

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0052a.3
(242 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
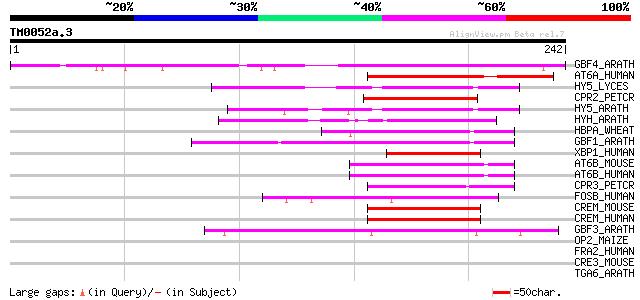
Score E
Sequences producing significant alignments: (bits) Value
GBF4_ARATH (P42777) G-box binding factor 4 (AtbZIP40) 190 3e-48
AT6A_HUMAN (P18850) Cyclic-AMP-dependent transcription factor AT... 56 9e-08
HY5_LYCES (Q9SM50) Transcription factor HY5 (LeHY5) (tHY5) 53 8e-07
CPR2_PETCR (Q99090) Light-inducible protein CPRF-2 52 2e-06
HY5_ARATH (O24646) Transcription factor HY5 (LONG HYPOCOL5 prote... 50 5e-06
HYH_ARATH (Q8W191) Transcription factor HY5-like (HY5 homolog) 49 1e-05
HBPA_WHEAT (P23922) Transcription factor HBP-1a (Histone-specifi... 47 4e-05
GBF1_ARATH (P42774) G-box binding factor 1 (AtbZIP41) 45 1e-04
XBP1_HUMAN (P17861) X box binding protein-1 (XBP-1) (TREB5 protein) 44 3e-04
AT6B_MOUSE (O35451) Cyclic-AMP-dependent transcription factor AT... 44 3e-04
AT6B_HUMAN (Q99941) Cyclic-AMP-dependent transcription factor AT... 44 3e-04
CPR3_PETCR (Q99091) Light-inducible protein CPRF-3 44 5e-04
FOSB_HUMAN (P53539) Protein fosB (G0/G1 switch regulatory protei... 43 6e-04
CREM_MOUSE (P27699) cAMP responsive element modulator 43 6e-04
CREM_HUMAN (Q03060) cAMP responsive element modulator 43 6e-04
GBF3_ARATH (P42776) G-box binding factor 3 (AtbZIP55) 43 8e-04
OP2_MAIZE (P12959) Opaque-2 regulatory protein 42 0.002
FRA2_HUMAN (P15408) Fos-related antigen 2 41 0.002
CRE3_MOUSE (Q61817) Cyclic-AMP responsive element binding protei... 41 0.002
TGA6_ARATH (Q39140) Transcription factor TGA6 (AtbZIP45) 40 0.004
>GBF4_ARATH (P42777) G-box binding factor 4 (AtbZIP40)
Length = 270
Score = 190 bits (482), Expect = 3e-48
Identities = 130/289 (44%), Positives = 169/289 (57%), Gaps = 66/289 (22%)
Query: 1 MASSKMMMSSSKSLDHQQPSSPPPSSMPQFTTTNFI------DHA-----------DSTL 43
MAS K+M SS+ L + SS SS P +++ + DH+ D T
Sbjct: 1 MASFKLMSSSNSDLSRRNSSSA--SSSPSIRSSHHLRPNPHADHSRISFAYGGGVNDYTF 58
Query: 44 TDPYVP--------------------RTVDDVWQEIIAGDHH--QCKEEIPDEMMTLEDF 81
P ++VDDVW+EI++G+ KEE P+++MTLEDF
Sbjct: 59 ASDSKPFEMAIDVDRSIGDRNSVNNGKSVDDVWKEIVSGEQKTIMMKEEEPEDIMTLEDF 118
Query: 82 LVKAGAVDGDDEDDEVKMSMPLTERLS--GVFSLD-----GSSFQGMEGVEGGSASVGGF 134
L KA +G ++ +VK+ TERL+ G ++ D SSFQ +EG GG
Sbjct: 119 LAKAEMDEGASDEIDVKIP---TERLNNDGSYTFDFPMQRHSSFQMVEGSMGGGV----- 170
Query: 135 GNGVEVVEGTRGKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVK 194
TRGKRGR ++E +DKAA QRQ+RMIKNRESAARSRERKQAYQVELE+LA K
Sbjct: 171 ---------TRGKRGRVMMEAMDKAAAQRQKRMIKNRESAARSRERKQAYQVELETLAAK 221
Query: 195 LEEENDKLLKEKAERTKERFKQLMEEVIPVVEKRRPP-RPLRRINSMQW 242
LEEEN++LLKE E TKER+K+LME +IPV EK RPP RPL R +S++W
