

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
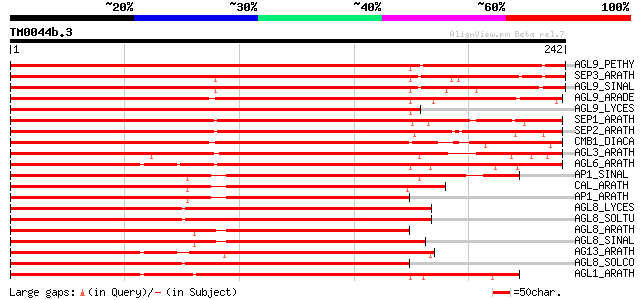
Query= TM0044b.3
(242 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AGL9_PETHY (Q03489) Agamous-like MADS box protein AGL9 homolog (... 404 e-112
SEP3_ARATH (O22456) Developmental protein SEPALLATA3 (Agamous-li... 357 1e-98
AGL9_SINAL (O04067) Agamous-like MADS box protein AGL9 homolog (... 352 6e-97
AGL9_ARADE (Q38694) Agamous-like MADS box protein AGL9 homolog (... 341 1e-93
AGL9_LYCES (Q42464) Agamous-like MADS box protein AGL9 homolog (... 317 1e-86
SEP1_ARATH (P29382) Developmental protein SEPALLATA1 (Agamous-li... 299 4e-81
SEP2_ARATH (P29384) Developmental protein SEPALLATA2 (Agamous-li... 298 1e-80
CMB1_DIACA (Q39685) MADS box protein CMB1 264 1e-70
AGL3_ARATH (P29383) Agamous-like MADS box protein AGL3 228 1e-59
AGL6_ARATH (P29386) Agamous-like MADS box protein AGL6 200 2e-51
AP1_SINAL (Q41276) Floral homeotic protein APETALA1 (MADS C) 191 1e-48
CAL_ARATH (Q39081) Transcription factor CAULIFLOWER (Agamous-lik... 187 1e-47
AP1_ARATH (P35631) Floral homeotic protein APETALA1 (Agamous-lik... 186 6e-47
AGL8_LYCES (Q40170) Agamous-like MADS box protein AGL8 homolog (... 185 7e-47
AGL8_SOLTU (Q42429) Agamous-like MADS box protein AGL8 homolog (... 183 4e-46
AGL8_ARATH (Q38876) Agamous-like MADS box protein AGL8 (Floral h... 181 1e-45
AGL8_SINAL (Q41274) Agamous-like MADS box protein AGL8 homolog (... 181 2e-45
AG13_ARATH (Q38837) Agamous-like MADS box protein AGL13 179 4e-45
AGL8_SOLCO (O22328) Agamous-like MADS box protein AGL8 homolog 175 8e-44
AGL1_ARATH (P29381) Agamous-like MADS box protein AGL1 (Protein ... 167 2e-41
>AGL9_PETHY (Q03489) Agamous-like MADS box protein AGL9 homolog
(Floral homeotic protein FBP2) (Floral binding protein
2)
Length = 241
Score = 404 bits (1038), Expect = e-112
Identities = 207/243 (85%), Positives = 220/243 (90%), Gaps = 3/243 (1%)
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS
Sbjct: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLGP 120
SSMLKTLERYQKCNYGAPE N+STREALE+SSQQEYLKLKARYEALQRSQRNL+GEDLGP
Sbjct: 61 SSMLKTLERYQKCNYGAPETNISTREALEISSQQEYLKLKARYEALQRSQRNLLGEDLGP 120
Query: 121 LNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRL-EGYQLN 179
LNSKELESLERQLD SLKQIRSTRTQ MLDQL DLQRKEH L+EANR+L+QRL EG LN
Sbjct: 121 LNSKELESLERQLDMSLKQIRSTRTQLMLDQLQDLQRKEHALNEANRTLKQRLMEGSTLN 180
Query: 180 SLQLNPGVEDMGYGRHPAQTHGDAFYHPIECEPTLQIGYQPDPVSVVTAGPSMNNNYMAG 239
LQ +D+GYGR QT GD F+HP+ECEPTLQIGYQ DP++V AGPS+ NNYMAG
Sbjct: 181 -LQWQQNAQDVGYGRQATQTQGDGFFHPLECEPTLQIGYQNDPITVGGAGPSV-NNYMAG 238
Query: 240 WLP 242
