

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0044a.9
(464 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
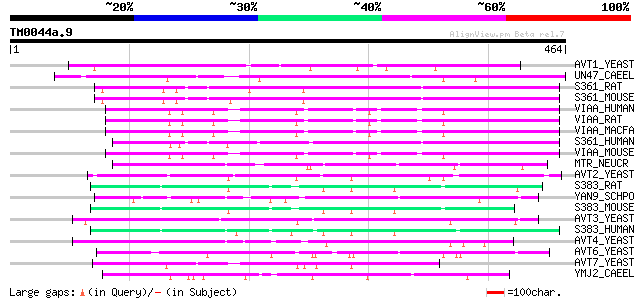
Score E
Sequences producing significant alignments: (bits) Value
AVT1_YEAST (P47082) Vacuolar amino acid transporter 1 126 1e-28
UN47_CAEEL (P34579) Vesicular GABA transporter (Uncoordinated pr... 103 9e-22
S361_RAT (Q924A5) Proton-coupled amino acid transporter 1 (Proto... 92 3e-18
S361_MOUSE (Q8K4D3) Proton-coupled amino acid transporter 1 (Pro... 92 4e-18
VIAA_HUMAN (Q9H598) Vesicular inhibitory amino acid transporter ... 91 6e-18
VIAA_RAT (O35458) Vesicular inhibitory amino acid transporter (G... 89 2e-17
VIAA_MACFA (Q95KE2) Vesicular inhibitory amino acid transporter ... 89 2e-17
S361_HUMAN (Q7Z2H8) Proton-coupled amino acid transporter 1 (Pro... 89 2e-17
VIAA_MOUSE (O35633) Vesicular inhibitory amino acid transporter ... 89 3e-17
MTR_NEUCR (P38680) N amino acid transport system protein (Methyl... 77 1e-13
AVT2_YEAST (P39981) Vacuolar amino acid transporter 2 71 5e-12
S383_RAT (Q9JHZ9) System N amino acid transporter 1 (SN1) (N-sys... 71 7e-12
YAN9_SCHPO (Q10074) Hypothetical protein C3H1.09c in chromosome I 70 1e-11
S383_MOUSE (Q9DCP2) System N amino acid transporter 1 (SN1) (N-s... 69 3e-11
AVT3_YEAST (P36062) Vacuolar amino acid transporter 3 69 3e-11
S383_HUMAN (Q99624) System N amino acid transporter 1 (SN1) (N-s... 69 3e-11
AVT4_YEAST (P50944) Vacuolar amino acid transporter 4 68 4e-11
AVT6_YEAST (P40074) Vacuolar amino acid transporter 6 56 2e-07
AVT7_YEAST (P40501) Vacuolar amino acid transporter 7 50 1e-05
YMJ2_CAEEL (P34479) Hypothetical protein F59B2.2 in chromosome III 47 1e-04
>AVT1_YEAST (P47082) Vacuolar amino acid transporter 1
Length = 602
Score = 126 bits (316), Expect = 1e-28
Identities = 104/397 (26%), Positives = 184/397 (46%), Gaps = 25/397 (6%)
Query: 50 NNIEESAVARDTNVDAEHDA--KANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAI 107
N+++ V R VD + S+ ++ N + +LIG+G L+ P ++ GW +
Sbjct: 183 NDMDSIVVKRVEGVDGKVVTLLAGQSTAPQTIFNSINVLIGIGLLALPLGLKYAGWVIGL 242
Query: 108 LLIALGVMCTY-TCHLLGKCLKKNPKLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVS 166
++A+ + T+ T LL +CL +P L SY D+G AFG KGR L + L +D+ S VS
Sbjct: 243 TMLAIFALATFCTAELLSRCLDTDPTLISYADLGYAAFGTKGRALISALFTLDLLGSGVS 302
Query: 167 YTISLHDNLTTVLHLKQLHLAKLSAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMS 226
I D+L + K+ + I+T V +P L ++S + LS+ G ++
Sbjct: 303 LVILFGDSLNALFPQYSTTFFKIVSFFIVTPP---VFIPLSVLSNISLLGILSTTGTVL- 358
Query: 227 VVIFVSVAATAVFGGVRVNHEIPF--LRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPS 284
V+ + ++ G + E + L ++ G+ +GGH VFPNL M+ P
Sbjct: 359 VICCCGLYKSSSPGSLVNPMETSMWPIDLKHLCLSIGLLSACWGGHAVFPNLKTDMRHPD 418
Query: 285 KFTKV--SILSFTTVTALYTIMGYMGAKMFG----PEVNSQVTLSMPSKQIVTKIALWAT 338
KF + T+VT + T + +G MFG E+ V L+ + V +
Sbjct: 419 KFKDCLKTTYKITSVTDIGTAV--IGFLMFGNLVKDEITKNVLLTEGYPKFVYGLISALM 476
Query: 339 VLTPMTKYALEFAPVA--------IQLEHKLPSSMSGRTKMILRGIIGSSLLLVILALAL 390
+ P+ K L P+ +Q + S++ R L+ + +V + +A+
Sbjct: 477 TIIPIAKTPLNARPIVSVLDVLMNVQHIDEAASAIKRRAAKGLQVFNRIFINVVFVLIAI 536
Query: 391 TVPYFEHVLSLTGSLVSVAISLIFPCAFYIKICRGQI 427
P F+ +++ G+ + I LI PC FY+++C+ I
Sbjct: 537 NFPEFDKIIAFLGAGLCFTICLILPCWFYLRLCKTTI 573
>UN47_CAEEL (P34579) Vesicular GABA transporter (Uncoordinated
protein 47) (Protein unc-47)
Length = 486
Score = 103 bits (257), Expect = 9e-22
Identities = 101/445 (22%), Positives = 193/445 (42%), Gaps = 35/445 (7%)
Query: 38 VEESQLCKCDHINNIEESAVARDTNVDAEHDAKANSSLAHSVMNMVGMLIGLGQLSTPYA 97
++ES + + ++I + A+D D +A S + N+ + G+ + P A
Sbjct: 57 IQESVVSEQPQKDDINKQEEAKD---DGHGEASEPISALQAAWNVTNAIQGMFIVGLPIA 113
Query: 98 VEQGGWASAILLIALGVMCTYTCHLLGKCLKKN--PKLRSYVDIGNHAFGAKGRFLAAIL 155
V+ GGW S ++ + +C +T LL +CL +N K ++Y +I + G+++ A
Sbjct: 114 VKVGGWWSIGAMVGVAYVCYWTGVLLIECLYENGVKKRKTYREIADFYKPGFGKWVLAAQ 173
Query: 156 IYMDIFMSLVSYTISLHDNLTTVLHLKQLHLAKLSAPQILTVGAVLVALPSL----WLRD 211
+ ++ + + Y + D L + P + G +++ SL +L D
Sbjct: 174 L-TELLSTCIIYLVLAADLLQSCF------------PSVDKAGWMMITSASLLTCSFLDD 220
Query: 212 LSSISFLSSVGVLMSVVI-FVSVAATAVFGGVRVNHEIPF-LRLHNIPSISGIYIFSYGG 269
L +S LS + +++ + V F I F L ++ +P+I G+ +F Y
Sbjct: 221 LQIVSRLSFFNAISHLIVNLIMVLYCLSFVSQWSFSTITFSLNINTLPTIVGMVVFGYTS 280
