

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
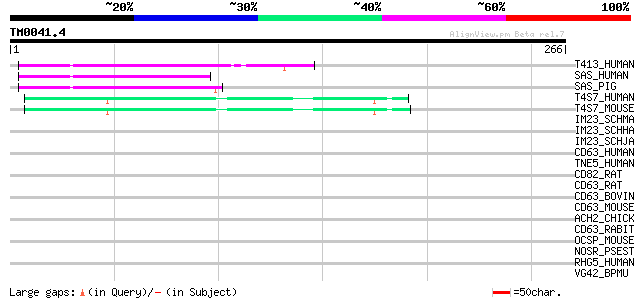
Query= TM0041.4
(266 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
T413_HUMAN (O95857) Transmembrane 4 superfamily member 13 (Tetra... 50 6e-06
SAS_HUMAN (Q12999) Sarcoma amplified sequence 50 7e-06
SAS_PIG (Q29257) Sarcoma amplified sequence (Fragment) 49 1e-05
T4S7_HUMAN (O14817) Transmembrane 4 superfamily member 7 (Novel ... 47 5e-05
T4S7_MOUSE (Q9DCK3) Transmembrane 4 superfamily member 7 45 1e-04
IM23_SCHMA (P19331) 23 kDa integral membrane protein (SM23) 37 0.050
IM23_SCHHA (Q26499) 23 kDa integral membrane protein (Sh23) 37 0.050
IM23_SCHJA (P27591) 23 kDa integral membrane protein (SJ23) 36 0.085
CD63_HUMAN (P08962) CD63 antigen (Melanoma-associated antigen ME... 35 0.14
TNE5_HUMAN (O75954) Tetraspan NET-5 34 0.42
CD82_RAT (O70352) CD82 antigen (Metastasis suppressor homolog) 34 0.42
CD63_RAT (P28648) CD63 antigen (AD1 antigen) 33 0.55
CD63_BOVIN (Q9XSK2) CD63 antigen 33 0.55
CD63_MOUSE (P41731) CD63 antigen 32 1.2
ACH2_CHICK (P09480) Neuronal acetylcholine receptor protein, alp... 32 1.2
CD63_RABIT (Q28709) CD63 antigen 32 1.6
OCSP_MOUSE (Q8VCF5) Oculospanin 32 2.1
NOSR_PSEST (Q00790) Regulatory protein nosR 32 2.1
RHG5_HUMAN (Q13017) Rho-GTPase-activating protein 5 (p190-B) 31 2.7
VG42_BPMU (Q9T1V6) Protein gp42 30 4.7
>T413_HUMAN (O95857) Transmembrane 4 superfamily member 13
(Tetraspan NET-6) (UNQ260/PRO296)
Length = 204
Score = 50.1 bits (118), Expect = 6e-06
Identities = 39/148 (26%), Positives = 70/148 (46%), Gaps = 10/148 (6%)
Query: 5 SNNLIGLLNFLTLLLSIPILVTGVWLSKQPGTDCERWLERPIIALGVFLLLVSLAGLVGA 64
S N + LN L L+S+ ++ W G + +IA+G+FL L++L GL+GA
Sbjct: 9 SKNCLCALNLLYTLVSLLLIGIAAW-GIGFGLISSLRVVGVVIAVGIFLFLIALVGLIGA 67
Query: 65 CCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSNWLQK 124
LL+ Y++++ L+ ++ F+ + A+ + G+ L G+ N +Q+
Sbjct: 68 VKHHQVLLFFYMIILLLVFIVQFSVSCACLALNQEQQGQLLE-VGWN--NTASARNDIQR 124
Query: 125 RVNSTG------NWNRIRSCLGSGKLCS 146
+N G N + SC+ S CS
Sbjct: 125 NLNCCGFRSVNPNDTCLASCVKSDHSCS 152
>SAS_HUMAN (Q12999) Sarcoma amplified sequence
Length = 210
Score = 49.7 bits (117), Expect = 7e-06
Identities = 29/92 (31%), Positives = 51/92 (54%), Gaps = 1/92 (1%)
Query: 5 SNNLIGLLNFLTLLLSIPILVTGVWLSKQPGTDCERWLERPIIALGVFLLLVSLAGLVGA 64
S N + LN + +L+S+ ++ W K G + +IA+GVFLLL+++AGLVGA
Sbjct: 9 SKNALCALNVVYMLVSLLLIGVAAW-GKGLGLVSSIHIIGGVIAVGVFLLLIAVAGLVGA 67
Query: 65 CCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAV 96
LL+ Y++++ L+ + F + A+
Sbjct: 68 VNHHQVLLFFYMIILGLVFIFQFVISCSCLAI 99