WLP
Sbjct: 239 WLP 241
>SEP3_ARATH (O22456) Developmental protein SEPALLATA3 (Agamous-like
MADS box protein AGL9)
Length = 251
Score = 357 bits (917), Expect = 1e-98
Identities = 193/248 (77%), Positives = 213/248 (85%), Gaps = 9/248 (3%)
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS
Sbjct: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREAL--ELSSQQEYLKLKARYEALQRSQRNLMGEDL 118
SSML+TLERYQKCNYGAPE NV +REAL ELSSQQEYLKLK RY+ALQR+QRNL+GEDL
Sbjct: 61 SSMLRTLERYQKCNYGAPEPNVPSREALAVELSSQQEYLKLKERYDALQRTQRNLLGEDL 120
Query: 119 GPLNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRL-EGYQ 177
GPL++KELESLERQLDSSLKQIR+ RTQFMLDQL+DLQ KE ML+E N++LR RL +GYQ
Sbjct: 121 GPLSTKELESLERQLDSSLKQIRALRTQFMLDQLNDLQSKERMLTETNKTLRLRLADGYQ 180
Query: 178 LNSLQLNPGVEDMG-YGR--HPAQTHGDAFYHPIECEPTLQIGYQPDPVSVVTAGPSMNN 234
+ LQLNP E++ YGR H Q H AF+ P+ECEP LQIGYQ + AGPS+ N
Sbjct: 181 M-PLQLNPNQEEVDHYGRHHHQQQQHSQAFFQPLECEPILQIGYQGQQ-DGMGAGPSV-N 237
Query: 235 NYMAGWLP 242
NYM GWLP
Sbjct: 238 NYMLGWLP 245
>AGL9_SINAL (O04067) Agamous-like MADS box protein AGL9 homolog
(MADS D)
Length = 254
Score = 352 bits (902), Expect = 6e-97
Identities = 189/250 (75%), Positives = 210/250 (83%), Gaps = 10/250 (4%)
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS
Sbjct: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREAL--ELSSQQEYLKLKARYEALQRSQRNLMGEDL 118
SSM++TLERYQKCNYG PE NV +REAL ELSSQQEYLKLK RY+ALQR+QRNL+GEDL
Sbjct: 61 SSMIRTLERYQKCNYGPPEPNVPSREALAVELSSQQEYLKLKERYDALQRTQRNLLGEDL 120
Query: 119 GPLNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRL-EGYQ 177
GPL++KELE LERQLDSSLKQIR+ RTQFMLDQL+DLQ KE ML+E N++LR RL +GYQ
Sbjct: 121 GPLSTKELELLERQLDSSLKQIRALRTQFMLDQLNDLQSKERMLNETNKTLRLRLADGYQ 180
Query: 178 LNSLQLNPGVED--MGYGRHPAQTHGD---AFYHPIECEPTLQIGYQPDPVSVVTAGPSM 232
+ LQLNP ED + YGRH Q + AF+ P+ECEP LQ+GYQ + AGPS
Sbjct: 181 M-PLQLNPNQEDHHVDYGRHDQQQQQNSHHAFFQPLECEPILQMGYQGQQDHGMEAGPS- 238
Query: 233 NNNYMAGWLP 242
NNYM GWLP
Sbjct: 239 ENNYMLGWLP 248
>AGL9_ARADE (Q38694) Agamous-like MADS box protein AGL9 homolog
(OM1)
Length = 250
Score = 341 bits (874), Expect = 1e-93
Identities = 181/245 (73%), Positives = 203/245 (81%), Gaps = 7/245 (2%)
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRGRVELK IENKINRQVTFAKRR LLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS
Sbjct: 1 MGRGRVELKMIENKINRQVTFAKRRKRLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLGP 120
+SMLKTLE+YQKCN+G+PE+ + +RE SSQQEYLKLK R EALQRSQRNL+GEDLGP
Sbjct: 61 TSMLKTLEKYQKCNFGSPESTIISRET--QSSQQEYLKLKNRVEALQRSQRNLLGEDLGP 118
Query: 121 LNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRL-EGYQLN 179