Query: 270 HIVFPNLYKAMKDPSKFTKVSILSFTTVTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQI 329
HI PNL MK+P++F + S + G +G FG +++ S+P++
Sbjct: 281 HIFLPNLEGNMKNPAQFNVMLKWSHIAAAVFKVVFGMLGFLTFGELTQEEISNSLPNQSF 340
Query: 330 VTKIALWATVLTPMTKYALEFAPVAIQLEHKL--------PSSMSGRTKMILRGIIGSSL 381
+ L V+ + Y L F L++ L +S K + + +
Sbjct: 341 KILVNL-ILVVKALLSYPLPFYAAVQLLKNNLFLGYPQTPFTSCYSPDKSLREWAVTLRI 399
Query: 382 LLVILAL--ALTVPYFEHVLSLTGSLVSVAISLIFPCAFYIKICRGQISKPLFVLNISII 439
+LV+ L AL+VPY ++ L G++ +S I+P F++ I ++ + II
Sbjct: 400 ILVLFTLFVALSVPYLVELMGLVGNITGTMLSFIWPALFHLYIKEKTLNNFEKRFDQGII 459
Query: 440 TFGFLLGVVGTISSSKLLVEKILAA 464
G + + G SS L+ I +A
Sbjct: 460 IMGCSVCISGVYFSSMELLRAINSA 484
>S361_RAT (Q924A5) Proton-coupled amino acid transporter 1
(Proton/amino acid transporter 1) (Solute carrier family
36 member 1) (Lysosomal amino acid transporter 1)
(LYAAT-1) (Neutral amino acid/proton symporter)
Length = 475
Score = 92.0 bits (227), Expect = 3e-18
Identities = 103/421 (24%), Positives = 184/421 (43%), Gaps = 36/421 (8%)
Query: 72 NSSLA--HSVMNMVGMLIGLGQLSTPYAVEQGGWASAIL-LIALGVMCTYTCHLLGKCL- 127
NSS+ ++++++ IG G L P AV+ G L L+ +G++ + +L KC
Sbjct: 43 NSSMTWFQTLIHLLKGNIGTGLLGLPLAVKNAGLLLGPLSLLVIGIVAVHCMGILVKCAH 102
Query: 128 ----KKNPKLRSYVD-------------IGNHAFGAKGRFLAAILIYMDIFMSLVSYTIS 170
+ N Y D I NH+ + R + L+ + V Y +
Sbjct: 103 HLCRRLNKPFLDYGDTVMYGLECSPSTWIRNHSHWGR-RIVDFFLVVTQLGFCCV-YFVF 160
Query: 171 LHDNLTTVLHLKQLHLAKLSAPQILTVGA-------VLVALPSLWLRD-LSSISFLSSVG 222
L DN V+ + + + + +L LP L L + ++ LS
Sbjct: 161 LADNFKQVIEAANGTTTNCNNNETVILTPTMDSRLYMLTFLPFLVLLSFIRNLRILSIFS 220
Query: 223 VLMSVVIFVSVAATAVFGGVRV---NHEIPFLRLHNIPSISGIYIFSYGGHIVFPNLYKA 279
+L ++ +FVS+ F R+ +H P G IF++ G V L
Sbjct: 221 LLANISMFVSLIMIYQFIVQRIPDPSHLPLVAPWKTYPLFFGTAIFAFEGIGVVLPLENK 280
Query: 280 MKDPSKFTKVSILSFTTVTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATV 339
MKD KF + L +T LY +G +G FG ++ +TL++P+ + + L ++
Sbjct: 281 MKDSQKFPLILYLGMAIITVLYISLGSLGYLQFGADIKGSITLNLPNCWLYQSVKLLYSI 340
Query: 340 LTPMTKYALEFAPVAIQLEHKLPSSMSGRTKMILRGIIGSSLLLVILALALTVPYFEHVL 399
T YAL+F A + + S + R ++++ ++++ V LA+ +P + V+
Sbjct: 341 GIFFT-YALQFYVAAEIIIPAIVSRVPERFELVVDLSARTAMVCVTCVLAVLIPRLDLVI 399
Query: 400 SLTGSLVSVAISLIFPCAFYIKICRGQISKPLFVLNISIIT-FGFLLGVVGTISSSKLLV 458
SL GS+ S A++LI P + G+ PL + ++I+ GF+ VVGT S L+
Sbjct: 400 SLVGSVSSSALALIIPPLLEVTTYYGEGISPLTITKDALISILGFVGFVVGTYESLWELI 459
Query: 459 E 459
+
Sbjct: 460 Q 460
>S361_MOUSE (Q8K4D3) Proton-coupled amino acid transporter 1
(Proton/amino acid transporter 1) (Solute carrier family
36 member 1)
Length = 475
Score = 91.7 bits (226), Expect = 4e-18
Identities = 104/421 (24%), Positives = 185/421 (43%), Gaps = 36/421 (8%)
Query: 72 NSSLA--HSVMNMVGMLIGLGQLSTPYAVEQGGWASAIL-LIALGVMCTYTCHLLGKCL- 127
NSS+ ++++++ IG G L P AV+ G L L+ +G++ + +L KC
Sbjct: 43 NSSMTWFQTLIHLLKGNIGTGLLGLPLAVKNAGLLLGPLSLLVIGIVAVHCMGILVKCAH 102
Query: 128 ----KKNPKLRSYVD-------------IGNHAFGAKGRFLAAILIYMDIFMSLVSYTIS 170
+ N Y D + NH+ + R + LI + V Y +
Sbjct: 103 HLCRRLNKPFLDYGDTVMYGLECSPSTWVRNHSHWGR-RIVDFFLIVTQLGFCCV-YFVF 160
Query: 171 LHDNLTTVLHLKQ-------LHLAKLSAPQILTVGAVLVALPSLWLRD-LSSISFLSSVG 222
L DN V+ ++ + P + + +L LP L L + ++ LS
Sbjct: 161 LADNFKQVIEAANGTTTNCNNNVTVIPTPTMDSRLYMLSFLPFLVLLSFIRNLRVLSIFS 220
Query: 223 VLMSVVIFVSVAATAVFGGVRV---NHEIPFLRLHNIPSISGIYIFSYGGHIVFPNLYKA 279
+L ++ +FVS+ F R+ +H P G IF++ G V L
Sbjct: 221 LLANISMFVSLIMIYQFIVQRIPDPSHLPLVAPWKTYPLFFGTAIFAFEGIGVVLPLENK 280
Query: 280 MKDPSKFTKVSILSFTTVTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATV 339
MKD KF + L +T LY +G +G FG + +TL++P+ + + L ++
Sbjct: 281 MKDSQKFPLILYLGMAIITVLYISLGSLGYLQFGANIKGSITLNLPNCWLYQSVKLLYSI 340
Query: 340 LTPMTKYALEFAPVAIQLEHKLPSSMSGRTKMILRGIIGSSLLLVILALALTVPYFEHVL 399
T YAL+F A + + S + ++++ + ++++ V LA+ +P + V+
Sbjct: 341 GIFFT-YALQFYVAAEIIIPAIVSRVPEHFELMVDLCVRTAMVCVTCVLAILIPRLDLVI 399
Query: 400 SLTGSLVSVAISLIFPCAFYIKICRGQISKPLFVLNISIIT-FGFLLGVVGTISSSKLLV 458
SL GS+ S A++LI P + G+ PL V ++I+ GF+ VVGT S L+
Sbjct: 400 SLVGSVSSSALALIIPPLLEVVTYYGEGISPLTVTKDALISILGFVGFVVGTYESLCELI 459
Query: 459 E 459
+
Sbjct: 460 Q 460
>VIAA_HUMAN (Q9H598) Vesicular inhibitory amino acid transporter
(GABA and glycine transporter) (Vesicular GABA
transporter) (hVIAAT)
Length = 525
Score = 90.9 bits (224), Expect = 6e-18