>SAS_PIG (Q29257) Sarcoma amplified sequence (Fragment)
Length = 109
Score = 49.3 bits (116), Expect = 1e-05
Identities = 31/99 (31%), Positives = 55/99 (55%), Gaps = 2/99 (2%)
Query: 5 SNNLIGLLNFLTLLLSIPILVTGVWLSKQPGTDCERWLERPIIALGVFLLLVSLAGLVGA 64
S N + LN + +L+ + ++ W +K G + +IA+GVFLLL+++AGLVGA
Sbjct: 9 SKNALCALNVVYMLVGLLLIGVAAW-AKGLGLVSSIHIIGGVIAVGVFLLLIAVAGLVGA 67
Query: 65 CCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVT-NKGAG 102
LL+ Y++++ L+ + F + A+ +K AG
Sbjct: 68 VNHHQVLLFFYMIILGLVFIFQFGISCSCLAINLSKQAG 106
>T4S7_HUMAN (O14817) Transmembrane 4 superfamily member 7 (Novel
antigen 2) (NAG-2) (Tetraspanin 4) (Tspan-4)
Length = 238
Score = 47.0 bits (110), Expect = 5e-05
Identities = 45/192 (23%), Positives = 76/192 (39%), Gaps = 24/192 (12%)
Query: 8 LIGLLNFLTLLLSIPILVTGVWLSKQPGTDCERWLERP-------IIALGVFLLLVSLAG 60
L+ N L L +L G+WL+ G+ P +I G F++ + G
Sbjct: 12 LMFAFNLLFWLGGCGVLGVGIWLAATQGSFATLSSSFPSLSAANLLIITGAFVMAIGFVG 71
Query: 61 LVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSN 120
+GA LL + L++ L+ +L I FA T+K + ++ + G +
Sbjct: 72 CLGAIKENKCLLLTFFLLLLLVFLLEATIAILFFAYTDK-----IDRYAQQDLKKGLHLY 126
Query: 121 WLQKRVNSTGNWNRIRSCLGSGKLCSEFRNRFVNDTAEQFYAENLSALQSGCC-KPSNDC 179
Q V T W+ I+ ++FR V++ + F N + + CC + S C
Sbjct: 127 GTQGNVGLTNAWSIIQ---------TDFRCCGVSNYTDWFEVYNATRVPDSCCLEFSESC 177
Query: 180 GFTYQGPSVWNK 191
G P W K
Sbjct: 178 GL--HAPGTWWK 187
>T4S7_MOUSE (Q9DCK3) Transmembrane 4 superfamily member 7
Length = 238
Score = 45.4 bits (106), Expect = 1e-04
Identities = 42/193 (21%), Positives = 77/193 (39%), Gaps = 24/193 (12%)
Query: 8 LIGLLNFLTLLLSIPILVTGVWLSKQPGTDCERWLERP-------IIALGVFLLLVSLAG 60
L+ N L L +L G+WL+ G P +I G F++ + G
Sbjct: 12 LMFAFNLLFWLGGCGVLGVGIWLAATQGNFATLSSSFPSLSAANLLIVTGTFVMAIGFVG 71
Query: 61 LVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSN 120
+GA LL + +++ L+ +L + FA ++K + ++ + G +
Sbjct: 72 CIGALKENKCLLLTFFVLLLLVFLLEATIAVLFFAYSDK-----IDSYAQQDLKKGLHLY 126
Query: 121 WLQKRVNSTGNWNRIRSCLGSGKLCSEFRNRFVNDTAEQFYAENLSALQSGCC-KPSNDC 179
Q V T W+ I+ ++FR V++ + F N + + CC + S+ C
Sbjct: 127 GTQGNVGLTNAWSIIQ---------TDFRCCGVSNYTDWFEVYNATRVPDSCCLEFSDSC 177
Query: 180 GFTYQGPSVWNKT 192
G P W K+
Sbjct: 178 GL--HEPGTWWKS 188
>IM23_SCHMA (P19331) 23 kDa integral membrane protein (SM23)
Length = 218
Score = 37.0 bits (84), Expect = 0.050
Identities = 28/93 (30%), Positives = 48/93 (51%), Gaps = 9/93 (9%)
Query: 2 VRLSNNLIGLLNFLTLLLSIPILVTGVWLS---KQPGTDCER-WLERPI--IALGVFLLL 55
+R + + +LN + LL S+ ++ G ++ Q G + + W PI I +GV +L+