L SKELE LERQLDSSL+QIRSTRTQFMLDQL+DLQR+E ML EAN++L++R E Q N
Sbjct: 119 LGSKELEQLERQLDSSLRQIRSTRTQFMLDQLADLQRREQMLCEANKTLKRRFEESSQAN 178
Query: 180 SLQL--NPGVEDMGYGRHPAQTHGDAFYHPIECEPTLQIGYQPDPVSVVTAGPSMNNNYM 237
Q+ +GYGR PAQ HG+AFYHP+ECEPTLQIGY D +++ TA S NNYM
Sbjct: 179 QQQVWDPSNTHAVGYGRQPAQHHGEAFYHPLECEPTLQIGYHSD-ITMATATASTVNNYM 237
Query: 238 -AGWL 241
GWL
Sbjct: 238 PPGWL 242
>AGL9_LYCES (Q42464) Agamous-like MADS box protein AGL9 homolog
(TM5)
Length = 224
Score = 317 bits (813), Expect = 1e-86
Identities = 164/180 (91%), Positives = 171/180 (94%), Gaps = 1/180 (0%)
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRGRVELKRIE KINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS
Sbjct: 1 MGRGRVELKRIEGKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLGP 120
SSMLKTLERYQKCNYGAPE N+STREALE+SSQQEYLKLK RYEALQRSQRNL+GEDLGP
Sbjct: 61 SSMLKTLERYQKCNYGAPEPNISTREALEISSQQEYLKLKGRYEALQRSQRNLLGEDLGP 120
Query: 121 LNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRL-EGYQLN 179
LNSKELESLERQLD SLKQIRSTRTQ MLDQL+D QRKEH L+EANR+L+QRL EG QLN
Sbjct: 121 LNSKELESLERQLDMSLKQIRSTRTQLMLDQLTDYQRKEHALNEANRTLKQRLMEGSQLN 180
>SEP1_ARATH (P29382) Developmental protein SEPALLATA1 (Agamous-like
MADS box protein AGL2)
Length = 251
Score = 299 bits (766), Expect = 4e-81
Identities = 163/254 (64%), Positives = 190/254 (74%), Gaps = 17/254 (6%)
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS
Sbjct: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLGP 120
S+MLKTL+RYQKC+YG+ E N + LE +S +EYLKLK RYE LQR QRNL+GEDLGP
Sbjct: 61 SNMLKTLDRYQKCSYGSIEVNNKPAKELE-NSYREYLKLKGRYENLQRQQRNLLGEDLGP 119
Query: 121 LNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRLE---GYQ 177
LNSKELE LERQLD SLKQ+RS +TQ+MLDQLSDLQ KE ML E NR+L +L+ G +
Sbjct: 120 LNSKELEQLERQLDGSLKQVRSIKTQYMLDQLSDLQNKEQMLLETNRALAMKLDDMIGVR 179
Query: 178 LNSL----QLNPGVEDMGYGRHPAQTHGDAFYHPIECEPTLQIGYQPDPV------SVVT 227
+ + G +++ Y H AQ+ G Y P+EC PTLQ+GY +PV +
Sbjct: 180 SHHMGGGGGWEGGEQNVTYAHHQAQSQG--LYQPLECNPTLQMGYD-NPVCSEQITATTQ 236
Query: 228 AGPSMNNNYMAGWL 241
A N Y+ GW+
Sbjct: 237 AQAQQGNGYIPGWM 250
>SEP2_ARATH (P29384) Developmental protein SEPALLATA2 (Agamous-like
MADS box protein AGL4)
Length = 250
Score = 298 bits (762), Expect = 1e-80
Identities = 160/252 (63%), Positives = 189/252 (74%), Gaps = 14/252 (5%)
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEV+LI+FSNRGKLYEFCS+
Sbjct: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVSLIVFSNRGKLYEFCST 60
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLGP 120
S+MLKTLERYQKC+YG+ E N + LE +S +EYLKLK RYE LQR QRNL+GEDLGP
