

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0051a.1
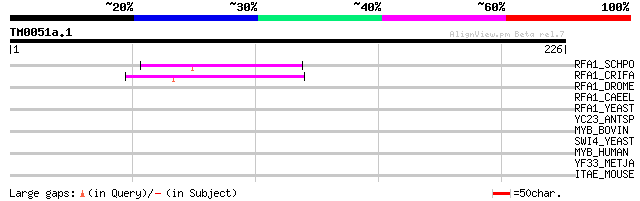
(226 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
RFA1_SCHPO (Q92372) Replication factor-A protein 1 (Single-stran... 45 2e-04
RFA1_CRIFA (Q23696) Replication factor A 51 kDa subunit (RP-A) (... 45 2e-04
RFA1_DROME (Q24492) Replication protein A 70 kDa DNA-binding sub... 39 0.013
RFA1_CAEEL (Q19537) Probable replication factor A 73 kDa subunit... 35 0.11
RFA1_YEAST (P22336) Replication factor-A protein 1 (RF-A) (Singl... 31 2.7
YC23_ANTSP (P46314) Hypothetical 30.3 kDa protein ycf23 (ORF 277) 30 4.7
MYB_BOVIN (P46200) Myb proto-oncogene protein (C-myb) 30 4.7
SWI4_YEAST (P25302) Regulatory protein SWI4 (Cell-cycle box fact... 30 6.1
MYB_HUMAN (P10242) Myb proto-oncogene protein (C-myb) 30 6.1
YF33_METJA (Q58928) Hypothetical protein MJ1533 29 7.9
ITAE_MOUSE (Q60677) Integrin alpha-E precursor (Integrin alpha M... 29 7.9
>RFA1_SCHPO (Q92372) Replication factor-A protein 1 (Single-stranded
DNA-binding protein p68 subunit)
Length = 609
Score = 44.7 bits (104), Expect = 2e-04
Identities = 22/67 (32%), Positives = 33/67 (48%), Gaps = 1/67 (1%)
Query: 54 ECNSAAYSDGAQYYCEKCNK-HVTPTIRYKLLLNVADDTASATFTVFYREGCYLMNKKGN 112
+CN + G + CEKCNK + P RY + + V D T VF G +M+K +
Sbjct: 481 DCNKKVFDQGGSWRCEKCNKEYDAPQYRYIITIAVGDHTGQLWLNVFDDVGKLIMHKTAD 540
Query: 113 EILEKME 119
E+ + E
Sbjct: 541 ELNDLQE 547
>RFA1_CRIFA (Q23696) Replication factor A 51 kDa subunit (RP-A)
(RF-A) (Replication factor-A protein 1) (Single-stranded
DNA-binding protein P51 subunit)
Length = 467
Score = 44.7 bits (104), Expect = 2e-04
Identities = 22/76 (28%), Positives = 36/76 (46%), Gaps = 3/76 (3%)
Query: 48 WYYHDCECNSAAYSDGAQ---YYCEKCNKHVTPTIRYKLLLNVADDTASATFTVFYREGC 104
WY CN +GAQ + CEKC+ V PT RY + + V D+ + T+F G
Sbjct: 309 WYDACPTCNKKVTEEGAQGDRFRCEKCDATVVPTQRYLVSIQVTDNVSQVWLTLFNEAGV 368
Query: 105 YLMNKKGNEILEKMEK 120
+ +E+ + ++
Sbjct: 369 EFFGMEASELKRRAQE 384
>RFA1_DROME (Q24492) Replication protein A 70 kDa DNA-binding
subunit (RP-A) (RF-A) (Replication factor-A protein 1)
(Single-stranded DNA-binding protein) (DmRPA1)
Length = 603
Score = 38.5 bits (88), Expect = 0.013
Identities = 24/77 (31%), Positives = 39/77 (50%), Gaps = 7/77 (9%)
Query: 49 YYHDC---ECNSAAYSDGA-QYYCEKCNKHVTPTIRYKLLLN--VADDTASATFTVFYRE 102
+Y C +CN +G Q+ CEKCN + P +Y+LL+N + D T++ + F
Sbjct: 460 FYRACPQSDCNKKVVDEGNDQFRCEKCNA-LFPNFKYRLLINMSIGDWTSNRWVSSFNEV 518
Query: 103 GCYLMNKKGNEILEKME 119
G L+ E+ E +E
Sbjct: 519 GEQLLGHTSQEVGEALE 535
>RFA1_CAEEL (Q19537) Probable replication factor A 73 kDa subunit
(RP-A) (RF-A) (Replication factor-A protein 1)
Length = 655
Score = 35.4 bits (80), Expect = 0.11
Identities = 24/89 (26%), Positives = 37/89 (40%), Gaps = 9/89 (10%)
Query: 36 YATIK----KIESTNDWYYHDCE---CNSAAYSDGAQYYCEKCNKHVTP-TIRYKLLLNV 87
YAT+K ++ TN Y C C + Y CEKCNK++ Y + +
Sbjct: 498 YATVKAMITRVNPTNA-LYRGCASEGCQKKLVGENGDYRCEKCNKNMNKFKWLYMMQFEL 556
Query: 88 ADDTASATFTVFYREGCYLMNKKGNEILE 116
+D+T T F ++ K E+ E
Sbjct: 557 SDETGQVYVTAFGDSAAKIVGKSAAELGE 585
>RFA1_YEAST (P22336) Replication factor-A protein 1 (RF-A)
(Single-stranded DNA-binding protein) (DNA binding
protein BUF2) (Replication protein A 69 kDa DNA-binding
subunit)
Length = 621
Score = 30.8 bits (68), Expect = 2.7
Identities = 25/120 (20%), Positives = 50/120 (40%), Gaps = 7/120 (5%)