Sbjct: 8 MRCLKSCVFVLNIICLLCSLVLIGAGAYVEVKFSQYGDNLHKVWQAAPIAIIVVGVIILI 67
Query: 56 VSLAGLVGACCRVSWLLWVYLLVMFLLIVLVFA 88
VS G GA +L++Y F L+VL+ A
Sbjct: 68 VSFLGCCGAIKENVCMLYMYA---FFLVVLLIA 97
>IM23_SCHHA (Q26499) 23 kDa integral membrane protein (Sh23)
Length = 218
Score = 37.0 bits (84), Expect = 0.050
Identities = 28/93 (30%), Positives = 48/93 (51%), Gaps = 9/93 (9%)
Query: 2 VRLSNNLIGLLNFLTLLLSIPILVTGVWLS---KQPGTDCER-WLERPI--IALGVFLLL 55
+R + + +LN + LL S+ ++ G ++ Q G + + W PI I +GV +L+
Sbjct: 8 MRCLKSCVFVLNIICLLCSLVLIGAGAYVEVKFSQYGDNLHKVWQAAPIAIIVVGVIILI 67
Query: 56 VSLAGLVGACCRVSWLLWVYLLVMFLLIVLVFA 88
VS G GA +L++Y F LI+L+ A
Sbjct: 68 VSFLGCCGAIKENVCMLYMYA---FFLIILLIA 97
>IM23_SCHJA (P27591) 23 kDa integral membrane protein (SJ23)
Length = 218
Score = 36.2 bits (82), Expect = 0.085
Identities = 29/98 (29%), Positives = 49/98 (49%), Gaps = 9/98 (9%)
Query: 2 VRLSNNLIGLLNFLTLLLSIPILVTGVWL----SKQPGTDCERWLERPI--IALGVFLLL 55
+R + + +LN + LL S+ ++ G ++ S+ + W PI I +GV +L+
Sbjct: 8 MRCLKSCVFILNIICLLCSLVLIGAGAYVEVKFSQYEANLHKVWQAAPIAIIVVGVVILI 67
Query: 56 VSLAGLVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFA 93
VS G GA +L++Y F LIVL+ A + A
Sbjct: 68 VSFLGCCGAIKENVCMLYMYA---FFLIVLLIAELVAA 102
>CD63_HUMAN (P08962) CD63 antigen (Melanoma-associated antigen
ME491) (Lysosome-associated membrane glycoprotein 3)
(LAMP-3) (Ocular melanoma-associated antigen) (OMA81H)
(Granulophysin)
Length = 237
Score = 35.4 bits (80), Expect = 0.14
Identities = 25/94 (26%), Positives = 46/94 (48%), Gaps = 14/94 (14%)
Query: 46 IIALGVFLLLVSLAGLVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEAL 105
IIA+GVFL LV+ G GA C+ ++ L + + LI+LV A+
Sbjct: 56 IIAVGVFLFLVAFVGCCGA-CKENYCLMITFAIFLSLIMLV-------------EVAAAI 101
Query: 106 SGRGYREYRLGDYSNWLQKRVNSTGNWNRIRSCL 139
+G +R+ + +++N ++++ + N S L
Sbjct: 102 AGYVFRDKVMSEFNNNFRQQMENYPKNNHTASIL 135
>TNE5_HUMAN (O75954) Tetraspan NET-5
Length = 239
Score = 33.9 bits (76), Expect = 0.42
Identities = 36/178 (20%), Positives = 66/178 (36%), Gaps = 22/178 (12%)
Query: 11 LLNFLTLLLSIPILVTGVWLSKQPGTDCERWLERP-------IIALGVFLLLVSLAGLVG 63
L N + L +L G+WLS G P +IA+G +++ G +G
Sbjct: 15 LFNLIFWLCGCGLLGVGIWLSVSQGNFATFSPSFPSLSAANLVIAIGTIVMVTGFLGCLG 74
Query: 64 ACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSNWLQ 123
A LL + +V+ ++++ I F +K ++ ++ + G +
Sbjct: 75 AIKENKCLLLSFFIVLLVILLAELILLILFFVYMDK-----VNENAKKDLKEGLLLYHTE 129
Query: 124 KRVNSTGNWNRIRSCLGSGKLCSEFRNRFVNDTAEQFYAENLSALQSGCC-KPSNDCG 180
V WN I+ +E R V D + + + + CC + S CG
Sbjct: 130 NNVGLKNAWNIIQ---------AEMRCCGVTDYTDWYPVLGENTVPDRCCMENSQGCG 178