Sbjct: 61 SNMLKTLERYQKCSYGSIEVNNKPAKELE-NSYREYLKLKGRYENLQRQQRNLLGEDLGP 119
Query: 121 LNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRLEG----- 175
LNSKELE LERQLD SLKQ+R +TQ+MLDQLSDLQ KEH+L +ANR+L +LE
Sbjct: 120 LNSKELEQLERQLDGSLKQVRCIKTQYMLDQLSDLQGKEHILLDANRALSMKLEDMIGVR 179
Query: 176 -YQLNSLQLNPGVEDMGYGRHPAQTHGDAFYHPIECEPTLQIGYQ----PDPVSVVTAGP 230
+ + +++ YG HP Q H Y +EC+PTLQIGY + ++V G
Sbjct: 180 HHHIGGGWEGGDQQNIAYG-HP-QAHSQGLYQSLECDPTLQIGYSHPVCSEQMAVTVQGQ 237
Query: 231 S-MNNNYMAGWL 241
S N Y+ GW+
Sbjct: 238 SQQGNGYIPGWM 249
>CMB1_DIACA (Q39685) MADS box protein CMB1
Length = 233
Score = 264 bits (675), Expect = 1e-70
Identities = 150/245 (61%), Positives = 177/245 (72%), Gaps = 17/245 (6%)
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALI+FSNRGKLYEFCS+
Sbjct: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIVFSNRGKLYEFCST 60
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLGP 120
S M KTLERYQ+C+YG+ E + ++E SS QEYLKLKA+ + LQRS RNL+GEDLG
Sbjct: 61 SCMNKTLERYQRCSYGSLETSQPSKET--ESSYQEYLKLKAKVDVLQRSHRNLLGEDLGE 118
Query: 121 LNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRLEGYQLNS 180
L++KELE LE QLD SL+QIRS +TQ MLDQL+DLQ+KE ML E+NR+L+ +LE S
Sbjct: 119 LSTKELEQLEHQLDKSLRQIRSIKTQHMLDQLADLQKKEEMLFESNRALKTKLE-ESCAS 177
Query: 181 LQLNPGVEDMGYGRHPAQTHGDAFYH--PIECEPTLQIGYQPDPVSVVTAGPSMNN--NY 236
+ N V R P GD F+ P+ C LQIGY + A S N +
Sbjct: 178 FRPNWDV------RQP----GDGFFEPLPLPCNNNLQIGYNEATQDQMNATTSAQNVHGF 227
Query: 237 MAGWL 241
GW+
Sbjct: 228 AQGWM 232
>AGL3_ARATH (P29383) Agamous-like MADS box protein AGL3
Length = 258
Score = 228 bits (581), Expect = 1e-59
Identities = 133/271 (49%), Positives = 175/271 (64%), Gaps = 44/271 (16%)
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRG+VELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAE+AL+IFSNRGKLYEFCSS
Sbjct: 1 MGRGKVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEIALLIFSNRGKLYEFCSS 60
Query: 61 -SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLG 119
S M +T+++Y+K +Y + N S ++ + Q+YLKLK+R E LQ SQR+L+GE+L
Sbjct: 61 PSGMARTVDKYRKHSYATMDPNQSAKDLQD--KYQDYLKLKSRVEILQHSQRHLLGEELS 118
Query: 120 PLNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRLEGYQ-- 177
++ ELE LERQ+D+SL+QIRST+ + MLDQLSDL+ KE ML E NR LR++LE
Sbjct: 119 EMDVNELEHLERQVDASLRQIRSTKARSMLDQLSDLKTKEEMLLETNRDLRRKLEDSDAA 178
Query: 178 -----------------------LNSLQLNPGVEDMGYGRHPAQTHGDAFYHPIECEPTL 214
++S Q NP +++ G F+ P++ L
Sbjct: 179 LTQSFWGSSAAEQQQQHQQQQQGMSSYQSNPPIQEAG------------FFKPLQGNVAL 226
Query: 215 QIG--YQPDPVSVV-TAGPSMN-NNYMAGWL 241
Q+ Y +P + +A S N N + GW+
Sbjct: 227 QMSSHYNHNPANATNSATTSQNVNGFFPGWM 257