Sbjct: 222 LEEENEQLLKEIEESTKERYKKLMEVLIPVDEKPRPPSRPLSRSHSLEW 270
>AT6A_HUMAN (P18850) Cyclic-AMP-dependent transcription factor ATF-6
alpha (Activating transcription factor 6 alpha)
(ATF6-alpha)
Length = 670
Score = 55.8 bits (133), Expect = 9e-08
Identities = 31/81 (38%), Positives = 49/81 (60%), Gaps = 5/81 (6%)
Query: 157 DKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAERTKERFKQ 216
D A +RQ+RMIKNRESA +SR++K+ Y + LE+ EN++L KE K+
Sbjct: 303 DIAVLRRQQRMIKNRESACQSRKKKKEYMLGLEARLKAALSENEQLKKENG-----TLKR 357
Query: 217 LMEEVIPVVEKRRPPRPLRRI 237
++EV+ ++ + P P RR+
Sbjct: 358 QLDEVVSENQRLKVPSPKRRV 378
>HY5_LYCES (Q9SM50) Transcription factor HY5 (LeHY5) (tHY5)
Length = 158
Score = 52.8 bits (125), Expect = 8e-07
Identities = 38/134 (28%), Positives = 66/134 (48%), Gaps = 19/134 (14%)
Query: 89 DGDDEDDEVKMSMPLTERLSGVFSLDGSSFQGMEGVEGGSASVGGFGNGVEVVEGTRGKR 148
+G + DDE++ + +G S G G SA GT+ KR
Sbjct: 32 EGMESDDEIRRVPEMGGEATGTTSASGRDGVSAAGQAQPSA-------------GTQRKR 78
Query: 149 GRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAE 208
GR + +K R +R+++NR SA ++RERK+AY ++LE+ +LE +N +L E+
Sbjct: 79 GRSPADKENK----RLKRLLRNRVSAQQARERKKAYLIDLEARVKELETKNAEL--EERL 132
Query: 209 RTKERFKQLMEEVI 222
T + Q++ ++
Sbjct: 133 STLQNENQMLRHIL 146
>CPR2_PETCR (Q99090) Light-inducible protein CPRF-2
Length = 401
Score = 51.6 bits (122), Expect = 2e-06
Identities = 27/50 (54%), Positives = 33/50 (66%)
Query: 155 NLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLK 204
N D + +R RRM+ NRESA RSR RKQA+ ELE+ +L EN LLK
Sbjct: 193 NGDPSDAKRVRRMLSNRESARRSRRRKQAHMTELETQVSQLRVENSSLLK 242
>HY5_ARATH (O24646) Transcription factor HY5 (LONG HYPOCOL5 protein)
(AtbZIP56)
Length = 168
Score = 50.1 bits (118), Expect = 5e-06
Identities = 42/132 (31%), Positives = 66/132 (49%), Gaps = 17/132 (12%)
Query: 96 EVKMSMPLTERLSGVFSLDGSSF-QGMEGVEGGSASVGGFGNGVEVVEGTRG----KRGR 150
E+K + E + V G + + G E GSA+ G E + T G KRGR
Sbjct: 29 EIKEGIESDEEIRRVPEFGGEAVGKETSGRESGSAT------GQERTQATVGESQRKRGR 82
Query: 151 PVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAERT 210
E +K R +R+++NR SA ++RERK+AY ELE+ LE +N +L E+ T
Sbjct: 83 TPAEKENK----RLKRLLRNRVSAQQARERKKAYLSELENRVKDLENKNSEL--EERLST 136
Query: 211 KERFKQLMEEVI 222
+ Q++ ++
Sbjct: 137 LQNENQMLRHIL 148
>HYH_ARATH (Q8W191) Transcription factor HY5-like (HY5 homolog)
Length = 149
Score = 48.5 bits (114), Expect = 1e-05
Identities = 38/121 (31%), Positives = 60/121 (49%), Gaps = 12/121 (9%)
Query: 92 DEDDEVKMSMPLTERLSGVFSLDGSSFQGMEGVEGGSASVGGFGNGVEVVEGTRGKRGRP 151
+E DE + +P E L S+ G+ E NGV + RG+ P
Sbjct: 22 EESDEELLMVPDMEAAGSTCVLSSSADDGVNNPELDQTQ-----NGVSTAKRRRGRN--P 74