Identities = 100/414 (24%), Positives = 182/414 (43%), Gaps = 55/414 (13%)
Query: 81 NMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTYTCHLLGKCLKKNPK-------L 133
N+ + G+ L PYA+ GG+ L+I V+C YT +L CL + +
Sbjct: 124 NVTNAIQGMFVLGLPYAILHGGYLGLFLIIFAAVVCCYTGKILIACLYEENEDGEVVRVR 183
Query: 134 RSYVDIGNHA----FGAKGRFLAAILIYMDIFMSLVSYTI---SLHDNLTTVLHLKQLHL 186
SYV I N F G + + +++ M+ + Y + +L N L + Q
Sbjct: 184 DSYVAIANACCAPRFPTLGGRVVNVAQIIELVMTCILYVVVSGNLMYNSFPGLPVSQ--- 240
Query: 187 AKLSAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAV-----FGG 241
+ ++ A V LP +L++L ++S S + L VI + V A + +
Sbjct: 241 ------KSWSIIATAVLLPCAFLKNLKAVSKFSLLCTLAHFVINILVIAYCLSRARDWAW 294
Query: 242 VRVNHEIPFLRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTTVTA-- 299
+V ++ + P GI +FSY I P+L M+ PS+F ++++T + A
Sbjct: 295 EKVKF---YIDVKKFPISIGIIVFSYTSQIFLPSLEGNMQQPSEFH--CMMNWTHIAACV 349
Query: 300 ---LYTIMGYMGAKMFGPEVNSQVTLSMP-SKQIVTKIALWATVLTPMTKYALEFAPVAI 355
L+ ++ Y+ + E +T ++P S + V I L A L Y L F
Sbjct: 350 LKGLFALVAYL---TWADETKEVITDNLPGSIRAVVNIFLVAKALL---SYPLPFFAAVE 403
Query: 356 QLEHKL---------PSSMSGRTKMILRGI-IGSSLLLVILALALTVPYFEHVLSLTGSL 405
LE L P+ SG ++ G+ + +L++ L +A+ VP+F ++ LTGSL
Sbjct: 404 VLEKSLFQEGSRAFFPACYSGDGRLKSWGLTLRCALVVFTLLMAIYVPHFALLMGLTGSL 463
Query: 406 VSVAISLIFPCAFYIKICRGQISKPLFVLNISIITFGFLLGVVGTISSSKLLVE 459
+ + P F++++ ++ +++I G + V G + S + L+E
Sbjct: 464 TGAGLCFLLPSLFHLRLLWRKLLWHQVFFDVAIFVIGGICSVSGFVHSLEGLIE 517
>VIAA_RAT (O35458) Vesicular inhibitory amino acid transporter (GABA
and glycine transporter) (Vesicular GABA transporter)
(rGVAT)
Length = 525
Score = 89.4 bits (220), Expect = 2e-17
Identities = 99/414 (23%), Positives = 181/414 (42%), Gaps = 55/414 (13%)
Query: 81 NMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTYTCHLLGKCLKKNPK-------L 133
N+ + G+ L PYA+ GG+ L+I V+C YT +L CL + +
Sbjct: 124 NVTNAIQGMFVLGLPYAILHGGYLGLFLIIFAAVVCCYTGKILIACLYEENEDGEVVRVR 183
Query: 134 RSYVDIGNHA----FGAKGRFLAAILIYMDIFMSLVSYTI---SLHDNLTTVLHLKQLHL 186
SYV I N F G + + +++ M+ + Y + +L N L + Q
Sbjct: 184 DSYVAIANACCAPRFPTLGGRVVNVAQIIELVMTCILYVVVSGNLMYNSFPGLPVSQ--- 240
Query: 187 AKLSAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAV-----FGG 241
+ ++ A V LP +L++L ++S S + L VI + V A + +
Sbjct: 241 ------KSWSIIATAVLLPCAFLKNLKAVSKFSLLCTLAHFVINILVIAYCLSRARDWAW 294
Query: 242 VRVNHEIPFLRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTTVTA-- 299
+V ++ + P GI +FSY I P+L M+ PS+F ++++T + A
Sbjct: 295 EKVKF---YIDVKKFPISIGIIVFSYTSQIFLPSLEGNMQQPSEFH--CMMNWTHIAACV 349
Query: 300 ---LYTIMGYMGAKMFGPEVNSQVTLSMP-SKQIVTKIALWATVLTPMTKYALEFAPVAI 355
L+ ++ Y+ + E +T ++P S + V I L A L Y L F
Sbjct: 350 LKGLFALVAYL---TWADETKEVITDNLPGSIRAVVNIFLVAKALL---SYPLPFFAAVE 403
Query: 356 QLEHKL---------PSSMSGRTKMILRGI-IGSSLLLVILALALTVPYFEHVLSLTGSL 405
LE L P+ G ++ G+ + +L++ L +A+ VP+F ++ LTGSL
Sbjct: 404 VLEKSLFQEGSRAFFPACYGGDGRLKSWGLTLRCALVVFTLLMAIYVPHFALLMGLTGSL 463
Query: 406 VSVAISLIFPCAFYIKICRGQISKPLFVLNISIITFGFLLGVVGTISSSKLLVE 459
+ + P F++++ ++ +++I G + V G + S + L+E
Sbjct: 464 TGAGLCFLLPSLFHLRLLWRKLLWHQVFFDVAIFVIGGICSVSGFVHSLEGLIE 517
>VIAA_MACFA (Q95KE2) Vesicular inhibitory amino acid transporter
(GABA and glycine transporter) (Vesicular GABA
transporter) (QccE-21148)
Length = 525
Score = 89.4 bits (220), Expect = 2e-17
Identities = 99/414 (23%), Positives = 181/414 (42%), Gaps = 55/414 (13%)
Query: 81 NMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTYTCHLLGKCLKKNPK-------L 133
N+ + G+ L PYA+ GG+ L+I V+C YT +L CL + +
Sbjct: 124 NVTNAIQGMFVLGLPYAILHGGYLGLFLIIFAAVVCCYTGKILIACLYEENEDGEVVRVR 183
Query: 134 RSYVDIGNHA----FGAKGRFLAAILIYMDIFMSLVSYTI---SLHDNLTTVLHLKQLHL 186
SYV I N F G + + +++ M+ + Y + +L N L + Q
Sbjct: 184 DSYVAIANACCAPRFPTLGGRVVNVAQIIELVMTCILYVVVSGNLMYNSFPGLPVSQ--- 240
Query: 187 AKLSAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAV-----FGG 241
+ ++ A V LP +L++L ++S S + L VI + V A + +
Sbjct: 241 ------KSWSIIATAVLLPCAFLKNLKAVSKFSLLCTLAHFVINILVIAYCLSRARDWAW 294
Query: 242 VRVNHEIPFLRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTTVTA-- 299
+V ++ + P GI +FSY I P+L M+ PS+F ++++T + A
Sbjct: 295 EKVKF---YIDVKKFPISIGIIVFSYTSQIFLPSLEGNMQQPSEFH--CMMNWTHIAACV 349
Query: 300 ---LYTIMGYMGAKMFGPEVNSQVTLSMP-SKQIVTKIALWATVLTPMTKYALEFAPVAI 355
L+ ++ Y+ + E +T ++P S + V I L A L Y L F