>AGL6_ARATH (P29386) Agamous-like MADS box protein AGL6
Length = 252
Score = 200 bits (509), Expect = 2e-51
Identities = 119/254 (46%), Positives = 163/254 (63%), Gaps = 16/254 (6%)
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRGRVE+KRIENKINRQVTF+KRRNGLLKKAYELSVLCDAEVALIIFS+RGKLYEF S
Sbjct: 1 MGRGRVEMKRIENKINRQVTFSKRRNGLLKKAYELSVLCDAEVALIIFSSRGKLYEF-GS 59
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLGP 120
+ T+ERY +C Y +N E + S QE KLK++YE+L R+ RNL+GEDLG
Sbjct: 60 VGIESTIERYNRC-YNCSLSNNKPEETTQ-SWCQEVTKLKSKYESLVRTNRNLLGEDLGE 117
Query: 121 LNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRL--EGYQL 178
+ KEL++LERQL+++L R +TQ M++++ DL++KE L + N+ L+ + EG+
Sbjct: 118 MGVKELQALERQLEAALTATRQRKTQVMMEEMEDLRKKERQLGDINKQLKIKFETEGHAF 177
Query: 179 NSLQ---LNPGVEDMGYGRHPAQTHGDAFYHPIEC--EPTLQIGYQP------DPVSVVT 227
+ Q N G + + + ++C EP LQIG+Q + SV
Sbjct: 178 KTFQDLWANSAASVAGDPNNSEFPVEPSHPNVLDCNTEPFLQIGFQQHYYVQGEGSSVSK 237
Query: 228 AGPSMNNNYMAGWL 241
+ + N++ GW+
Sbjct: 238 SNVAGETNFVQGWV 251
>AP1_SINAL (Q41276) Floral homeotic protein APETALA1 (MADS C)
Length = 254
Score = 191 bits (485), Expect = 1e-48
Identities = 106/229 (46%), Positives = 156/229 (67%), Gaps = 20/229 (8%)
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRGRV+LKRIENKINRQVTF+KRR GLLKKA+E+SVLCDAEVAL++FS++GKL+E+ +
Sbjct: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALVVFSHKGKLFEYSTD 60
Query: 61 SSMLKTLERYQKCNYG-----APEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMG 115
S M K LERY++ +Y APE++V+T ++ EY +LKA+ E L+R+QR+ +G
Sbjct: 61 SCMEKILERYERYSYAERQLIAPESDVNTNWSM------EYNRLKAKIELLERNQRHYLG 114
Query: 116 EDLGPLNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRLEG 175
EDL ++SKEL++LE+QLD++LK IRS + Q M D +++LQRKE + E N L ++++
Sbjct: 115 EDLQAMSSKELQNLEQQLDTALKHIRSRKNQLMHDSINELQRKEKAIQEQNSMLSKQIKE 174
Query: 176 YQ--LNSLQLNPGVEDMGYGRHPAQTHGDAFYHPIECEPTLQIGYQPDP 222
+ L + Q ++ G+ P P + + + +QP P
Sbjct: 175 REKILRAQQEQWDQQNHGHNMPPPPP-------PQQIQHPYMLSHQPSP 216
>CAL_ARATH (Q39081) Transcription factor CAULIFLOWER (Agamous-like
MADS box protein AGL10)
Length = 253
Score = 187 bits (476), Expect = 1e-47
Identities = 103/202 (50%), Positives = 144/202 (70%), Gaps = 18/202 (8%)
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRGRVELKRIENKINRQVTF+KRR GLLKKA E+SVLCDAEV+LI+FS++GKL+E+ S
Sbjct: 1 MGRGRVELKRIENKINRQVTFSKRRTGLLKKAQEISVLCDAEVSLIVFSHKGKLFEYSSE 60
Query: 61 SSMLKTLERYQKCNYG-----APEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMG 115
S M K LERY++ +Y AP+++V+T ++ EY +LKA+ E L+R+QR+ +G
Sbjct: 61 SCMEKVLERYERYSYAERQLIAPDSHVNTNWSM------EYSRLKAKIELLERNQRHYLG 114