Query: 152 VVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAERTK 211
V DK + +R+++NR SA ++RERK+ Y +LES A +L+ ND+L ++ + T
Sbjct: 75 V----DKEYRSL-KRLLRNRVSAQQARERKKVYVSDLESRANELQNNNDQLEEKISTLTN 129
Query: 212 E 212
E
Sbjct: 130 E 130
>HBPA_WHEAT (P23922) Transcription factor HBP-1a (Histone-specific
transcription factor HBP1)
Length = 349
Score = 47.0 bits (110), Expect = 4e-05
Identities = 32/86 (37%), Positives = 47/86 (54%), Gaps = 3/86 (3%)
Query: 137 GVEVVEGTRGK--RGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVK 194
G V RGK G E D+ ++Q+R + NRESA RSR RKQA EL A
Sbjct: 227 GSSPVPAMRGKVPSGSARGEQWDERELKKQKRKLSNRESARRSRLRKQAECEELGQRAEA 286
Query: 195 LEEENDKLLKEKAERTKERFKQLMEE 220
L+ EN L+ + +R K+ +++L+ +
Sbjct: 287 LKSENSS-LRIELDRIKKEYEELLSK 311
>GBF1_ARATH (P42774) G-box binding factor 1 (AtbZIP41)
Length = 315
Score = 45.4 bits (106), Expect = 1e-04
Identities = 42/141 (29%), Positives = 61/141 (42%), Gaps = 2/141 (1%)
Query: 80 DFLVKAGAVDGDDEDDEVKMSMPLTERLSGVFSLDGSSFQGMEGVEGGSASVGGFGNGVE 139
D V AG+ D +DE+ + + + G D SS Q G GS + G
Sbjct: 143 DESVTAGSSDENDENANQQEQGSIRKPSFGQMLADASS-QSTTGEIQGSVPMKPVAPGTN 201
Query: 140 VVEGTRGKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEEN 199
+ G + V D+ +RQ+R NRESA RSR RKQA +L+ L EN
Sbjct: 202 LNIGMDLWSSQAGVPVKDERELKRQKRKQSNRESARRSRLRKQAECEQLQQRVESLSNEN 261
Query: 200 DKLLKEKAERTKERFKQLMEE 220
L+++ +R +L E
Sbjct: 262 QS-LRDELQRLSSECDKLKSE 281
>XBP1_HUMAN (P17861) X box binding protein-1 (XBP-1) (TREB5 protein)
Length = 260
Score = 44.3 bits (103), Expect = 3e-04
Identities = 23/41 (56%), Positives = 28/41 (68%)
Query: 165 RRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKE 205
RR +KNR +A +R+RK+A ELE V LEEEN KLL E
Sbjct: 74 RRKLKNRVAAQTARDRKKARMSELEQQVVDLEEENQKLLLE 114
>AT6B_MOUSE (O35451) Cyclic-AMP-dependent transcription factor ATF-6
beta (Activating transcription factor 6 beta)
(ATF6-beta) (cAMP responsive element binding
protein-like 1) (cAMP response element binding
protein-related protein) (Creb-rp)
Length = 699
Score = 43.9 bits (102), Expect = 3e-04
Identities = 26/72 (36%), Positives = 40/72 (55%), Gaps = 1/72 (1%)
Query: 149 GRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAE 208
G +D +RQ+RMIKNRESA +SR +K+ Y LE+ + +N +L +E A
Sbjct: 311 GNSCPPEVDAKLLKRQQRMIKNRESACQSRRKKKEYLQGLEARLQAVLADNQQLRRENA- 369
Query: 209 RTKERFKQLMEE 220
+ R + L+ E
Sbjct: 370 ALRRRLEALLAE 381
>AT6B_HUMAN (Q99941) Cyclic-AMP-dependent transcription factor ATF-6
beta (Activating transcription factor 6 beta)
(ATF6-beta) (cAMP responsive element binding
protein-like 1) (cAMP response element binding
protein-related protein) (Creb-rp) (G13 protein)
Length = 703
Score = 43.9 bits (102), Expect = 3e-04