>CD82_RAT (O70352) CD82 antigen (Metastasis suppressor homolog)
Length = 266
Score = 33.9 bits (76), Expect = 0.42
Identities = 26/99 (26%), Positives = 44/99 (44%), Gaps = 11/99 (11%)
Query: 2 VRLSNNLIGLLNFLTLLLSIPILVTGVWLSK---------QPGTDCERWLERPIIALGVF 52
V+++ + L N L +L IL GVW+ Q + + I +G
Sbjct: 6 VKVTKYFLFLFNLLFFILGAVILGFGVWILADKSSFISVLQTSSSSLQVGAYVFIGVGAI 65
Query: 53 LLLVSLAGLVGACCRVSWLLWVYLLVMFLLIVLVFAFTI 91
+L+ G +GA V LL +Y +FLL++L+ T+
Sbjct: 66 TMLMGFLGCIGAVNEVRCLLGLYF--VFLLLILIAQVTV 102
>CD63_RAT (P28648) CD63 antigen (AD1 antigen)
Length = 237
Score = 33.5 bits (75), Expect = 0.55
Identities = 23/83 (27%), Positives = 40/83 (47%), Gaps = 14/83 (16%)
Query: 46 IIALGVFLLLVSLAGLVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEAL 105
IIA+G FL LV+ G GA C+ ++ L + + LI+LV A+
Sbjct: 56 IIAVGAFLFLVAFVGCCGA-CKENYCLMITFAIFLSLIMLV-------------EVAVAI 101
Query: 106 SGRGYREYRLGDYSNWLQKRVNS 128
+G +R+ ++S QK++ +
Sbjct: 102 AGYVFRDQVKSEFSKSFQKQMQN 124
>CD63_BOVIN (Q9XSK2) CD63 antigen
Length = 236
Score = 33.5 bits (75), Expect = 0.55
Identities = 18/58 (31%), Positives = 30/58 (51%)
Query: 46 IIALGVFLLLVSLAGLVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGE 103
IIA+G FL LV+ G GAC L+ + + + L++++ A I + +K E
Sbjct: 56 IIAVGAFLFLVAFVGCCGACKENYCLMITFAIFLSLIMLVEVAAAIAGYVFRDKVRSE 113
>CD63_MOUSE (P41731) CD63 antigen
Length = 237
Score = 32.3 bits (72), Expect = 1.2
Identities = 17/41 (41%), Positives = 25/41 (60%), Gaps = 1/41 (2%)
Query: 46 IIALGVFLLLVSLAGLVGACCRVSWLLWVYLLVMFLLIVLV 86
IIA+G FL LV+ G GA C+ ++ L + + LI+LV
Sbjct: 56 IIAVGAFLFLVAFVGCCGA-CKENYCLMITFAIFLSLIMLV 95
>ACH2_CHICK (P09480) Neuronal acetylcholine receptor protein,
alpha-2 chain precursor
Length = 528
Score = 32.3 bits (72), Expect = 1.2
Identities = 57/255 (22%), Positives = 94/255 (36%), Gaps = 67/255 (26%)
Query: 46 IIALGVFLLLVSLAGLVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEAL 105
+++L VFLLL++ ++ + V L+ YLL + + L T+F V ++
Sbjct: 279 LLSLTVFLLLIT--EIIPSTSLVIPLIGEYLLFTMIFVTLSIIITVFVLNVHHRSPSTHT 336
Query: 106 SGRGYREYRLGDYSNWL-QKR-------VNSTGNWNRIRSCLGSGK--LCSEFRNRFVND 155
R + LG WL KR +TG ++ + L + + L ++ +++ +
Sbjct: 337 MPHWVRSFFLGFIPRWLFMKRPPLLLPAEGTTGQYDPPGTRLSTSRCWLETDVDDKWEEE 396
Query: 156 TAEQ----------------------------FYAENLSALQSGCCKP---------SND 178
E+ + E + SG P +D
Sbjct: 397 EEEEEEEEEEEEEEKAYPSRVPSGGSQGTQCHYSCERQAGKASGGPAPQVPLKGEEVGSD 456
Query: 179 CGFTYQGPSVWNKTEGVNYSNPDCHAWDNDPNVLCFNCQSCKAGLLQNIKTDWKKVA-VV 237
G T PS+ EGV Y A D D ++K DWK VA V+
Sbjct: 457 QGLTL-SPSILRALEGVQYIADHLRAEDAD----------------FSVKEDWKYVAMVI 499