Query: 116 EDLGPLNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQR--- 172
E+L P++ K+L++LE+QL+++LK IRS + Q M + L+ LQRKE + E N L ++
Sbjct: 115 EELEPMSLKDLQNLEQQLETALKHIRSRKNQLMNESLNHLQRKEKEIQEENSMLTKQIKE 174
Query: 173 ----LEGYQLNSLQLNPGVEDM 190
L Q QLN V+D+
Sbjct: 175 RENILRTKQTQCEQLNRSVDDV 196
>AP1_ARATH (P35631) Floral homeotic protein APETALA1 (Agamous-like
MADS box protein AGL7)
Length = 256
Score = 186 bits (471), Expect = 6e-47
Identities = 94/179 (52%), Positives = 138/179 (76%), Gaps = 11/179 (6%)
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRGRV+LKRIENKINRQVTF+KRR GLLKKA+E+SVLCDAEVAL++FS++GKL+E+ +
Sbjct: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALVVFSHKGKLFEYSTD 60
Query: 61 SSMLKTLERYQKCNYG-----APEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMG 115
S M K LERY++ +Y APE++V+T ++ EY +LKA+ E L+R+QR+ +G
Sbjct: 61 SCMEKILERYERYSYAERQLIAPESDVNTNWSM------EYNRLKAKIELLERNQRHYLG 114
Query: 116 EDLGPLNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRLE 174
EDL ++ KEL++LE+QLD++LK IR+ + Q M + +++LQ+KE + E N L ++++
Sbjct: 115 EDLQAMSPKELQNLEQQLDTALKHIRTRKNQLMYESINELQKKEKAIQEQNSMLSKQIK 173
>AGL8_LYCES (Q40170) Agamous-like MADS box protein AGL8 homolog
(TM4)
Length = 227
Score = 185 bits (470), Expect = 7e-47
Identities = 97/184 (52%), Positives = 137/184 (73%), Gaps = 1/184 (0%)
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRGRV+LKRIENKINRQVTF+KRR+GLLKKA+E+SVLCDAEV LI+FS +GKL+E+ +
Sbjct: 1 MGRGRVQLKRIENKINRQVTFSKRRSGLLKKAHEISVLCDAEVGLIVFSTKGKLFEYAND 60
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLGP 120
S M + LERY++ ++ A + V T +S E+ KLKAR E LQR+Q++ +GEDL
Sbjct: 61 SCMERILERYERYSF-AEKQLVPTDHTSPVSWTLEHRKLKARLEVLQRNQKHYVGEDLES 119
Query: 121 LNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRLEGYQLNS 180
L+ KEL++LE QLDS+LK IRS + Q M + +S LQ+K+ L E N L ++++ + ++
Sbjct: 120 LSMKELQNLEHQLDSALKHIRSRKNQLMHESISVLQKKDRALQEQNNQLSKKVKEREKSA 179
Query: 181 LQLN 184
Q++
Sbjct: 180 QQIS 183
>AGL8_SOLTU (Q42429) Agamous-like MADS box protein AGL8 homolog
(POTM1-1)
Length = 250
Score = 183 bits (464), Expect = 4e-46
Identities = 98/184 (53%), Positives = 132/184 (71%), Gaps = 1/184 (0%)
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRGRV+LKRIENKINRQVTF+KRR+GLLKKA+E+SVLCDAEV LI+FS +GKL+E+ +
Sbjct: 1 MGRGRVQLKRIENKINRQVTFSKRRSGLLKKAHEISVLCDAEVGLIVFSTKGKLFEYAND 60
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLGP 120
S M + LERY++ ++ A V T S E+ KLKAR E LQR+Q++ +GEDL
Sbjct: 61 SCMERLLERYERYSF-AERQLVPTDHTSPGSWTLEHAKLKARLEVLQRNQKHYVGEDLES 119
Query: 121 LNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRLEGYQLNS 180
