

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0048.9
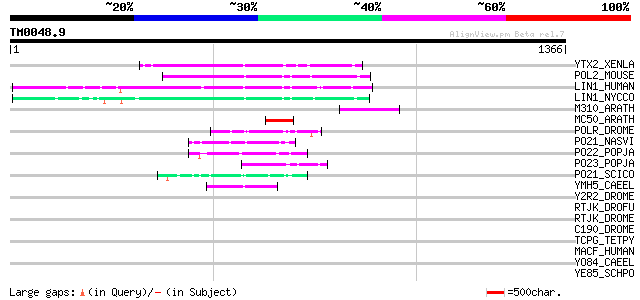
(1366 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 164 2e-39
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 157 1e-37
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 157 2e-37
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 153 4e-36
M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310... 122 5e-27
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 82 1e-14
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 75 9e-13
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 69 9e-11
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 67 3e-10
PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type... 61 2e-08
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 57 4e-07
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 53 6e-06
Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I retro... 43 0.007
RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile elem... 41 0.020
RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile elem... 39 0.074
C190_DROME (Q9VJE5) Restin homolog (Cytoplasmic linker protein 1... 39 0.13
TCPG_TETPY (P54408) T-complex protein 1, gamma subunit (TCP-1-ga... 37 0.28
MACF_HUMAN (Q9UPN3) Microtubule-actin crosslinking factor 1, iso... 36 0.82
YO84_CAEEL (P34620) Hypothetical protein ZK1236.4 in chromosome III 35 1.8
YE85_SCHPO (O14301) Hypothetical WD-repeat protein C9G1.05 in ch... 34 3.1
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 164 bits (415), Expect = 2e-39
Identities = 151/555 (27%), Positives = 248/555 (44%), Gaps = 32/555 (5%)
Query: 320 EEVQTEQVVRATREAEKNLDVLLKQEEIVWSQRSRANWLKHGDRNTKFFHSKATQRRKRN 379
+ +Q E + R +EA +N++ + V RSR L DR ++FF++ ++ R
Sbjct: 332 QALQCEYLER--KEALRNMEQRQARGAFV---RSRMQLLCDMDRGSRFFYALEKKKGNRK 386
Query: 380 LIEEIQDDNGRKFVRDADIARVLTSYFQEIFSTCNPTGIEEVATLVDGR--VTDAHRRIL 437
I + ++G I S++Q +FS +P + L DG V++ + L
Sbjct: 387 QITCLFAEDGTPLEDPEAIRDRARSFYQNLFSP-DPISPDACEELWDGLPVVSERRKERL 445
Query: 438 STPFTREDVEEALFQMHPTKAPGLDGFPALFYQKFWPIVGDDISEFCLQVLQGDISPGMV 497
TP T +++ +AL M K+PGLDG F+Q FW +G D + + P
Sbjct: 446 ETPITLDELSQALRLMPHNKSPGLDGLTIEFFQFFWDTLGPDFHRVLTEAFKKGELPLSC 505
Query: 498 NQTLIVLIPKVKKAVFANQYRPISLCNVIFKIISKTIANRMKLILHDLISEAQSAFVPGR 557
+ ++ L+PK +RP+SL + +KI++K I+ R+K +L ++I QS VPGR
Sbjct: 506 RRAVLSLLPKKGDLRLIKNWRPVSLLSTDYKIVAKAISLRLKSVLAEVIHPDQSYTVPGR 565
Query: 558 LITDNALIAFECFHYMKKRISGRSGMMALKLDMSKAYDRVEWPFLRSVLTHMGFPVHWVN 617
I DN + + H+ ++ +G S + L LD KA+DRV+ +L L F +V
Sbjct: 566 TIFDNVFLIRDLLHFARR--TGLS-LAFLSLDQEKAFDRVDHQYLIGTLQAYSFGPQFVG 622
Query: 618 LVMNCVTTVNFAVMLNGNPQPTFAPHRGLRQGDPLSPYLFILCGEAFSALIQKSISTSAL 677
+ + V +N + A RG+RQG PLS L+ L E F L++K ++
Sbjct: 623 YLKTMYASAECLVKINWSLTAPLAFGRGVRQGCPLSGQLYSLAIEPFLCLLRKRLT---- 678
Query: 678 HGIKISRSAPVISHLLFADDSVLFARATVQEAECIKNILATYERASGQKINLEKSMLSVS 737
G+ + + +ADD +L A+ V + E + Y AS +IN KS S
Sbjct: 679 -GLVLKEPDMRVVLSAYADDVILVAQDLV-DLERAQECQEVYAAASSARINWSKS----S 732
Query: 738 RNVPENCFVDL--QLLLGVKAVDSYDKYLGLPTIIGK-SKTQIFRFVKERVWKKLKGWK- 793
+ + VD + KYLG+ + +Q F ++E V +L WK
Sbjct: 733 GLLEGSLKVDFLPPAFRDISWESKIIKYLGVYLSAEEYPVSQNFIELEECVLTRLGKWKG 792
Query: 794 -EQTLSRAGREVLIKSVAQAIPSYVMSCFVLPDGLCNEIESMISRFYWGGDASKRGIHWV 852