Sbjct: 350 LKGLFALVAYL---TWADETKEVITDNLPGSIRAVVNIFLVAKALL---SYPLPFFAAVE 403
Query: 356 QLEHKL---------PSSMSGRTKMILRGI-IGSSLLLVILALALTVPYFEHVLSLTGSL 405
LE L P+ G ++ G+ + +L++ L +A+ VP+F ++ LTGSL
Sbjct: 404 VLEKSLFQEGSRAFFPACYGGDGRLKSWGLTLRCALVVFTLLMAIYVPHFALLMGLTGSL 463
Query: 406 VSVAISLIFPCAFYIKICRGQISKPLFVLNISIITFGFLLGVVGTISSSKLLVE 459
+ + P F++++ ++ +++I G + V G + S + L+E
Sbjct: 464 TGAGLCFLLPSLFHLRLLWRKLLWHQVFFDVAIFVIGGICSVSGFVHSLEGLIE 517
>S361_HUMAN (Q7Z2H8) Proton-coupled amino acid transporter 1
(Proton/amino acid transporter 1) (Solute carrier family
36 member 1)
Length = 476
Score = 89.0 bits (219), Expect = 2e-17
Identities = 102/409 (24%), Positives = 173/409 (41%), Gaps = 44/409 (10%)
Query: 87 IGLGQLSTPYAVEQGGWASA-ILLIALGVMCTYTCHLLGKCLKKNPKL--RSYVDIG--- 140
IG G L P AV+ G I L+ +G++ + +L KC + +S+VD G
Sbjct: 61 IGTGLLGLPLAVKNAGIVMGPISLLIIGIVAVHCMGILVKCAHHFCRRLNKSFVDYGDTV 120
Query: 141 -------------NHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLH------- 180
NHA + R + LI + V Y + L DN V+
Sbjct: 121 MYGLESSPCSWLRNHAHWGR-RVVDFFLIVTQLGFCCV-YFVFLADNFKQVIEAANGTTN 178
Query: 181 ---------LKQLHLAKLSAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFV 231
L ++L L +LV + +L R LS S L+++ +L+S+V+
Sbjct: 179 NCHNNETVILTPTMDSRLYMLSFLPFLVLLVFIRNL--RALSIFSLLANITMLVSLVMIY 236
Query: 232 SVAATAVFGGVRVNHEIPFLRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSI 291
+ + P+ P G IFS+ G + L MKDP KF +
Sbjct: 237 QFIVQRIPDPSHLPLVAPW---KTYPLFFGTAIFSFEGIGMVLPLENKMKDPRKFPLILY 293
Query: 292 LSFTTVTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEFA 351
L VT LY +G +G FG + +TL++P+ + + L ++ T YAL+F
Sbjct: 294 LGMVIVTILYISLGCLGYLQFGANIQGSITLNLPNCWLYQSVKLLYSIGIFFT-YALQFY 352
Query: 352 PVAIQLEHKLPSSMSGRTKMILRGIIGSSLLLVILALALTVPYFEHVLSLTGSLVSVAIS 411
A + S ++++ + + L+ + LA+ +P + V+SL GS+ S A++
Sbjct: 353 VPAEIIIPFFVSRAPEHCELVVDLFVRTVLVCLTCILAILIPRLDLVISLVGSVSSSALA 412
Query: 412 LIFPCAFYIKICRGQISKPLFVLNISIIT-FGFLLGVVGTISSSKLLVE 459
LI P + + PL + ++I+ GF+ VVGT + L++
Sbjct: 413 LIIPPLLEVTTFYSEGMSPLTIFKDALISILGFVGFVVGTYEALYELIQ 461
>VIAA_MOUSE (O35633) Vesicular inhibitory amino acid transporter
(GABA and glycine transporter) (Vesicular GABA
transporter) (mVIAAT) (mVGAT)
Length = 525
Score = 88.6 bits (218), Expect = 3e-17
Identities = 98/414 (23%), Positives = 181/414 (43%), Gaps = 55/414 (13%)
Query: 81 NMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTYTCHLLGKCLKKNPK-------L 133
N+ + G+ L PYA+ GG+ L+I V+C YT +L CL + +
Sbjct: 124 NVTNAIQGMFVLGLPYAILHGGYLGLFLIIFAAVVCCYTGKILIACLYEENEDGEVVRVR 183
Query: 134 RSYVDIGNHA----FGAKGRFLAAILIYMDIFMSLVSYTI---SLHDNLTTVLHLKQLHL 186
SYV I N F G + + +++ M+ + Y + +L N L + Q
Sbjct: 184 DSYVAIANACCAPRFPTLGGRVVNVAQIIELVMTCILYVVVSGNLMYNSFPGLPVSQ--- 240
Query: 187 AKLSAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAV-----FGG 241
+ ++ A V LP +L++L ++S S + L VI + V A + +
Sbjct: 241 ------KSWSIIATAVLLPCAFLKNLKAVSKFSLLCTLAHFVINILVIAYCLSRARDWAW 294
Query: 242 VRVNHEIPFLRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTTVTA-- 299
+V ++ + P GI +FSY I P+L M+ PS+F ++++T + A
Sbjct: 295 EKVKF---YIDVKKFPISIGIIVFSYTSQIFLPSLEGNMQQPSEFH--CMMNWTHIAACV 349
Query: 300 ---LYTIMGYMGAKMFGPEVNSQVTLSMP-SKQIVTKIALWATVLTPMTKYALEFAPVAI 355
L+ ++ Y+ + E +T ++P S + V + L A L Y L F
Sbjct: 350 LKGLFALVAYL---TWADETKEVITDNLPGSIRAVVNLFLVAKALL---SYPLPFFAAVE 403
Query: 356 QLEHKL---------PSSMSGRTKMILRGI-IGSSLLLVILALALTVPYFEHVLSLTGSL 405
LE L P+ G ++ G+ + +L++ L +A+ VP+F ++ LTGSL
Sbjct: 404 VLEKSLFQEGSRAFFPACYGGDGRLKSWGLTLRCALVVFTLLMAIYVPHFALLMGLTGSL 463
Query: 406 VSVAISLIFPCAFYIKICRGQISKPLFVLNISIITFGFLLGVVGTISSSKLLVE 459
+ + P F++++ ++ +++I G + V G + S + L+E
Sbjct: 464 TGAGLCFLLPSLFHLRLLWRKLLWHQVFFDVAIFVIGGICSVSGFVHSLEGLIE 517
>MTR_NEUCR (P38680) N amino acid transport system protein
(Methyltryptophan resistance protein)
Length = 470
Score = 76.6 bits (187), Expect = 1e-13
Identities = 83/387 (21%), Positives = 165/387 (42%), Gaps = 36/387 (9%)
Query: 87 IGLGQLSTPYAVEQGGWASAILL-IALGVMCTYTCHLLGKCLKKNPKLRSYVDIGNHAFG 145
I LG LS P A G ++L + +G++C YT H++G+ K+P++ Y D+G FG
Sbjct: 67 IALGSLSLPGAFATLGMVPGVILSVGMGLICIYTAHVIGQTKLKHPEIAHYADVGRVMFG 126
Query: 146 AKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLHLKQLHLAKLSAPQILTVGAVLVALP 205
G + + + + + + S+ ++ T+ L + + L+A+P
