number of Sequences: 164201
Number of extensions: 2585202
Number of successful extensions: 65911
Number of sequences better than 10.0: 1449
Number of HSP's better than 10.0 without gapping: 1003
Number of HSP's successfully gapped in prelim test: 465
Number of HSP's that attempted gapping in prelim test: 10819
Number of HSP's gapped (non-prelim): 13299
length of query: 243
length of database: 59,974,054
effective HSP length: 107
effective length of query: 136
effective length of database: 42,404,547
effective search space: 5767018392
effective search space used: 5767018392
T: 11
A: 40
X1: 17 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0052b.4