+ LS GR ++I + + Y + C +I+ + F W G HWV
Sbjct: 793 FAKVLSMRGRALVINQLVASQIWYRLICLSPTQEFIAKIQRRLLDFLWIGK------HWV 846
Query: 853 KWKTMCQPKHVGGLG 867
P GG G
Sbjct: 847 SAGVSSLPLKEGGQG 861
Score = 34.7 bits (78), Expect = 1.8
Identities = 19/59 (32%), Positives = 30/59 (50%), Gaps = 2/59 (3%)
Query: 1057 WKRFWKAPSMPRVREVSWRVCTGALPVRTKLRQRGVDVDSSCPLCGLEDETVDHLFLGC 1115
W+ F+ + ++SW+V GAL L R D ++CP CG + E+V H + C
Sbjct: 1122 WRAFYSSLVPRPTGDLSWKVLHGALSTGEYL-ARFTDSPAACPFCG-KGESVFHAYFTC 1178
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 157 bits (398), Expect = 1e-37
Identities = 134/523 (25%), Positives = 241/523 (45%), Gaps = 27/523 (5%)
Query: 376 RKRNLIEEIQDDNGRKFVRDADIARVLTSYFQEIFSTCNPTGIEEVATLVDG----RVTD 431
R + LI +I+++ G +I + S+++ ++ST ++E+ +D ++
Sbjct: 411 RDKILINKIRNEKGDITTDPEEIQNTIRSFYKRLYST-KLENLDEMDKFLDRYQVPKLNQ 469
Query: 432 AHRRILSTPFTREDVEEALFQMHPTKAPGLDGFPALFYQKFWPIVGDDISEFCLQVLQGD 491
L++P + +++E + + K+PG DGF A FYQ F + + + ++
Sbjct: 470 DQVDHLNSPISPKEIEAVINSLPTKKSPGPDGFSAEFYQTFKEDLIPILHKLFHKIEVEG 529
Query: 492 ISPGMVNQTLIVLIPKVKK-AVFANQYRPISLCNVIFKIISKTIANRMKLILHDLISEAQ 550
P + I LIPK +K +RPISL N+ KI++K +ANR++ + +I Q
Sbjct: 530 TLPNSFYEATITLIPKPQKDPTKIENFRPISLMNIDAKILNKILANRIQEHIKAIIHPDQ 589
Query: 551 SAFVPGRLITDNALIAFECFHYMKKRISGRSGMMALKLDMSKAYDRVEWPFLRSVLTHMG 610
F+PG N + HY+ K M + LD KA+D+++ PF+ VL G
Sbjct: 590 VGFIPGMQGWFNIRKSINVIHYINKLKD--KNHMIISLDAEKAFDKIQHPFMIKVLERSG 647
Query: 611 FPVHWVNLVMNCVTTVNFAVMLNGNPQPTFAPHRGLRQGDPLSPYLFILCGEAFSALIQK 670
++N++ + + +NG G RQG PLSPYLF + E + I++
Sbjct: 648 IQGPYLNMIKAIYSKPVANIKVNGEKLEAIPLKSGTRQGCPLSPYLFNIVLEVLARAIRQ 707
Query: 671 SISTSALHGIKISRSAPVISHLLFADDSVLFARATVQEAECIKNILATYERASGQKINLE 730
+ GI+I + IS L ADD +++ + N++ ++ G KIN
Sbjct: 708 Q---KEIKGIQIGKEEVKIS--LLADDMIVYISDPKNSTRELLNLINSFGEVVGYKINSN 762
Query: 731 KSMLSV-SRNVPENCFVDLQLLLGVKAVDSYDKYLGLPTIIGKSK---TQIFRFVKERVW 786
KSM + ++N + +++ V + KYLG+ T+ + K + F+ +K+ +
Sbjct: 763 KSMAFLYTKN--KQAEKEIRETTPFSIVTNNIKYLGV-TLTKEVKDLYDKNFKSLKKEIK 819
Query: 787 KKLKGWKEQTLSRAGREVLIKS--VAQAIPSYVMSCFVLPDGLCNEIESMISRFYWGGDA 844
+ L+ WK+ S GR ++K + +AI + +P NE+E I +F W
Sbjct: 820 EDLRRWKDLPCSWIGRINIVKMAILPKAIYRFNAIPIKIPTQFFNELEGAICKFVWNNKK 879
Query: 845 SKRGIHWVKWKTMCQPKHVGGLGFRDFKSFNLALVAKNWWRIY 887
+ +K K + GG+ D K + A+V K W Y
Sbjct: 880 PRIAKSLLKDK-----RTSGGITMPDLKLYYRAIVIKTAWYWY 917
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 157 bits (396), Expect = 2e-37
Identities = 202/922 (21%), Positives = 387/922 (41%), Gaps = 74/922 (8%)
Query: 8 LLSWNCRGLGNLEAVRALRGLIHSQAPDIVFLMETKLFSFEMQRHRGMGGLSNIFPVQCG 67
+L+ N GL L I SQ P + + ET L + R + + G NI+ G
Sbjct: 9 ILTLNVNGLNAPIKRHRLANWIKSQDPSVCCIQETHLTCRDTHRLK-IKGWRNIYQAN-G 66
Query: 68 RNRAGGLCLLW--RSEVEVTIINASL--HHILFT-VVHSDSVTVPMQVYALYGFPELHLK 122
+ + G+ +L +++ + T I H+I+ + + +T+ + +YA +K
Sbjct: 67 KQKKAGVAILVSDKTDFKPTKIKRDKEGHYIMVKGSIQQEELTI-LNIYAPNTGAPRFIK 125
Query: 123 DRTWEMIRSVRPMAPIPWLCIGDFNDILSPSDKLGGAP--PDLARLQVATQACADCGLQR 180
++ R + I +GDFN LS D+ D+ L A + R
Sbjct: 126 QVLSDLQRDLDSHTII----MGDFNTPLSTLDRSTRQKINKDIQELNSALHQADLIDIYR 181
Query: 181 VDFSGPSFTWTNNRVGPGRVDERLDYALINQAWVNLWPVSSVSHLIRHQSDHNPVVLHCG 240
P T P + D+ L ++ ++ + + + SDH+ + L
Sbjct: 182 T--LHPKSTEYTFFSAPHHTYSKTDHILGSKTLLSKCKRTEI--ITNCLSDHSAIKLELR 237