Sbjct: 127 RWGYEIISFMFVLQLIFIVGSHVLTGTIMWGTITD-NGNGTCSLVFGIVSAIILFLLAIP 185
Query: 206 SLWLRDLSSISFLSSVGVLMSVVIFVSVAATAVFGGVRVNHE------IPF-------LR 252
S + ++ +G + V I ++ T + G+R +H+ +P+ L
Sbjct: 186 -------PSFAEVAILGYIDFVSICAAILITMIATGIRSSHQEGGLAAVPWSCWPKEDLS 238
Query: 253 L-HNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTTVTALYTIMGYMGAKM 311
L ++S I +F+Y + + M PS + K + +YT+ G +
Sbjct: 239 LAEGFIAVSNI-VFAYSFAMCQFSFMDEMHTPSDYKKSIVALGLIEIFIYTVTGGVVYAF 297
Query: 312 FGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEFAPVAIQLEHKL-PSSMSGRTK 370
GPEV S LS + ++ K+A + ++ V+ L ++ P+++
Sbjct: 298 VGPEVQSPALLS--AGPLLAKVAFGIALPVIFISGSINTVVVSRYLIERIWPNNVIRYVN 355
Query: 371 -----MILRGIIGSSLLLVILALALTVPYFEHVLSLTGSLVSVAISLIFPCAFYIKICRG 425
M+ G + L+ +A +P+F +L++ +L S FP Y KI R
Sbjct: 356 TPAGWMVWLG-FDFGITLIAWVIAEAIPFFSDLLAICSALFISGFSFYFPALMYFKITRN 414
Query: 426 QI---SKPLFVLNISIITFGFLLGVVG 449
K F+ ++++ F +G++G
Sbjct: 415 DAKSQGKKYFLDALNMLCFVIGMGILG 441
>AVT2_YEAST (P39981) Vacuolar amino acid transporter 2
Length = 480
Score = 71.2 bits (173), Expect = 5e-12
Identities = 90/421 (21%), Positives = 178/421 (41%), Gaps = 37/421 (8%)
Query: 66 EHDAKANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAIL-LIALGVMCTYTCHLLG 124
E+D K SS+ + MN+ ++G G ++ P+A++ G +L +ALG + +T L+
Sbjct: 62 ENDKK--SSMRMAFMNLANSILGAGIITQPFAIKNAGILGGLLSYVALGFIVDWTLRLIV 119
Query: 125 KCLKKNPKLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLHL--- 181
L K R+Y H G KG+ L + F + Y I + D + VL
Sbjct: 120 INLTLAGK-RTYQGTVEHVMGKKGKLLILFTNGLFAFGGCIGYCIIIGDTIPHVLRAIFS 178
Query: 182 ---KQLHLAKLSAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAV 238
+H L I+ + ++ P R++ ++S S + V+ ++I ++V
Sbjct: 179 QNDGNVHFW-LRRNVIIVMVTTFISFPLSMKRNIEALSKASFLAVISMIIIVLTVVIRGP 237
Query: 239 -----FGGVRVNHEIPFLRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPS--KFTKVSI 291
+ G + F++ S+S + F+ H ++ +M++ S KFT+++
Sbjct: 238 MLPYDWKGHSLKLSDFFMKATIFRSLS-VISFALVCHHNTSFIFFSMRNRSVAKFTRLTH 296
Query: 292 LSFTTVTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALE-- 349
+S +MGY G +F + V S P IA +T + +E
Sbjct: 297 ISIIISVICCALMGYSGFAVFKEKTKGNVLNSFPGTDTAINIARLCFGFNMLTTFPMEIF 356
Query: 350 -FAPVAIQLEHKL--------PSSMSGRTKMILRGIIGSSLLLVILALALTVPYFEHVLS 400
V L H+ + +SG+ + +I SSL+ + + ++LT +
Sbjct: 357 VLRDVVGNLLHECNLIKNYDEHTQLSGKQHV----VITSSLVFITMGISLTTCNLGALFE 412
Query: 401 LTGSLVSVAISLIFPCAFYIKICRGQISKPLFVLNISIITFGFLLGVVGTISSSKLLVEK 460
L G+ + ++ I P + + + S + I FGF++ + ISS++ +++
Sbjct: 413 LIGATTASTMAYILPPYTNLLLTSKKKSWKERLPFYLCICFGFMIMI---ISSTQTIIDA 469
Query: 461 I 461
+
Sbjct: 470 V 470
>S383_RAT (Q9JHZ9) System N amino acid transporter 1 (SN1) (N-system
amino acid transporter 1) (Solute carrier family 38,
member 3)
Length = 504
Score = 70.9 bits (172), Expect = 7e-12
Identities = 92/433 (21%), Positives = 166/433 (38%), Gaps = 66/433 (15%)
Query: 68 DAKANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAI-LLIALGVMCTYTCHLLGKC 126
D + +S SV N+ ++G G L YA+ G + LL A+ ++ +Y+ HLL K
Sbjct: 60 DFEGKTSFGMSVFNLSNAIMGSGILGLAYAMANTGIILFLFLLTAVALLSSYSIHLLLKS 119
Query: 127 LKKNPKLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLHL----- 181
+R+Y +G AFG G+ AA+ I + ++ SY + L V+
Sbjct: 120 -SGIVGIRAYEQLGYRAFGTPGKLAAALAITLQNIGAMSSYLYIIKSELPLVIQTFLNLE 178
Query: 182 KQLHLAKLSAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAVFGG 241
K + + ++ + +V++ LP +R L + + S G +S ++F +A
Sbjct: 179 KPTPVWYMDGNYLVILVSVIIILPLALMRQLGYLGY--SSGFSLSCMVFFLIAV------ 230
Query: 242 VRVNHEIPFLRLHNIPSISG---------------------------------------- 261
+ ++P HN+ + +G
Sbjct: 231 IYKKFQVPCPLAHNLVNATGNFSHMVVVEEKSQLQSEPDTAEAFCTPSYFTLNSQTAYTI 290
Query: 262 -IYIFSYGGHIVFPNLYKAMKDPS--KFTKVSILSFTTVTALYTIMGYMGAKMFGPEVNS 318
I F++ H +Y +KDPS K +S LS + +Y + G F V S
Sbjct: 291 PIMAFAFVCHPEVLPIYTELKDPSKRKMQHISNLSIAVMYVMYFLAALFGYLTFYDGVES 350
Query: 319 QV--TLSMPSKQIVTKIALWATVLTPMT-KYALEFAPVAIQLEHKLPSSMSGRTKMILRG 375
++ T S V + + VL +T + PV ++ L + +
Sbjct: 351 ELLHTYSKVDPFDVLILCVRVAVLIAVTLTVPIVLFPVRRAIQQML--FQNQEFSWLRHV 408
Query: 376 IIGSSLLLVILALALTVPYFEHVLSLTGSLVSVAISLIFPCAFYIKIC---RGQISKPLF 432
+I + LL I L + P + + G+ + + IFP FY +I + +
Sbjct: 409 LIATGLLTCINLLVIFAPNILGIFGIIGATSAPCLIFIFPAIFYFRIMPTEKEPVRSTPK 468
Query: 433 VLNISIITFGFLL 445
+L + GFLL
Sbjct: 469 ILALCFAAVGFLL 481