LN KEL++LE QLDS+LK IRS + Q M + +S LQ+++ L E N L ++++ +
Sbjct: 120 LNMKELQNLEHQLDSALKHIRSRKNQLMHESISVLQKQDRALQEQNNQLSKKVKEREKEV 179
Query: 181 LQLN 184
Q N
Sbjct: 180 AQQN 183
>AGL8_ARATH (Q38876) Agamous-like MADS box protein AGL8 (Floral
homeotic protein AGL8) (Transcription factor FRUITFULL)
Length = 242
Score = 181 bits (460), Expect = 1e-45
Identities = 97/177 (54%), Positives = 132/177 (73%), Gaps = 7/177 (3%)
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRGRV+LKRIENKINRQVTF+KRR+GLLKKA+E+SVLCDAEVALI+FS++GKL+E+ +
Sbjct: 1 MGRGRVQLKRIENKINRQVTFSKRRSGLLKKAHEISVLCDAEVALIVFSSKGKLFEYSTD 60
Query: 61 SSMLKTLERYQKCNYGAPE---ANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGED 117
S M + LERY + Y + +VS E L E+ KLKAR E L++++RN MGED
Sbjct: 61 SCMERILERYDRYLYSDKQLVGRDVSQSENWVL----EHAKLKARVEVLEKNKRNFMGED 116
Query: 118 LGPLNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRLE 174
L L+ KEL+SLE QLD+++K IRS + Q M + +S LQ+K+ L + N SL ++++
Sbjct: 117 LDSLSLKELQSLEHQLDAAIKSIRSRKNQAMFESISALQKKDKALQDHNNSLLKKIK 173
>AGL8_SINAL (Q41274) Agamous-like MADS box protein AGL8 homolog
(MADS B)
Length = 241
Score = 181 bits (458), Expect = 2e-45
Identities = 96/184 (52%), Positives = 136/184 (73%), Gaps = 7/184 (3%)
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRGRV+LKRIENKINRQVTF+KRR+GLLKKA+E+SVLCDAEVAL+IFS++GKL+E+ +
Sbjct: 1 MGRGRVQLKRIENKINRQVTFSKRRSGLLKKAHEISVLCDAEVALVIFSSKGKLFEYSTD 60
Query: 61 SSMLKTLERYQKCNYGAPE---ANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGED 117
S M K LERY + Y + ++S E L E+ KLKAR E L++++RN MGED
Sbjct: 61 SCMEKILERYDRYLYSDKQLVGRDISQSENWVL----EHAKLKARVEVLEKNKRNFMGED 116
Query: 118 LGPLNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRLEGYQ 177
L L+ KEL+SLE QL +++K IRS + Q M + +S LQ+K+ +L + N +L ++++ +
Sbjct: 117 LDSLSLKELQSLEHQLHAAIKSIRSRKNQAMFESISALQKKDKVLQDHNNALLKKIKERE 176
Query: 178 LNSL 181
N++
Sbjct: 177 KNTV 180
>AG13_ARATH (Q38837) Agamous-like MADS box protein AGL13
Length = 244
Score = 179 bits (455), Expect = 4e-45
Identities = 99/190 (52%), Positives = 134/190 (70%), Gaps = 11/190 (5%)
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRG+VE+KRIENKI RQVTF+KR++GLLKKAYELSVLCDAEV+LIIFS GKLYEF S+
Sbjct: 1 MGRGKVEVKRIENKITRQVTFSKRKSGLLKKAYELSVLCDAEVSLIIFSTGGKLYEF-SN 59
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREALELSS--QQEYLKLKARYEALQRSQRNLMGEDL 118
+ +T+ERY +C + N+ + LE + +QE KLK +YE+L R+ RNL+GEDL
Sbjct: 60 VGVGRTIERYYRC-----KDNLLDNDTLEDTQGLRQEVTKLKCKYESLLRTHRNLVGEDL 114
Query: 119 GPLNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRLEGYQL 178
++ KEL++LERQL+ +L R +TQ M++Q+ +L+RKE L + N L+ E +