Query: 241 SRRTETQRHRTRM----FRFEEVWLESGEECAEI-----ISEGWSNTNLSILSRINLVGR 291
++ TQ H T + W+ + E AEI +E T ++ V R
Sbjct: 238 IKKL-TQNHSTTWKLNNLLLNDYWVHN-EMKAEIKKFFETNENKDTTYQNLWDTAKAVCR 295
Query: 292 S-LDSWGREKYGELPKRIKEARAFLQRLQEEVQTEQVVRATREAEK---NLDVLLKQEEI 347
+ K + +I + L+ L+++ QT +E K L + Q+ +
Sbjct: 296 GKFIALNAHKRKQERSKIDTLISQLKELEKQEQTNSKASRRQEIIKIRAELKEIETQKTL 355
Query: 348 VWSQRSRANWLKHGDRNTKFFHSKATQRRKRNLIEEIQDDNGRKFVRDADIARVLTSYFQ 407
SR+ + + ++ + ++R++N I+ I++D G +I + Y++
Sbjct: 356 QKINESRSWFFEKINKIDRPLARLIKKKREKNQIDTIKNDRGDITTDPTEIQTTIREYYK 415
Query: 408 EIFSTCNPTGIEEVATLVDG----RVTDAHRRILSTPFTREDVEEALFQMHPTKAPGLDG 463
+++ +EE+ +D R+ L+ P T ++E + + K+PG +G
Sbjct: 416 HLYAN-KLENLEEMDKFLDTYTLPRLNQEEVESLNRPITSSEIEAIINSLPNKKSPGPEG 474
Query: 464 FPALFYQKFWPIVGDDISEFCLQVLQG----DISPGMVNQTLIVLIPKVKKAVFANQ-YR 518
F A FYQ++ +++ F L++ Q I P + I+LIPK + + +R
Sbjct: 475 FTAEFYQRY----KEELVPFLLKLFQSIEKEGILPNSFYEASIILIPKPGRDTTKKENFR 530
Query: 519 PISLCNVIFKIISKTIANRMKLILHDLISEAQSAFVPGRLITDNALIAFECFHYMKKRIS 578
PISL N+ KI++K +AN+++ + LI Q F+P N + ++ + +
Sbjct: 531 PISLMNIDAKILNKILANQIQQHIKKLIHHDQVGFIPAMQGWFNIRKSINIIQHINR--T 588
Query: 579 GRSGMMALKLDMSKAYDRVEWPFLRSVLTHMGFPVHWVNLVMNCVTTVNFAVMLNGNPQP 638
+ M + +D KA+D+++ PF+ L +G ++ ++ ++LNG
Sbjct: 589 KDTNHMIISIDAEKAFDKIQQPFMLKPLNKLGIDGTYLKIIRAIYDKPTANIILNGQKLE 648
Query: 639 TFAPHRGLRQGDPLSPYLFILCGEAFSALIQKSISTSALHGIKISRSAPVISHLLFADDS 698
G RQG PLSP L + E + I++ + GI++ + +S LFADD
Sbjct: 649 APPLKTGTRQGCPLSPLLPNIVLEVLARAIRQE---KEIKGIQLGKEEVKLS--LFADDM 703
Query: 699 VLFARATVQEAECIKNILATYERASGQKINLEKS---MLSVSRNVPENCFVDLQLLLGVK 755
+++ + A+ + +++ + + SG KIN++KS + + +R +L + K
Sbjct: 704 IVYLENPIVSAQNLLKLISNFSKVSGYKINVQKSQAFLYTNNRQTESQIMSELPFTIASK 763
Query: 756 AVDSYDKYLGLPTIIGKSKTQIFRFVKERVWKKLK----GWKEQTLSRAGREVLIKS--V 809
+ KYLG+ + + +F+ + + ++K WK S GR ++K +
Sbjct: 764 RI----KYLGIQ--LTRDVKDLFKENYKPLLNEIKEDTNKWKNIPCSWVGRINIVKMAIL 817
Query: 810 AQAIPSYVMSCFVLPDGLCNEIESMISRFYWGGDASKRGIHWVKWKTMCQPKHVGGLGFR 869
+ I + LP E+E +F W +++ H K T+ Q GG+
Sbjct: 818 PKVIYRFNAIPIKLPMTFFTELEKTTLKFIW----NQKRAHIAK-STLSQKNKAGGITLP 872
Query: 870 DFKSFNLALVAKNWWRIYNYPD 891
DFK + A V K W Y D
Sbjct: 873 DFKLYYKATVTKTAWYWYQNRD 894
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 153 bits (386), Expect = 4e-36
Identities = 193/930 (20%), Positives = 374/930 (39%), Gaps = 100/930 (10%)
Query: 6 MRLLSWNCRGLGNLEAVRALRGLIHSQAPDIVFLMETKLFSFEMQRHRGMGGLSNIFPVQ 65
+ + S N GL L I PDI + E+ L + R + + G S+IF
Sbjct: 7 LSIFSINVNGLNCPLKRHRLADWIQKLKPDICCIQESHLTLKDKYRLK-VKGWSSIFQAN 65
Query: 66 CGRNRAGGLCLLWRSEV---EVTIINASLHHILFTVVHS--DSVTVPMQVYALYGFPELH 120
G+ + G+ +L+ + I H +F ++ D +++ + +YA P +
Sbjct: 66 -GKQKKAGIAILFADAIGFKPTKIRKDKDGHFIFVKGNTQYDEISI-INIYA----PNHN 119
Query: 121 LKDRTWEMIRSVRPMAPIPWLCIGDFNDILSPSDKLGGAPPDLARLQVATQACADCGLQR 180
E + + + + +GDFN L+ D+ L + + +Q
Sbjct: 120 APQFIRETLTDMSNLISSTSIVVGDFNTPLAVLDRSSKKKLSKEILDLNST------IQH 173
Query: 181 VDFSGPSFTWTNNRVGPGRVDERLDYALINQAWVNLWPVSSVSHLIRHQS---------- 230
+D + T+ N+ +Y + A S + H++ H+S
Sbjct: 174 LDLTDIYRTFHPNKT---------EYTFFSSAHGTY---SKIDHILGHKSNLSKFKKIEI 221
Query: 231 ------DHNPVVLHCGSRR---TETQRHRTRMFRFEEVWL--ESGEECAEIISEG----- 274
DH+ + + + R T T+ + ++ W+ E +E + + +