>YAN9_SCHPO (Q10074) Hypothetical protein C3H1.09c in chromosome I
Length = 656
Score = 69.7 bits (169), Expect = 1e-11
Identities = 91/387 (23%), Positives = 170/387 (43%), Gaps = 33/387 (8%)
Query: 72 NSSLAHSVMNMVGMLIGLGQLSTPYAVEQGG--WASAILLIALGVMCTYTCHLLGKCLKK 129
N+S +V+ ++ +G G L P A + GG ++SA LLI +GV+ LL + K
Sbjct: 274 NASNGKAVLLLLKSFVGTGVLFLPKAFKLGGLVFSSATLLI-VGVLSHICFLLLIQTRMK 332
Query: 130 NPKLRSYVDIGNHAFGAKGRF--LAAILI----YMDIFMSLVSYTISLHDNLTTVLHLKQ 183
P S+ DIG +G RF LA+I++ + ++S V+ T+ + + H ++
Sbjct: 333 VPG--SFGDIGGTLYGPHMRFAILASIVVSQIGFSSAYISFVASTLQACVKVISTTH-RE 389
Query: 184 LHLAKLSAPQILTVGAVLVALPSLWLRDLSSIS---FLSSVGVLMSVVI--FVSVAATAV 238
HLA Q L V +P +R +S +S ++ V +L+ ++ F V A
Sbjct: 390 YHLAVFIFIQFL------VFVPLSLVRKISKLSATALIADVFILLGILYLYFWDVITLAT 443
Query: 239 FGGVRVNHEIPFLRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTTVT 298
G ++ + G+ IF+Y G + + + M P K+ ++
Sbjct: 444 KGIA----DVAMFNKTDFSLFIGVAIFTYEGICLILPIQEQMAKPKNLPKLLTGVMAAIS 499
Query: 299 ALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEFAPVAIQLE 358
L+ +G + FG +V + V L+MP + T I + + + L+ P +E
Sbjct: 500 LLFISIGLLSYAAFGSKVKTVVILNMP-ESTFTVIIQFLYAIAILLSTPLQLFPAIAIIE 558
Query: 359 HKLPSSMSGRTKMILRGIIGSSLLLVILALALT---VPYFEHVLSLTGSLVSVAISLIFP 415
+ + R + I +L+VILA+ ++ + +S+ GS+ + + ++P
Sbjct: 559 QGIFTRSGKRNRKIKWRKNYLRVLIVILAILISWAGSSRLDLFVSMVGSVCCIPLIYMYP 618
Query: 416 CAFYIKICRGQISKPLFVLNISIITFG 442
+ K C + L L+I + T G
Sbjct: 619 PMLHYKACAN--NWILRTLDIFMFTIG 643
>S383_MOUSE (Q9DCP2) System N amino acid transporter 1 (SN1)
(N-system amino acid transporter 1) (Solute carrier
family 38, member 3) (mNAT)
Length = 505
Score = 68.9 bits (167), Expect = 3e-11
Identities = 87/408 (21%), Positives = 156/408 (37%), Gaps = 64/408 (15%)
Query: 68 DAKANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAI-LLIALGVMCTYTCHLLGKC 126
D + +S SV N+ ++G G L YA+ G + LL A+ ++ +Y+ HLL K
Sbjct: 60 DFEGKTSFGMSVFNLSNAIMGSGILGLAYAMANTGIILFLFLLTAVALLSSYSIHLLLKS 119
Query: 127 LKKNPKLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLHL----- 181
+R+Y +G AFG G+ AA+ I + ++ SY + L V+
Sbjct: 120 -SGIVGIRAYEQLGYRAFGTPGKLAAALAITLQNIGAMSSYLYIIKSELPLVIQTFLNLE 178
Query: 182 KQLHLAKLSAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAVFGG 241
K + + ++ + +V + LP +R L + + S G +S ++F +A
Sbjct: 179 KPASVWYMDGNYLVILVSVTIILPLALMRQLGYLGY--SSGFSLSCMVFFLIAV------ 230
Query: 242 VRVNHEIPFLRLHNIPSISG---------------------------------------- 261
+ ++P HN+ + +G
Sbjct: 231 IYKKFQVPCPLAHNLANATGNFSHMVVAEEKAQLQGEPDTAAEAFCTPSYFTLNSQTAYT 290
Query: 262 --IYIFSYGGHIVFPNLYKAMKDPS--KFTKVSILSFTTVTALYTIMGYMGAKMFGPEVN 317
I F++ H +Y +KDPS K +S LS + +Y + G F V
Sbjct: 291 IPIMAFAFVCHPEVLPIYTELKDPSKRKMQHISNLSIAVMYVMYFLAALFGYLTFYDGVE 350
Query: 318 SQV--TLSMPSKQIVTKIALWATVLTPMT-KYALEFAPVAIQLEHKLPSSMSGRTKMILR 374
S++ T S V + + VL +T + PV ++ L + +
Sbjct: 351 SELLHTYSKVDPFDVLILCVRVAVLIAVTLTVPIVLFPVRRAIQQML--FQNQEFSWLRH 408
Query: 375 GIIGSSLLLVILALALTVPYFEHVLSLTGSLVSVAISLIFPCAFYIKI 422
+I + LL I L + P + + G+ + + IFP FY +I
Sbjct: 409 VLIATGLLTCINLLVIFAPNILGIFGIIGATSAPCLIFIFPAIFYFRI 456
>AVT3_YEAST (P36062) Vacuolar amino acid transporter 3
Length = 692
Score = 68.9 bits (167), Expect = 3e-11
Identities = 90/405 (22%), Positives = 163/405 (40%), Gaps = 18/405 (4%)
Query: 53 EESAVARDTN--VDAEHDAKAN-SSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILL 109
EE A+ ++ V EH + SS +V+ ++ +G G L P A GGW + L
Sbjct: 274 EEEALETESTQLVSREHGRHPHKSSTVKAVLLLLKSFVGTGVLFLPKAFHNGGWGFSALC 333
Query: 110 IALGVMCTYTCHLLGKCLKKNPKLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTI 169
+ + +Y C + K + Y D+G +G K +F I + +YT+
Sbjct: 334 LLSCALISYGCFVSLITTKDKVGVDGYGDMGRILYGPKMKFAILSSIALSQIGFSAAYTV 393
Query: 170 SLHDNLTTVLHLKQLHLAKLSAPQILTVGA-VLVALPSLWLRDLSSISFLSSVGVLMSVV 228
NL V HL S + A VL+ +P R+++ +S + + L ++
Sbjct: 394 FTATNL-QVFSENFFHLKPGSISLATYIFAQVLIFVPLSLTRNIAKLSGTALIADLFILL 452
Query: 229 IFVSVAATAVF----GGVRVNHEIPFLRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPS 284
V V +++ GV + + F + + G IF++ G + + ++MK P
Sbjct: 453 GLVYVYVYSIYYIAVNGVASDTMLMFNKA-DWSLFIGTAIFTFEGIGLLIPIQESMKHPK 511
Query: 285 KFTKVSILSFTTVTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMT 344
F V ++ G + FG +V + V L+ P T L +
Sbjct: 512 HFRPSLSAVMCIVAVIFISCGLLCYAAFGSDVKTVVLLNFPQDTSYTLTVQLLYALAILL 571