Identities = 26/72 (36%), Positives = 40/72 (55%), Gaps = 1/72 (1%)
Query: 149 GRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAE 208
G +D +RQ+RMIKNRESA +SR +K+ Y LE+ + +N +L +E A
Sbjct: 314 GNSCPPEVDAKLLKRQQRMIKNRESACQSRRKKKEYLQGLEARLQAVLADNQQLRRENA- 372
Query: 209 RTKERFKQLMEE 220
+ R + L+ E
Sbjct: 373 ALRRRLEALLAE 384
>CPR3_PETCR (Q99091) Light-inducible protein CPRF-3
Length = 296
Score = 43.5 bits (101), Expect = 5e-04
Identities = 26/64 (40%), Positives = 37/64 (57%), Gaps = 1/64 (1%)
Query: 157 DKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAERTKERFKQ 216
D+ +RQRR NRESA RSR RKQA EL+ L +EN ++L++ +R E +
Sbjct: 193 DERELKRQRRKQSNRESARRSRLRKQAKSDELQERLDNLSKEN-RILRKNLQRISEACAE 251
Query: 217 LMEE 220
+ E
Sbjct: 252 VTSE 255
>FOSB_HUMAN (P53539) Protein fosB (G0/G1 switch regulatory protein
3)
Length = 338
Score = 43.1 bits (100), Expect = 6e-04
Identities = 33/112 (29%), Positives = 54/112 (47%), Gaps = 9/112 (8%)
Query: 111 FSLDGSSFQ--GMEGVEGGSAS------VGGFGNGVEVVEGTRGKRGRPVVENLDKAAQQ 162
+ + G+S+ GM G G AS G +G R + RP E L ++
Sbjct: 97 YDMPGTSYSTPGMSGYSSGGASGSGGPSTSGTTSGPGPARPARARPRRPREETLTPEEEE 156
Query: 163 RQR-RMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAERTKER 213
++R R +N+ +AA+ R R++ L++ +LEEE +L E AE KE+
Sbjct: 157 KRRVRRERNKLAAAKCRNRRRELTDRLQAETDQLEEEKAELESEIAELQKEK 208
>CREM_MOUSE (P27699) cAMP responsive element modulator
Length = 341
Score = 43.1 bits (100), Expect = 6e-04
Identities = 21/49 (42%), Positives = 33/49 (66%)
Query: 157 DKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKE 205
++A ++R+ R++KNRE+A R RK+ Y LES LE +N KL++E
Sbjct: 280 EEATRKRELRLMKNREAAKECRRRKKEYVKCLESRVAVLEVQNKKLIEE 328
>CREM_HUMAN (Q03060) cAMP responsive element modulator
Length = 332
Score = 43.1 bits (100), Expect = 6e-04
Identities = 21/49 (42%), Positives = 33/49 (66%)
Query: 157 DKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKE 205
++A ++R+ R++KNRE+A R RK+ Y LES LE +N KL++E
Sbjct: 271 EEATRKRELRLMKNREAAKECRRRKKEYVKCLESRVAVLEVQNKKLIEE 319
>GBF3_ARATH (P42776) G-box binding factor 3 (AtbZIP55)
Length = 382
Score = 42.7 bits (99), Expect = 8e-04
Identities = 47/177 (26%), Positives = 74/177 (41%), Gaps = 23/177 (12%)
Query: 86 GAVDGDD------EDDEVKMSMPLTERLSGVFSLDGSSFQGMEGVEGGSASVGGFGNGVE 139
G+ DG D ++ ++K S T G + SSF + G + G+G
Sbjct: 169 GSTDGSDGNTTGADEPKLKRSREGTPTKDGKQLVQASSFHSVSPSSGDTGVKLIQGSGAI 228
Query: 140 VVEGTRGKRGRPVVENL----------DKAAQQRQRRMIKNRESAARSRERKQAYQVELE 189
+ G + ++L ++ +R+RR NRESA RSR RKQA EL
Sbjct: 229 LSPGVSANSNPFMSQSLAMVPPETWLQNERELKRERRKQSNRESARRSRLRKQAETEELA 288
Query: 190 SLAVKLEEENDKL------LKEKAERTKERFKQLMEEV-IPVVEKRRPPRPLRRINS 239