Sbjct: 222 IPCIFSDHHGIKVELNNNRNLHTHTKTWKLNNLMLKDTWVIDEIKKEITKFLEQNNNQDT 281
Query: 275 -----WSNTNLSILSRINLVGRSLDSWGREKYGELPKRIKEARAFLQRLQEEVQTEQVVR 329
W + + + L RE+ L +K+ L++E +
Sbjct: 282 NYQNLWDTAKAVLRGKFIALQAFLKKTEREEVNNLMGHLKQ-------LEKEEHSNPKPS 334
Query: 330 ATREAEK---NLDVLLKQEEIVWSQRSRANWLKHGDRNTKFFHSKATQRRKRNLIEEIQD 386
+E K L+ + + I +S++ + + ++ K + ++R ++LI I++
Sbjct: 335 RRKEITKIRAELNEIENKRIIQQINKSKSWFFEKINKIDKPLANLTRKKRVKSLISSIRN 394
Query: 387 DNGRKFVRDADIARVLTSYFQEIFSTCNPTGIEEVATLVDG----RVTDAHRRILSTPFT 442
N ++I ++L Y+++++S ++E+ ++ R++ +L+ P +
Sbjct: 395 GNDEITTDPSEIQKILNEYYKKLYSH-KYENLKEIDQYLEACHLPRLSQKEVEMLNRPIS 453
Query: 443 REDVEEALFQMHPTKAPGLDGFPALFYQKFWPIVGDDISEFCLQVLQGDISPGMVNQTLI 502
++ + + K+PG DGF + FYQ F + + + + I P + I
Sbjct: 454 SSEIASTIQNLPKKKSPGPDGFTSEFYQTFKEELVPILLNLFQNIEKEGILPNTFYEANI 513
Query: 503 VLIPKV-KKAVFANQYRPISLCNVIFKIISKTIANRMKLILHDLISEAQSAFVPGRLITD 561
LIPK K YRPISL N+ KI++K + NR++ + +I Q F+PG
Sbjct: 514 TLIPKPGKDPTRKENYRPISLMNIDAKILNKILTNRIQQHIKKIIHHDQVGFIPGSQGWF 573
Query: 562 NALIAFECFHYMKKRISGRSGMMALKLDMSKAYDRVEWPFLRSVLTHMGFPVHWVNLVMN 621
N + ++ K + M L +D KA+D ++ PF+ L +G ++ L+
Sbjct: 574 NIRKSINVIQHINKLKN--KDHMILSIDAEKAFDNIQHPFMIRTLKKIGIEGTFLKLIEA 631
Query: 622 CVTTVNFAVMLNGNPQPTFAPHRGLRQGDPLSPYLFILCGEAFSALIQKSISTSALHGIK 681
+ ++LNG +F G RQG PLSP LF + E + I++ A+ GI
Sbjct: 632 IYSKPTANIILNGVKLKSFPLRSGTRQGCPLSPLLFNIVMEVLAIAIREE---KAIKGIH 688
Query: 682 ISRSAPVISHLLFADDSVLFARATVQEAECIKNILATYERASGQKINLEKSM--LSVSRN 739
I +S LFADD +++ T + ++ Y SG KIN KS+ + + N
Sbjct: 689 IGSEEIKLS--LFADDMIVYLENTRDSTTKLLEVIKEYSNVSGYKINTHKSVAFIYTNNN 746
Query: 740 VPENCFVDLQLLLGVKAVDSYDKYLG--LPTIIGKSKTQIFRFVKERVWKKLKGWKEQTL 797
E D + V KYLG L + + + +++ + + + WK
Sbjct: 747 QAEKTVKD---SIPFTVVPKKMKYLGVYLTKDVKDLYKENYETLRKEIAEDVNKWKNIPC 803
Query: 798 SRAGREVLIKS--VAQAIPSYVMSCFVLPDGLCNEIESMISRFYWGGDASKRGIHWVKWK 855
S GR ++K + +AI ++ P ++E +I F W + K
Sbjct: 804 SWLGRINIVKMSILPKAIYNFNAIPIKAPLSYFKDLEKIILHFIWNQKKPQIA------K 857
Query: 856 TMCQPKH-VGGLGFRDFKSFNLALVAKNWW 884
T+ K+ GG+ D + + ++V K W
Sbjct: 858 TLLSNKNKAGGITLPDLRLYYKSIVIKTAW 887
>M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310
(ORF154)
Length = 154
Score = 122 bits (307), Expect = 5e-27
Identities = 62/150 (41%), Positives = 90/150 (59%), Gaps = 3/150 (2%)
Query: 812 AIPSYVMSCFVLPDGLCNEIESMISRFYWGGDASKRGIHWVKWKTMCQPKHV-GGLGFRD 870
A+P Y MSCF L LC ++ S ++ F+W +KR I WV W+ +C+ K GGLGFRD
Sbjct: 2 ALPVYAMSCFRLSKLLCKKLTSAMTEFWWSSCENKRKISWVAWQKLCKSKEDDGGLGFRD 61
Query: 871 FKSFNLALVAKNWWRIYNYPDSLLGKVFKSVYFS-SGRMECAKKGYRPSYAWSRILKTSA 929
FN AL+AK +RI + P +LL ++ +S YF S MEC+ G RPSYAW I+
Sbjct: 62 LGWFNQALLAKQSFRIIHQPHTLLSRLLRSRYFPHSSMMECSV-GTRPSYAWRSIIHGRE 120
Query: 930 MIQKGSCWRIGEGSRVRIWEDNWLANGPPV 959
++ +G IG+G ++W D W+ + P+
Sbjct: 121 LLSRGLLRTIGDGIHTKVWLDRWIMDETPL 150
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 81.6 bits (200), Expect = 1e-14
Identities = 38/68 (55%), Positives = 49/68 (71%)
Query: 631 MLNGNPQPTFAPHRGLRQGDPLSPYLFILCGEAFSALIQKSISTSALHGIKISRSAPVIS 690
++NG PQ P RGLRQGDPLSPYLFILC E S L +++ L GI++S ++P I+
Sbjct: 13 IINGAPQGLVTPSRGLRQGDPLSPYLFILCTEVLSGLCRRAQEQGRLPGIRVSNNSPRIN 72
Query: 691 HLLFADDS 698
HLLFADD+
Sbjct: 73 HLLFADDT 80
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 75.5 bits (184), Expect = 9e-13