Query: 345 KYALEFAPVAIQLEH-KLPSSMSG----RTKMILRGIIGSSLLLVILALALTVPYFEHVL 399
L+ P LE+ PS+ SG + K + + ++L + + + +
Sbjct: 572 STPLQLFPAIRILENWTFPSNASGKYNPKVKWLKNYFRCAIVVLTSILAWVGANDLDKFV 631
Query: 400 SLTGSLVSVAISLIFPCAFYIK--ICRGQISKPLFVLNISIITFG 442
SL GS + + I+P + K I G S+ +L++ +I FG
Sbjct: 632 SLVGSFACIPLIYIYPPLLHYKASILSG-TSRARLLLDLIVIVFG 675
>S383_HUMAN (Q99624) System N amino acid transporter 1 (SN1)
(N-system amino acid transporter 1) (Solute carrier
family 38, member 3)
Length = 504
Score = 68.6 bits (166), Expect = 3e-11
Identities = 100/441 (22%), Positives = 176/441 (39%), Gaps = 58/441 (13%)
Query: 68 DAKANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAI-LLIALGVMCTYTCHLLGKC 126
D + +S SV N+ ++G G L YA+ G + LL A+ ++ +Y+ HLL K
Sbjct: 61 DFEGKTSFGMSVFNLSNAIMGSGILGLAYAMANTGIILFLFLLTAVALLSSYSIHLLLKS 120
Query: 127 LKKNPKLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLHLKQLHL 186
+R+Y +G AFG G+ AA+ I + ++ SY + L V+ L+L
Sbjct: 121 -SGVVGIRAYEQLGYRAFGTPGKLAAALAITLQNIGAMSSYLYIIKSELPLVIQ-TFLNL 178
Query: 187 AK------LSAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVA------ 234
+ ++ ++ + +V + LP +R L + + S G +S ++F +A
Sbjct: 179 EEKTSDWYMNGNYLVILVSVTIILPLALMRQLGYLGY--SSGFSLSCMVFFLIAVIYKKF 236
Query: 235 ------------ATAVFGGVRVNHEIPFLRLHNIPSISG------------------IYI 264
T F V + E L++ P S I
Sbjct: 237 HVPCPLPPNFNNTTGNFSHVEIVKEKVQLQVE--PEASAFCTPSYFTLNSQTAYTIPIMA 294
Query: 265 FSYGGHIVFPNLYKAMKDPS--KFTKVSILSFTTVTALYTIMGYMGAKMFGPEVNSQV-- 320
F++ H +Y +KDPS K +S LS + +Y + G F V S++
Sbjct: 295 FAFVCHPEVLPIYTELKDPSKKKMQHISNLSIAVMYIMYFLAALFGYLTFYNGVESELLH 354
Query: 321 TLSMPSKQIVTKIALWATVLTPMT-KYALEFAPVAIQLEHKL-PSSMSGRTKMILRGIIG 378
T S V + + VLT +T + PV ++ L P+ + +L I
Sbjct: 355 TYSKVDPFDVLILCVRVAVLTAVTLTVPIVLFPVRRAIQQMLFPNQEFSWLRHVL---IA 411
Query: 379 SSLLLVILALALTVPYFEHVLSLTGSLVSVAISLIFPCAFYIKICRGQISKPLFVLNISI 438
LL I L + P + + G+ + + IFP FY +I + I
Sbjct: 412 VGLLTCINLLVIFAPNILGIFGVIGATSAPFLIFIFPAIFYFRIMPTEKEPARSTPKILA 471
Query: 439 ITFGFLLGVVGTISSSKLLVE 459
+ F L ++ T+S S ++++
Sbjct: 472 LCFAMLGFLLMTMSLSFIIID 492
>AVT4_YEAST (P50944) Vacuolar amino acid transporter 4
Length = 713
Score = 68.2 bits (165), Expect = 4e-11
Identities = 82/407 (20%), Positives = 165/407 (40%), Gaps = 48/407 (11%)
Query: 53 EESAV-ARDTNVDAEHDAKANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIA 111
E SA+ +R ++ AK +S + ++ IG G L P A GG ++ ++A
Sbjct: 276 ERSALLSRPDHMKVLPSAKGTTSTKKVFLILLKSFIGTGVLFLPNAFHNGGLFFSVSMLA 335
Query: 112 LGVMCTYTCHLLGKCLKKNPKLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISL 171
+ +Y C+ + K + + S+ DIG +G R + + + +Y I
Sbjct: 336 FFGIYSYWCYYILVQAKSSCGVSSFGDIGLKLYGPWMRIIILFSLVITQVGFSGAYMIFT 395
Query: 172 HDNLTTVLHLKQLHLAKLSAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFV 231
NL L H+ L ++ V ++ +P ++R++S +S S +L + I
Sbjct: 396 AKNLQAFLD-NVFHVGVLPLSYLM-VFQTIIFIPLSFIRNISKLSLPS---LLANFFIMA 450
Query: 232 SVAATAVFGGVRVNHEIPFLRLHNIPSISGIY--------------IFSYGGHIVFPNLY 277
+ +F R+ F L P++ +Y IF++ G + +
Sbjct: 451 GLVIVIIFTAKRL-----FFDLMGTPAMGVVYGLNADRWTLFIGTAIFAFEGIGLIIPVQ 505
Query: 278 KAMKDPSKFTKVSILSFTTVTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWA 337
+M++P KF V L T T L+ + +G +G V + + L++P I +
Sbjct: 506 DSMRNPEKFPLVLALVILTATILFISIATLGYLAYGSNVQTVILLNLPQSNIFVNLIQLF 565
Query: 338 TVLTPMTKYALEFAPVAIQLEHKLPSSMSG--------RTKMILRGIIG----------S 379
+ M L+ P +E+K + T++ LR G +
Sbjct: 566 YSIAIMLSTPLQLFPAIKIIENKFFPKFTKIYVKHDDLTTRVELRPNSGKLNWKIKWLKN 625
Query: 380 SLLLVILALALTVPYF-----EHVLSLTGSLVSVAISLIFPCAFYIK 421
+ +I+ + +++ YF + +S+ GSL + + I+P +++
Sbjct: 626 FIRSIIVIIVVSIAYFGSDNLDKFVSVIGSLACIPLVYIYPSMLHLR 672
>AVT6_YEAST (P40074) Vacuolar amino acid transporter 6
Length = 448
Score = 56.2 bits (134), Expect = 2e-07
Identities = 96/446 (21%), Positives = 181/446 (40%), Gaps = 80/446 (17%)
Query: 73 SSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTYTCHLLGKCLKKNPK 132
+S+ V+ ++ G G L+ PYA + G +++I L C + L +
Sbjct: 3 ASIRSGVLTLLHTACGAGILAMPYAFKPFGLIPGVIMIVLCGACAM------QSLFIQAR 56
Query: 133 LRSYVDIGNHAFGAKGRFLAAIL-IYMDIFMSL------VSYTISLHDNLTTVLHLKQLH 185
+ YV G +F A R + L I D+ +++ VSY I + D + ++ + +
Sbjct: 57 VAKYVPQGRASFSALTRLINPNLGIVFDLAIAIKCFGVGVSYMIVVGDLMPQIMSVWTRN 116
Query: 186 LAKLSAPQILTVGAVLVALPSLWLRDLSSISF-----LSSVGVLMSVVIFVSVAATAVFG 240
L+ +++ + P +L+ L+S+ + +SSV L +V+ VA +