Query: 238 NVIFLVFLIIVYSIG 252
+ IFL IIV +G
Sbjct: 500 DRIFLWMFIIVCLLG 514
>CD63_RABIT (Q28709) CD63 antigen
Length = 237
Score = 32.0 bits (71), Expect = 1.6
Identities = 17/58 (29%), Positives = 29/58 (49%)
Query: 46 IIALGVFLLLVSLAGLVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGE 103
IIA+G FL LV+ G G C L+ + + + L++++ A I + +K E
Sbjct: 56 IIAVGAFLFLVAFVGCCGTCKENYCLMITFAIFLSLIMLVEVAAAIAGYVFRDKVMSE 113
>OCSP_MOUSE (Q8VCF5) Oculospanin
Length = 331
Score = 31.6 bits (70), Expect = 2.1
Identities = 30/89 (33%), Positives = 42/89 (46%), Gaps = 8/89 (8%)
Query: 8 LIGLLNFLTLLLSIPILVTGVWLSKQPGTDCERW----LERPIIAL---GVFLLLVSLAG 60
LI L NFL L S+ L G+W + W P++ L G+ + +VSL+G
Sbjct: 77 LIFLSNFLFSLPSLLALAAGLWGLTVKRSQGIGWGGPVPTDPMLMLVLGGLVVSVVSLSG 136
Query: 61 LVGACCRVSWLL-WVYLLVMFLLIVLVFA 88
+GA C S LL W V+F L + A
Sbjct: 137 CLGAFCENSCLLHWYCGAVLFCLALEALA 165
>NOSR_PSEST (Q00790) Regulatory protein nosR
Length = 724
Score = 31.6 bits (70), Expect = 2.1
Identities = 18/53 (33%), Positives = 29/53 (53%), Gaps = 7/53 (13%)
Query: 73 WVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSNWLQKR 125
W +FLL++ +F ++ V GAG A++GR +RL D WL++R
Sbjct: 603 WFVAYAVFLLVINIFTRKVYCRYVCPLGAGLAITGR----FRLFD---WLKRR 648
>RHG5_HUMAN (Q13017) Rho-GTPase-activating protein 5 (p190-B)
Length = 1499
Score = 31.2 bits (69), Expect = 2.7
Identities = 22/59 (37%), Positives = 30/59 (50%), Gaps = 6/59 (10%)
Query: 140 GSGKLCSEFRNRFVNDTAEQFYAEN---LSALQSGCCKPSNDCGFTYQGPSVWNKTEGV 195
G GK C NRFV A+++Y E+ LS + G +ND F Y G + N +GV
Sbjct: 31 GVGKSC--LCNRFVRSKADEYYPEHTSVLSTIDFGGRVVNND-HFLYWGDIIQNSEDGV 86
>VG42_BPMU (Q9T1V6) Protein gp42
Length = 690
Score = 30.4 bits (67), Expect = 4.7
Identities = 14/57 (24%), Positives = 25/57 (43%)
Query: 14 FLTLLLSIPILVTGVWLSKQPGTDCERWLERPIIALGVFLLLVSLAGLVGACCRVSW 70
FLT ++ I +W +PG D W+++ I++L ++ L R W
Sbjct: 310 FLTTFRTVKISAQELWRWLKPGKDALAWVDQNIVSLKKLAAVLVSVWLANKALRAGW 366
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.326 0.141 0.471
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 34,028,015
Number of Sequences: 164201
Number of extensions: 1459622
Number of successful extensions: 4718
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 4695
Number of HSP's gapped (non-prelim): 41
length of query: 266
length of database: 59,974,054
effective HSP length: 108
effective length of query: 158
effective length of database: 42,240,346
effective search space: 6673974668
effective search space used: 6673974668
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0041.4