Identities = 75/282 (26%), Positives = 121/282 (42%), Gaps = 24/282 (8%)
Query: 494 PGMVNQTLIVLIPKVKKAVFANQYRPISLCNVIFKIISKTIANRMKLILHDLISEAQSAF 553
P + V IPK A +RPIS+ +V+ + ++ +A R+ ++ Q F
Sbjct: 392 PHSIRLARTVFIPKTVTAKRPQDFRPISVPSVLVRQLNAILATRLNSSIN--WDPRQRGF 449
Query: 554 VPGRLITDNALIAFECFHYMKKRISGRSGMMALKLDMSKAYDRVEWPFLRSVLTHMGFPV 613
+P DNA I + K RS +A LD+SKA+D + + L G P
Sbjct: 450 LPTDGCADNATIVDLVLRHSHKHF--RSCYIA-NLDVSKAFDSLSHASIYDTLRAYGAPK 506
Query: 614 HWVNLVMNCVTTVNFAVMLNGNPQPTFAPHRGLRQGDPLSPYLFILCGEAFSALIQKSIS 673
+V+ V N ++ +G F P RG++QGDPLSP LF L + + I
Sbjct: 507 GFVDYVQNTYEGGGTSLNGDGWSSEEFVPARGVKQGDPLSPILFNLVMDRLLRTLPSEI- 565
Query: 674 TSALHGIKISRSAPVISHLLFADDSVLFARATVQEAECIKNILATYERASGQKINLEKSM 733
G K+ + + + FADD VLFA T + + + + G K+N +K
Sbjct: 566 -----GAKVGNA--ITNAAAFADDLVLFAE-TRMGLQVLLDKTLDFLSIVGLKLNADKCF 617
Query: 734 LSVSRNVP---------ENCFVDLQLLLGVKAVDSYDKYLGL 766
+ P ++ +V + +K D + KYLG+
Sbjct: 618 TVGIKGQPKQKCTVLEAQSFYVGSSEIPSLKRTDEW-KYLGI 658
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 68.9 bits (167), Expect = 9e-11
Identities = 68/263 (25%), Positives = 108/263 (40%), Gaps = 19/263 (7%)
Query: 440 PFTREDVEEALFQMHPTKAPGLDGFPALFYQKFWPIVGDDISEFCLQVLQGDISPGMVNQ 499
P + ++++E + A G DG W + + I ++ P
Sbjct: 318 PISNDEIKEV--EACKRTAAGPDGMTTTA----WNSIDECIKSLFNMIMYHGQCPRRYLD 371
Query: 500 TLIVLIPKVKKAVFANQYRPISLCNVIFKIISKTIANRMKLILHDLISEAQSAFVPGRLI 559
+ VLIPK + +RP+S+ +V + + +ANR+ H L+ Q AF+ +
Sbjct: 372 SRTVLIPKEPGTMDPACFRPLSIASVALRHFHRILANRIGE--HGLLDTRQRAFIVADGV 429
Query: 560 TDNALIAFECFHYMKKRISGRSGMMALKLDMSKAYDRVEWPFLRSVLTHMGFPVHWVNLV 619
+N + + +I G + LD+ KA+D VE + L P+ N +
Sbjct: 430 AENTSLLSAMIKEARMKIKG---LYIAILDVKKAFDSVEHRSILDALRRKKLPLEMRNYI 486
Query: 620 MNCVTTVNFAVMLNGNPQPTFAPHRGLRQGDPLSPYLFILCGEAFSALIQKSISTSALHG 679
M + + P RG+RQGDPLSP LF +A L + +T L G
Sbjct: 487 MWVYRNSKTRLEVVKTKGRWIRPARGVRQGDPLSPLLFNCVMDA--VLRRLPENTGFLMG 544
Query: 680 IKISRSAPVISHLLFADDSVLFA 702
A I L+FADD VL A
Sbjct: 545 ------AEKIGALVFADDLVLLA 561
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 67.0 bits (162), Expect = 3e-10
Identities = 74/306 (24%), Positives = 126/306 (40%), Gaps = 41/306 (13%)
Query: 440 PFTREDVEEALFQMHPTKAPGLDGF----------PALFYQKFWPIVGDDISEFCLQVLQ 489
P RE+++ A+ P+ APG DG P F Q L +L+
Sbjct: 5 PIAREEIQCAIKGWKPS-APGSDGLTVQAITRTRLPRNFVQ--------------LHLLR 49
Query: 490 GDI-SPGMVNQTLIVLIPKVKKAVFANQYRPISLCNVIFKIISKTIANRMKLILHDLISE 548
G + +P +T LIPK + +RPI++ + + +++ + +A R++ + ++
Sbjct: 50 GHVPTPWTAMRT--TLIPKDGDLENPSNWRPITIASALQRLLHRILAKRLEAAVELHPAQ 107
Query: 549 AQSAFVPGRLITDNALIAFECFHYMKKRISGRSGMMALKLDMSKAYDRVEWPFLRSVLTH 608
A + G L+ L Y+ R R + LD+ KA+D V + L
Sbjct: 108 KGYARIDGTLVNSLLLDT-----YISSRREQRKTYNVVSLDVRKAFDTVSHSSICRALQR 162
Query: 609 MGFPVHWVNLVMNCVTTVNFAVMLN-GNPQPTFAPHRGLRQGDPLSPYLFILCGEAFSAL 667
+G N + ++ + + G+ RG++QGDPLSP+LF L
Sbjct: 163 LGIDEGTSNYITGSLSDSTTTIRVGPGSQTRKICIRRGVKQGDPLSPFLF---NAVLDEL 219
Query: 668 IQKSISTSALHGIKISRSAPVISHLLFADDSVLFARATVQEAECIKNILATYERASGQKI 727
+ ST + G PV L FADD +L V + + A + R G +
Sbjct: 220 LCSLQSTPGIGGTIGEEKIPV---LAFADDLLLLEDNDVLLPTTLATV-ANFFRLRGMSL 275
Query: 728 NLEKSM 733
N +KS+
Sbjct: 276 NAKKSV 281
>PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 606
Score = 60.8 bits (146), Expect = 2e-08