Sbjct: 117 AWLLNRNVQISLIMLFFVAPLSFLKKLNSLRYASMVAISSVAYLCVLVLLHYVAPSDEI- 175
Query: 241 GVRVNHEIPFL---RLH--NIPSISGIYIFSYGGHIVFPNLYKAMKD--PSKF---TKVS 290
+R+ I +L + H N+ + I++F+Y H N++ + + S+F K+
Sbjct: 176 -LRLKGRISYLLPPQSHDLNVLNTLPIFVFAYTCH---HNMFSIINEQRSSRFEHVMKIP 231
Query: 291 ILSFTTVTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEF 350
+++ + LY +G G FG + + + P + + + I A VL M + L+
Sbjct: 232 LIAISLALILYIAIGCAGYLTFGDNIIGNIIMLYP-QAVSSTIGRIAIVLLVMLAFPLQC 290
Query: 351 APVAIQLEHKL-----------------PSSMSGRTKMI--------------------- 372
P + L P+ + + +I
Sbjct: 291 HPARASIHQILQHFAEENVSISATSADEPTVATESSPLIRDSSLDLNEVIEEESIYQPKE 350
Query: 373 --LRG----IIGSSLLLVILALALTVPYFEHVLSLTGSLVSVAISLIFPCAFYIKICRGQ 426
LRG +I S+L+ +A++V VL++ G+ S +IS I P F K+ G
Sbjct: 351 TPLRGKSFIVITCSILVASYLVAISVSSLARVLAIVGATGSTSISFILPGLFGYKLI-GT 409
Query: 427 ISKPLFVLNISIITF-GFLLGVVGTI 451
K L I + G LL + G I
Sbjct: 410 EHKTAVPLTTKIFKYTGLLLFIWGLI 435
>AVT7_YEAST (P40501) Vacuolar amino acid transporter 7
Length = 490
Score = 50.1 bits (118), Expect = 1e-05
Identities = 65/304 (21%), Positives = 125/304 (40%), Gaps = 24/304 (7%)
Query: 70 KANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCT-YTCHLLGKCLK 128
+A SS S N+V ++G G L+ PY+ + G ++L L + + +L KC K
Sbjct: 2 EATSSALSSTANLVKTIVGAGTLAIPYSFKSDGVLVGVILTLLAAVTSGLGLFVLSKCSK 61
Query: 129 K--NPKLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLHLKQLHL 186
NP+ S+ + + +I + F +SY + + D + ++
Sbjct: 62 TLINPRNSSFFTLCMLTYPTLAPIFDLAMI-VQCFGVGLSYLVLIGDLFPGLFGGER--- 117
Query: 187 AKLSAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAVF---GGVR 243
+ + ++ +P ++ L + + S +G+ I + V + VF G
Sbjct: 118 ------NYWIIASAVIIIPLCLVKKLDQLKYSSILGLFALAYISILVFSHFVFELGKGEL 171
Query: 244 VN---HEIPFLRLHN---IPSISGIYIFSYGGHIVFPNLYKAMKDPS--KFTKVSILSFT 295
N ++I + ++H+ + S I IF++ G + + +KD S T V S +
Sbjct: 172 TNILRNDICWWKIHDFKGLLSTFSIIIFAFTGSMNLFPMINELKDNSMENITFVINNSIS 231
Query: 296 TVTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEFAPVAI 355
TAL+ I+G G FG E + L+ I I + + + L F P+ I
Sbjct: 232 LSTALFLIVGLSGYLTFGNETLGNLMLNYDPNSIWIVIGKFCLGSMLILSFPLLFHPLRI 291
Query: 356 QLEH 359
+ +
Sbjct: 292 AVNN 295
>YMJ2_CAEEL (P34479) Hypothetical protein F59B2.2 in chromosome III
Length = 460
Score = 46.6 bits (109), Expect = 1e-04
Identities = 74/369 (20%), Positives = 157/369 (42%), Gaps = 34/369 (9%)
Query: 78 SVMNMVGMLIGLGQLSTPYAVEQGG-WASAILLIALGVMCTYTCHLLGKCLKKNPKL--R 134
+V+ + + G S PYA + GG W S ++ + + Y H+L + + K R
Sbjct: 42 AVLTLSKSMFNAGCFSLPYAWKLGGLWVSFVMSFVIAGLNWYGNHILVRASQHLAKKSDR 101
Query: 135 SYVDIGNHAFGAKG----RFLA----AILIYMDI---FMSLVSYTISLHDNLTTVLHLKQ 183
S +D G+ A RFL A++ ++++ F L ++++ +++L
Sbjct: 102 SALDYGHFAKKVCDYSDIRFLRNNSKAVMYFVNVTILFYQLGMCSVAILFISDNLVNLVG 161
Query: 184 LHLAKLSAPQIL---TVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAVFG 240
HL Q++ TV + L +++ ++ +SF + L+S V FV AA +
Sbjct: 162 DHLGGTRHQQMILMATVSLFFILLTNMFT-EMRIVSFFA----LVSSVFFVIGAAVIMQY 216
Query: 241 GVRVNHEIPFL----RLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKF-TKVSILSFT 295
V+ ++ L ++ G+ ++++ G + + + +P+ F +LS T
Sbjct: 217 TVQQPNQWDKLPAATNFTGTITMIGMSMYAFEGQTMILPIENKLDNPAAFLAPFGVLSTT 276
Query: 296 TV--TALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEFAPV 353
+ TA T +G+ G FG + +T ++P + + + + ++ +L + ++ V
Sbjct: 277 MIICTAFMTALGFFGYTGFGDSIAPTITTNVPKEGLYSTVNVF-LMLQSLLGNSIAMYVV 335
Query: 354 AIQLEHKLPSSMSGRTKMILRGIIGSSL----LLVILALALTVPYFEHVLSLTGSLVSVA 409
+ R + + + +LV +A+ +P E ++ L G
Sbjct: 336 YDMFFNGFRRKFGARFPNVPKWLSDKGFRVFWVLVTYLMAVLIPKLEIMIPLVGVTSGAL 395
Query: 410 ISLIFPCAF 418
+LIFP F
Sbjct: 396 CALIFPPFF 404
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.324 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 48,328,465
Number of Sequences: 164201
Number of extensions: 1879656
Number of successful extensions: 6325
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 40
Number of HSP's that attempted gapping in prelim test: 6270
Number of HSP's gapped (non-prelim): 63
length of query: 464
length of database: 59,974,054
effective HSP length: 114
effective length of query: 350
effective length of database: 41,255,140
effective search space: 14439299000
effective search space used: 14439299000
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0044a.9