Sbjct: 115 EGMSIKELQTLERQLEGALSATRKQKTQVMMEQMEELRRKERELGDINNKLKLETEDHDF 174
Query: 179 NSLQ---LNP 185
Q LNP
Sbjct: 175 KGFQDLLLNP 184
>AGL8_SOLCO (O22328) Agamous-like MADS box protein AGL8 homolog
Length = 250
Score = 175 bits (444), Expect = 8e-44
Identities = 94/174 (54%), Positives = 127/174 (72%), Gaps = 1/174 (0%)
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRGRV+LKRIENKINRQVTF+KRR+GLLKKA+E+SVLCDAEV LI+FS +GKL+E+ +
Sbjct: 1 MGRGRVQLKRIENKINRQVTFSKRRSGLLKKAHEISVLCDAEVGLIVFSTKGKLFEYATD 60
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLGP 120
S M + LERY++ ++ A + V T S E KLKAR E LQR+++ +GEDL
Sbjct: 61 SCMERLLERYERYSF-AEKQLVPTDHTSPGSWTLENAKLKARLEVLQRNEKLYVGEDLES 119
Query: 121 LNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRLE 174
LN KEL++LE QL S+LK IRS + Q M + +S LQ+++ L E N L ++++
Sbjct: 120 LNMKELQNLEHQLASALKHIRSRKNQLMHESISVLQKQDRALQEQNNQLSKKVK 173
>AGL1_ARATH (P29381) Agamous-like MADS box protein AGL1 (Protein
Shatterproof 1)
Length = 248
Score = 167 bits (424), Expect = 2e-41
Identities = 100/230 (43%), Positives = 142/230 (61%), Gaps = 10/230 (4%)
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
+GRG++E+KRIEN NRQVTF KRRNGLLKKAYELSVLCDAEVAL+IFS RG+LYE+ ++
Sbjct: 16 LGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALVIFSTRGRLYEY-AN 74
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLGP 120
+S+ T+ERY+K A S EA QQE KL+ + +Q S R+++GE LG
Sbjct: 75 NSVRGTIERYKKACSDAVNP-PSVTEANTQYYQQEASKLRRQIRDIQNSNRHIVGESLGS 133
Query: 121 LNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRL-EGYQLN 179
LN KEL++LE +L+ + ++RS + + ++ ++ +Q++E L N LR ++ EG +LN
Sbjct: 134 LNFKELKNLEGRLEKGISRVRSKKNELLVAEIEYMQKREMELQHNNMYLRAKIAEGARLN 193
Query: 180 ------SLQLNPGVEDMGYGRHPAQTHGDAFYHPIE-CEPTLQIGYQPDP 222
S+ V + G H H + Y P+ EP Q Q P
Sbjct: 194 PDQQESSVIQGTTVYESGVSSHDQSQHYNRNYIPVNLLEPNQQFSGQDQP 243
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.132 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,485,945
Number of Sequences: 164201
Number of extensions: 1005701
Number of successful extensions: 3597
Number of sequences better than 10.0: 182
Number of HSP's better than 10.0 without gapping: 83
Number of HSP's successfully gapped in prelim test: 99
Number of HSP's that attempted gapping in prelim test: 3381
Number of HSP's gapped (non-prelim): 249
length of query: 242
length of database: 59,974,054
effective HSP length: 107
effective length of query: 135
effective length of database: 42,404,547
effective search space: 5724613845
effective search space used: 5724613845
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0044b.3