Identities = 62/226 (27%), Positives = 105/226 (46%), Gaps = 27/226 (11%)
Query: 571 HYMK-KRISGRSGMMALKLDMSKAYDRVEWPFLRSVLTHMGFPVHWVNLVMNCVTTVNFA 629
HY+K +R+ G++ + LD+ KA+D V P + + G + +M+ +T
Sbjct: 11 HYIKLRRLKGKT-YNVVSLDIRKAFDTVSHPAILRAMRAFGIDDGMQDFIMSTITDAYTN 69
Query: 630 VMLNGNPQPTFAPHRGLRQGDPLSPYLF-ILCGEAFSALIQKSISTSALHGIKISRSAPV 688
+++ G G++QGDPLSP LF I+ E + L + S K
Sbjct: 70 IVVGGRTTNKIYIRNGVKQGDPLSPVLFNIVLDELVTRLNDEQPGASMTPACK------- 122
Query: 689 ISHLLFADDSVLFARATVQEAECIKNILAT---YERASGQKINLEK----SMLSVS-RNV 740
I+ L FADD +L + + N LAT Y R G +N EK S +VS R+V
Sbjct: 123 IASLAFADDLLLLEDRDID----VPNSLATTCAYFRTRGMTLNPEKCASISAATVSGRSV 178
Query: 741 PE---NCFVDLQLLLGVKAVDSYDKYLGLP-TIIGKSKTQIFRFVK 782
P + +D + + + ++++ KYLGL + G +K ++ +
Sbjct: 179 PRSKPSFTIDGRYIKPLGGINTF-KYLGLTFSSTGVAKPTVYNLTR 223
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 56.6 bits (135), Expect = 4e-07
Identities = 90/386 (23%), Positives = 155/386 (39%), Gaps = 47/386 (12%)
Query: 365 TKFFHSKATQRRKRN--LIEEIQD-----------DNGRKFVRDADIARVLTSYFQEIFS 411
T +H Q+RK+ L++ + D K R + Y++++F+
Sbjct: 74 TTIYHGTNKQKRKQRYALVQSLYKKDISAAARVVLDENDKIATKIPPVRHMFDYWKDVFA 133
Query: 412 TCNPTGIEEVATLVDGRVTDAHRRILSTPFTREDVEEALFQMHPTKAPGLDGFPALFYQK 471
T G AT ++ H L P + +++ A + K G DG +
Sbjct: 134 T----GGGSAATNINRAPPAPHMETLWDPVSLIEIKSA--RASNEKGAGPDGVTP----R 183
Query: 472 FWPIVGDDISE--FCLQVLQGDISPGMVNQTLIVLIPKVKKAVFANQYRPISLCNVIFKI 529
W + D + + V G + P + + V PK++ +RP+S+C+VI +
Sbjct: 184 SWNALDDRYKRLLYNIFVFYGRV-PSPIKGSRTVFTPKIEGGPDPGVFRPLSICSVILRE 242
Query: 530 ISKTIANRMKLILHDLISEAQSAFVP--GRLITDNALIAFECFHYMKKRISGRSGMMALK 587
+K +A R + E Q+A++P G I + L A KR+ R +
Sbjct: 243 FNKILARR--FVSCYTYDERQTAYLPIDGVCINVSMLTAIIA---EAKRL--RKELHIAI 295
Query: 588 LDMSKAYDRVEWPFLRSVLTHMGFPVHWVNLVMNCVTTVNFAVMLNGNPQPTFAPHRGLR 647
LD+ KA++ V L +T G P V+ + + V + G + + G+
Sbjct: 296 LDLVKAFNSVYHSALIDAITEAGCPPGVVDYIADMYNNVITEMQFEGKCE-LASILAGVY 354
Query: 648 QGDPLSPYLFILCGE-AFSALIQKSISTSALHGIKISRSAPVISHLLFADDSVLFARATV 706
QGDPLS LF L E A AL + A ++++ SA ++DD +L A +
Sbjct: 355 QGDPLSGPLFTLAYEKALRALNNEGRFDIA--DVRVNASA-------YSDDGLLLAMTVI 405
Query: 707 QEAECIKNILATYERASGQKINLEKS 732
+ T + G +IN KS
Sbjct: 406 GLQHNLDKFGETLAKI-GLRINSRKS 430
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 52.8 bits (125), Expect = 6e-06
Identities = 44/176 (25%), Positives = 71/176 (40%), Gaps = 4/176 (2%)
Query: 485 LQVLQGDISPGMVNQTLIVLIPKVKKAVFANQYRPISLCNVIFKIISKTIANRMKLILHD 544
+Q P +I+ IPK + YRPISL + +I+ + I +R++
Sbjct: 653 MQSFSDSAIPNRWKHAVIIPIPKKGNPSSPSNYRPISLTDPFARIMERIICSRIRSEYSH 712
Query: 545 LISEAQSAFVPGRLITDNALIAFECFHYMKKRISGRSGMMALKLDMSKAYDRVEWPFLRS 604
L+S Q F+ R + + + +H + K + L D +KA+D+V P L
Sbjct: 713 LLSPHQHGFLNFRSCPSSLVRSISLYHSILK---NEKSLDILFFDFAKAFDKVSHPILLK 769
Query: 605 VLTHMGFPVHWVNLVMNCVTTVNFAVMLNGNPQPTFAP-HRGLRQGDPLSPYLFIL 659
L G + + F+V +N P G+ QG P LFIL
Sbjct: 770 KLALFGLDKLTCSWFKEFLHLRTFSVKINKFVSSNAYPISSGVPQGSVSGPLLFIL 825
>Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I
retrotransposable element R1DM (ORF 2)
Length = 1021
Score = 42.7 bits (99), Expect = 0.007
Identities = 50/229 (21%), Positives = 86/229 (36%), Gaps = 31/229 (13%)
Query: 388 NGRKFVRDADIARVLTSYFQEIFSTCNPTGIEEVATLVDGRVTDAHRRILSTPFTRE--D 445