L EN L L EK+++ + L++++ EKR P L R+ +
Sbjct: 289 RKVEALTAENMALRSELNQLNEKSDKLRGANATLLDKLKCSEPEKRVPANMLSRVKN 345
>OP2_MAIZE (P12959) Opaque-2 regulatory protein
Length = 453
Score = 41.6 bits (96), Expect = 0.002
Identities = 28/82 (34%), Positives = 40/82 (48%), Gaps = 9/82 (10%)
Query: 135 GNGVEVVEGTRGKRGRPVVENLD---------KAAQQRQRRMIKNRESAARSRERKQAYQ 185
G GV + + + P E++D ++R R+ NRESA RSR RK A+
Sbjct: 191 GLGVRLATSSSSRDPSPSDEDMDGEVEILGFKMPTEERVRKKESNRESARRSRYRKAAHL 250
Query: 186 VELESLAVKLEEENDKLLKEKA 207
ELE +L+ EN LL+ A
Sbjct: 251 KELEDQVAQLKAENSCLLRRIA 272
>FRA2_HUMAN (P15408) Fos-related antigen 2
Length = 326
Score = 41.2 bits (95), Expect = 0.002
Identities = 33/104 (31%), Positives = 53/104 (50%), Gaps = 10/104 (9%)
Query: 138 VEVVEGTRGKRGRPVVENLDKAAQQRQR-RMIKNRESAARSRERKQAYQVELESLAVKLE 196
++ + T G+R R E L ++++R R +N+ +AA+ R R++ +L++ +LE
Sbjct: 103 IKTIGTTVGRRRRD--EQLSPEEEEKRRIRRERNKLAAAKCRNRRRELTEKLQAETEELE 160
Query: 197 EENDKLLKEKAERTKERFKQLMEEV-------IPVVEKRRPPRP 233
EE L KE AE KE+ K V I E+R PP P
Sbjct: 161 EEKSGLQKEIAELQKEKEKLEFMLVAHGPVCKISPEERRSPPAP 204
>CRE3_MOUSE (Q61817) Cyclic-AMP responsive element binding protein 3
(Transcription factor LZIP)
Length = 404
Score = 41.2 bits (95), Expect = 0.002
Identities = 21/66 (31%), Positives = 42/66 (62%), Gaps = 1/66 (1%)
Query: 156 LDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAERTKERFK 215
+++ +R RR I+N+ +A SR++K+ Y V LES +K +N + L+ K +R +E+
Sbjct: 181 VEEQVLKRVRRKIRNKRAAQESRKKKKVYVVGLESRVLKYTAQN-RELQNKVQRLEEQNL 239
Query: 216 QLMEEV 221
L++++
Sbjct: 240 SLLDQL 245
>TGA6_ARATH (Q39140) Transcription factor TGA6 (AtbZIP45)
Length = 330
Score = 40.4 bits (93), Expect = 0.004
Identities = 20/52 (38%), Positives = 34/52 (64%)
Query: 157 DKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAE 208
DK Q+ RR+ +NRE+A +SR RK+AY +LE+ +KL + +L + + +
Sbjct: 41 DKLDQKTLRRLAQNREAARKSRLRKKAYVQQLENSRLKLTQLEQELQRARQQ 92
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.312 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,268,397
Number of Sequences: 164201
Number of extensions: 1253331
Number of successful extensions: 8007
Number of sequences better than 10.0: 431
Number of HSP's better than 10.0 without gapping: 181
Number of HSP's successfully gapped in prelim test: 254
Number of HSP's that attempted gapping in prelim test: 7082
Number of HSP's gapped (non-prelim): 1028
length of query: 242
length of database: 59,974,054
effective HSP length: 107
effective length of query: 135
effective length of database: 42,404,547
effective search space: 5724613845
effective search space used: 5724613845
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0052a.3