Query: 7 LYREVQEQCTILKSLPTKVQPQQRTIFIIYATIKKIESTNDWYYHDCE---CNSAAYS-- 61
L + + ++ TI ++ + ++ F +++ Y C CN
Sbjct: 440 LTKFIAQRITIARAQAENLGRSEKGDFFSVKAAISFLKVDNFAYPACSNENCNKKVLEQP 499
Query: 62 DGAQYYCEKCN-KHVTPTIRYKLLLNVADDTASATFTVFYREGCYLMNKKGNEILEKMEK 120
DG + CEKC+ + P RY L +++ D+T T+F + L+ N ++ E+
Sbjct: 500 DGT-WRCEKCDTNNARPNWRYILTISIIDETNQLWLTLFDDQAKQLLGVDANTLMSLKEE 558
>YC23_ANTSP (P46314) Hypothetical 30.3 kDa protein ycf23 (ORF 277)
Length = 277
Score = 30.0 bits (66), Expect = 4.7
Identities = 27/105 (25%), Positives = 40/105 (37%), Gaps = 18/105 (17%)
Query: 33 FIIYATIKKIESTNDW-------YYHDCECNSAAYSDGAQY-----------YCEKCNKH 74
FI IK I N++ H CE + A Y D A+ C
Sbjct: 12 FIQRKAIKVISGINNFNVGQIFKIIHACEISKATYVDVARNPKIVSFIKSISSIPVCVSS 71
Query: 75 VTPTIRYKLLLNVADDTASATFTVFYREGCYLMNKKGNEILEKME 119
+ P Y+ +L AD F FY G YL N + I+++++
Sbjct: 72 IDPRALYESVLAGADLVEIGNFDCFYTNGVYLSNDQIIAIVKQVK 116
>MYB_BOVIN (P46200) Myb proto-oncogene protein (C-myb)
Length = 640
Score = 30.0 bits (66), Expect = 4.7
Identities = 15/38 (39%), Positives = 19/38 (49%)
Query: 185 TPPPILEDSNTPIKRRSDEGSITEVVGSNDAEMFKKIK 222
TP ++ED IK+ SDE I N + KKIK
Sbjct: 490 TPSHLVEDLQDEIKQESDESGIVAEFQENGQPLLKKIK 527
>SWI4_YEAST (P25302) Regulatory protein SWI4 (Cell-cycle box factor,
chain SWI4) (ART1 protein)
Length = 1093
Score = 29.6 bits (65), Expect = 6.1
Identities = 21/64 (32%), Positives = 33/64 (50%), Gaps = 2/64 (3%)
Query: 158 NVLKICKDQQIISKFKTKTITKSVKRNTPPPILEDSNTPIKRRSDE-GSITEVVGSNDAE 216
N+ I +D I+ F T ++ K +K +TP ILE+S +RRS E N+ +
Sbjct: 772 NMNTIMEDLSNINSFVTSSVIKDIK-STPSKILENSPILYRRRSQSISDEKEKAKDNENQ 830
Query: 217 MFKK 220
+ KK
Sbjct: 831 VEKK 834
>MYB_HUMAN (P10242) Myb proto-oncogene protein (C-myb)
Length = 640
Score = 29.6 bits (65), Expect = 6.1
Identities = 15/38 (39%), Positives = 19/38 (49%)
Query: 185 TPPPILEDSNTPIKRRSDEGSITEVVGSNDAEMFKKIK 222
TP ++ED IK+ SDE I N + KKIK
Sbjct: 490 TPSHLVEDLQDVIKQESDESGIVAEFQENGPPLLKKIK 527
>YF33_METJA (Q58928) Hypothetical protein MJ1533
Length = 642
Score = 29.3 bits (64), Expect = 7.9
Identities = 32/122 (26%), Positives = 51/122 (41%), Gaps = 24/122 (19%)
Query: 26 QPQQRTIFI-----IYATIKKIES-------TNDWYYHDCECNSAAYSDGAQYYCEKCNK 73
+P + IF+ I A I+K+ T+DW + N A Y+ E +
Sbjct: 103 RPTREEIFLAKSGEIDAMIRKVAKETNSILLTSDWIQY----NLAKAQGIEAYFLEAAEE 158
Query: 74 HVTPTIRYKLLLNVADDTASATFTVFYREGCYLMNKKGNEILEKMEKIGSMEHPPQEIVD 133
V +L+L+ D T +V +EGC KKG K+ IG E +E+ D
Sbjct: 159 EV------ELVLDKYFD--EETMSVHLKEGCLPYAKKGKPGEVKLVPIGDKELTKEEMED 210
Query: 134 LV 135
++
Sbjct: 211 II 212
>ITAE_MOUSE (Q60677) Integrin alpha-E precursor (Integrin alpha M290)
Length = 1167
Score = 29.3 bits (64), Expect = 7.9
Identities = 17/45 (37%), Positives = 24/45 (52%), Gaps = 3/45 (6%)
Query: 136 HKEFLFKVEGKNICGRWFNPAFNVLKICKDQQIISKFKTKTITKS 180
HKEF F V G+N+ G F V +D QI+ + K +TK+
Sbjct: 976 HKEFFFNVHGENLFGAVFQLQICVPIKLQDFQIV---RVKNLTKT 1017
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,068,607
Number of Sequences: 164201
Number of extensions: 1096191
Number of successful extensions: 2886
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 2881
Number of HSP's gapped (non-prelim): 11
length of query: 226
length of database: 59,974,054
effective HSP length: 106
effective length of query: 120
effective length of database: 42,568,748
effective search space: 5108249760
effective search space used: 5108249760
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0051a.1