NG D ARVL F + + PT I E P E +
Sbjct: 396 NGELITDWGDCARVLLRNFFPVAESEAPTAIAE-----------------EVPPALEVFE 438
Query: 446 VEEALFQMHPTKAPGLDGFPALFYQKFWPIVGDDISEFCLQVLQGDISPGMVNQTLIVLI 505
V+ + ++ ++PGLDG + W + + ++ + ++ P +V +
Sbjct: 439 VDTCVARLKSRRSPGLDGINGTICKAVWRAIPEHLASLFSRCIRLGYFPAEWKCPRVVSL 498
Query: 506 PK--VKKAVFANQYRPISLCNVIFKIISKTIANRMKLILHDLISEAQSAFVPGRLITDNA 563
K K + YR I L V K++ + NR++ +L + Q F GR + D
Sbjct: 499 LKGPDKDKCEPSSYRGICLLPVFGKVLEAIMVNRVREVLPE-GCRWQFGFRQGRCVED-- 555
Query: 564 LIAFECFHYMKKRISGRSGMMALK--LDMSKAYDRVEWPFLRSVLTHMG 610
+ ++K + + L +D A+D VEW S L +G
Sbjct: 556 -----AWRHVKSSVGASAAQYVLGTFVDFKGAFDNVEWSAALSRLADLG 599
>RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 41.2 bits (95), Expect = 0.020
Identities = 51/228 (22%), Positives = 98/228 (42%), Gaps = 21/228 (9%)
Query: 440 PFTREDVEEALFQMHPTKAPGLDGFPALFYQKFWPIVGDDISEFCLQVLQGDISPGMV-- 497
P T E+V+E + ++ P KAPG D L + ++ D + + + + G
Sbjct: 437 PVTLEEVKELVSKLKPKKAPGED----LLDNRTIRLLPDQALLYLVLIFNSILRVGYFPK 492
Query: 498 ---NQTLIVLIPKVKKAVFANQYRPISLCNVIFKIISKTIANRMKLILHDLISEAQSAFV 554
++I+++ K+ + + YRP SL + K++ + I NR ++ + ++ A F
Sbjct: 493 ARPTASIIMILKPGKQPLDVDSYRPTSLLPSLGKMLERLILNR--ILTSEEVTRAIPKFQ 550
Query: 555 PG-RLITDNALIAFECFHYMKKRISGRSGMMALKLDMSKAYDRVEWPFL----RSVLTHM 609
G RL ++ + + + + LD+ +A+DRV P L +S+L+
Sbjct: 551 FGFRLQHGTPEQLHRVVNFALEALEKKEYAGSCFLDIQQAFDRVWHPGLLYKAKSLLSPQ 610
Query: 610 GFPVHWVNLVMNCVTTVNFAVMLNGNPQPTFAPHRGLRQGDPLSPYLF 657
F L+ + F+V +G G+ QG L P L+
Sbjct: 611 LF-----QLIKSFWEGRKFSVTADGCRSSVKFIEAGVPQGSVLGPTLY 653
>RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 39.3 bits (90), Expect = 0.074
Identities = 75/350 (21%), Positives = 128/350 (36%), Gaps = 49/350 (14%)
Query: 351 QRSRANWLKHGDRNTKFFHS-------KATQRRKRNLIEEIQDDNGRKFVRDADIARVLT 403
+R R + + GD + HS KA RRK+ I+ D+ G + + R+
Sbjct: 310 RRVRKEYARTGDPRMQQIHSRLANCLHKALARRKQAQIDTFLDNLGADASTNYSLWRITK 369
Query: 404 SYFQEIFSTC---NPTG---------IEEVATLVDGRVTDAH----------RRILSTPF 441
+ + NP+G E A ++ R T + L +PF
Sbjct: 370 RFKAQPTPKSAIKNPSGGWCRTSLEKTEVFANNLEQRFTPYNYAPESLCRQVEEYLESPF 429
Query: 442 ---------TREDVEEALFQMHPTKAPGLDGFPALFYQKFWPIVGDDISEFCLQVLQGDI 492
T E+V+ + ++ KAPG D L + ++ D +F + +
Sbjct: 430 QMSLPLSAVTLEEVKNLIAKLPLKKAPGED----LLDNRTIRLLPDQALQFLALIFNSVL 485
Query: 493 SPGMV----NQTLIVLIPKVKKA-VFANQYRPISLCNVIFKIISKTIANRMKLILHDLIS 547
G I++I K K + YRP SL + KI+ + I NR+ L D+
Sbjct: 486 DVGYFPKAWKSASIIMIHKTGKTPTDVDSYRPTSLLPSLGKIMERLILNRL-LTCKDVTK 544
Query: 548 EAQSAFVPGRLITDNALIAFECFHYMKKRISGRSGMMALKLDMSKAYDRVEWPFLRSVLT 607
RL ++ + + + + LD+ +A+DRV P L
Sbjct: 545 AIPKFQFGFRLQHGTPEQLHRVVNFALEAMENKEYAVGAFLDIQQAFDRVWHPGLLYKAK 604
Query: 608 HMGFPVHWVNLVMNCVTTVNFAVMLNGNPQPTFAPHRGLRQGDPLSPYLF 657
+ FP +V + + F V ++G G+ QG L P L+
Sbjct: 605 RL-FPPQLYLVVKSFLEERTFHVSVDGYKSSIKPIAAGVPQGSVLGPTLY 653
>C190_DROME (Q9VJE5) Restin homolog (Cytoplasmic linker protein 190)
(Microtubule binding protein 190) (d-CLIP-190)
Length = 1690
Score = 38.5 bits (88), Expect = 0.13
Identities = 35/165 (21%), Positives = 84/165 (50%), Gaps = 12/165 (7%)
Query: 298 REKYGELPKRIKEARAFLQRLQEEVQT--EQVVRATREAEKNLDVLLKQEEIVWS--QRS 353
++K+ EL +++K+A+ Q+LQ+E QT E++ + ++ D + ++EE+V + ++
Sbjct: 1188 QKKFEELEEKLKQAQQSEQKLQQESQTSKEKLTEIQQSLQELQDSVKQKEELVQNLEEKV 1247
Query: 354 RANWLKHGDRNTKFFHSKATQRRKRNLIEEIQD---DNGRKFVRDADIARVLTSYFQEIF 410
R + +NTK S K + ++E QD ++ +K + + A L+ Q++
Sbjct: 1248 RESSSIIEAQNTKLNESNVQLENKTSCLKETQDQLLESQKKEKQLQEEAAKLSGELQQVQ 1307
Query: 411 STCNP-----TGIEEVATLVDGRVTDAHRRILSTPFTREDVEEAL 450
+EE+ +++ ++ A ++ + T ++++E L
Sbjct: 1308 EANGDIKDSLVKVEELVKVLEEKLQAATSQLDAQQATNKELQELL 1352
>TCPG_TETPY (P54408) T-complex protein 1, gamma subunit
(TCP-1-gamma) (CCT-gamma)
Length = 559
Score = 37.4 bits (85), Expect = 0.28
Identities = 19/64 (29%), Positives = 34/64 (52%)
Query: 523 CNVIFKIISKTIANRMKLILHDLISEAQSAFVPGRLITDNALIAFECFHYMKKRISGRSG 582
C++I + SK + N M+ LHD ++ A++ FV +L+ I E +++K S G
Sbjct: 375 CSIILRGASKDVLNEMERNLHDCLAVAKNIFVNPKLVPGGGAIEMEVSSHLEKISSSIEG 434
Query: 583 MMAL 586
+ L
Sbjct: 435 LHQL 438
>MACF_HUMAN (Q9UPN3) Microtubule-actin crosslinking factor 1,
isoforms 1/2/3 (Actin cross-linking family protein 7)
(Macrophin 1) (Trabeculin-alpha) (620 kDa actin-binding
protein) (ABP620)
Length = 5430
Score = 35.8 bits (81), Expect = 0.82
Identities = 33/166 (19%), Positives = 65/166 (38%), Gaps = 22/166 (13%)
Query: 238 HCGSRRTET--QRHRTRMFRF------EEVWLESGEECAEIISEGWSNTNLSILSRINLV 289
+C + T + RH + +F E +WL EE E ++ WS+ N +I ++
Sbjct: 665 YCKLKETSSFRMRHLQSLHKFVSRATAELIWLNEKEE--EELAYDWSDNNSNISAK---- 718
Query: 290 GRSLDSWGREKYGELPKRIKEARAFLQRLQEEVQTEQVVRATREAEKNLDVLLKQEEIVW 349
R + EL ++E + + LQ+ + + + Q ++ W
Sbjct: 719 --------RNYFSELTMELEEKQDVFRSLQDTAELLSLENHPAKQTVEAYSAAVQSQLQW 770
Query: 350 SQRSRANWLKHGDRNTKFFHSKATQRRKRNLIEEIQDDNGRKFVRD 395
++ +H NT +F + R + + +QD RK+ D
Sbjct: 771 MKQLCLCVEQHVKENTAYFQFFSDARELESFLRNLQDSIKRKYSCD 816
>YO84_CAEEL (P34620) Hypothetical protein ZK1236.4 in chromosome III
Length = 364
Score = 34.7 bits (78), Expect = 1.8
Identities = 25/90 (27%), Positives = 37/90 (40%)
Query: 568 ECFHYMKKRISGRSGMMALKLDMSKAYDRVEWPFLRSVLTHMGFPVHWVNLVMNCVTTVN 627
EC + K I + LD SKA+D+V L LT + H + + +T +
Sbjct: 3 ECMNNWTKNIDEGAQTDVKYLDFSKAFDKVSHDILLDKLTSIKINKHLIRWLDVFLTNRS 62
Query: 628 FAVMLNGNPQPTFAPHRGLRQGDPLSPYLF 657
F V + G+ QG +SP LF
Sbjct: 63 FKVKVGNTLSEPKKTVCGVPQGSVISPVLF 92
>YE85_SCHPO (O14301) Hypothetical WD-repeat protein C9G1.05 in
chromosome I
Length = 595
Score = 33.9 bits (76), Expect = 3.1
Identities = 25/92 (27%), Positives = 48/92 (52%), Gaps = 6/92 (6%)
Query: 470 QKFWPIVGDDISEFCLQVLQGD---ISPGMVNQTLIVLIPK---VKKAVFANQYRPISLC 523
QK +P+VG+ + +Q++ D I+ GM + ++ I + K VF Y+PI +C
Sbjct: 347 QKAFPLVGESHTNQVMQMIMADDHVITIGMDDTLRVIDIKQGCFAKDNVFPTGYQPIGVC 406
Query: 524 NVIFKIISKTIANRMKLILHDLISEAQSAFVP 555
+V +I T+++ L +S A++ + P
Sbjct: 407 SVEDCLILVTVSDIQVLRSLTGVSTAKTIYQP 438
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.137 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 160,728,260
Number of Sequences: 164201
Number of extensions: 6803055
Number of successful extensions: 15081
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 15016
Number of HSP's gapped (non-prelim): 56
length of query: 1366
length of database: 59,974,054
effective HSP length: 123
effective length of query: 1243
effective length of database: 39,777,331
effective search space: 49443222433
effective search space used: 49443222433
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 72 (32.3 bits)
Lotus: description of TM0048.9


