

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
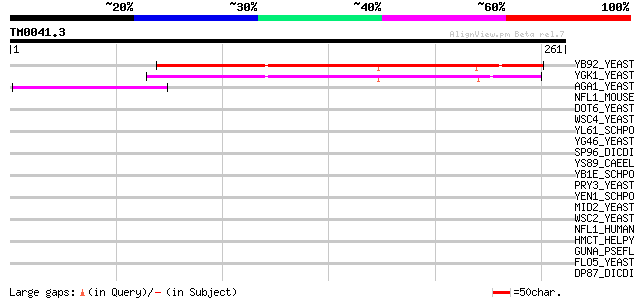
Query= TM0041.3
(261 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YB92_YEAST (P38331) Hypothetical 27.6 kDa protein in THI2-ALG7 i... 151 1e-36
YGK1_YEAST (P53144) Hypothetical 25.3 kDa protein in RPL28-SEH1 ... 140 3e-33
AGA1_YEAST (P32323) A-agglutinin attachment subunit precursor 43 7e-04
NFL1_MOUSE (Q61985) Nuclear factor erythroid 2 related factor 1 ... 41 0.003
DOT6_YEAST (P40059) Disrupter of telomere silencing protein 6 41 0.003
WSC4_YEAST (P38739) Cell wall integrity and stress response comp... 40 0.004
YL61_SCHPO (Q8TFG9) Hypothetical serine/threonine-rich protein P... 39 0.010
YG46_YEAST (P53301) Hypothetical 52.8 kDa protein in BUB1-HIP1 i... 39 0.010
SP96_DICDI (P14328) Spore coat protein SP96 39 0.010
YS89_CAEEL (Q09624) Hypothetical protein ZK945.9 in chromosome II 39 0.013
YB1E_SCHPO (P87179) Serine-rich protein C30B4.01c precursor 39 0.013
PRY3_YEAST (P47033) PRY3 protein (Pathogen related in Sc 3) 38 0.028
YEN1_SCHPO (O13695) Hypothetical serine-rich protein C11G7.01 in... 37 0.048
MID2_YEAST (P36027) Mating process protein MID2 (Serine-rich pro... 37 0.048
WSC2_YEAST (P53832) Cell wall integrity and stress response comp... 37 0.063
NFL1_HUMAN (Q14494) Nuclear factor erythroid 2 related factor 1 ... 36 0.082
HMCT_HELPY (Q59465) Cadmium, zinc and cobalt transporting ATPase... 36 0.11
GUNA_PSEFL (P10476) Endoglucanase A precursor (EC 3.2.1.4) (Endo... 35 0.14
FLO5_YEAST (P38894) Flocculation protein FLO5 precursor (Floccul... 35 0.14
DP87_DICDI (Q04503) Prespore protein DP87 precursor 35 0.14
>YB92_YEAST (P38331) Hypothetical 27.6 kDa protein in THI2-ALG7
intergenic region
Length = 238
Score = 151 bits (382), Expect = 1e-36
Identities = 79/187 (42%), Positives = 124/187 (66%), Gaps = 7/187 (3%)
Query: 70 PSASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKC 129
P + + FL++ +LK +R G++ +++ ESI+DHMYR+S++ ++ D V+RDKC
Sbjct: 43 PHPNYILAFLNVVQQLKIQRRTGYLDLGIKECESISDHMYRLSIITMLIKD-SRVNRDKC 101
Query: 130 VKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVL---GGGSRAKEVAELWA 186
V++A+VHDIAE++VGDITP D + KEEK+R E E + ++C L AKE+ + W
Sbjct: 102 VRIALVHDIAESLVGDITPVDPIGKEEKHRREWETIKYLCNALIKPYNEIAAKEIMDDWL 161
Query: 187 EYEANSSPEAKFVKDLDKVEMILQALEYEHEQ--GKDLDEFFQSTAGKFQTEIGKAWASE 244
YE +S EA++VKD+DK EM++Q EYE E K+ D+FF + A +T+ K W S+
Sbjct: 162 AYENVTSLEARYVKDIDKYEMLVQCFEYEREYKGTKNFDDFFGAVA-SIKTDEVKGWTSD 220
Query: 245 IVSRRKK 251
+V +R+K
Sbjct: 221 LVVQRQK 227
>YGK1_YEAST (P53144) Hypothetical 25.3 kDa protein in RPL28-SEH1
intergenic region
Length = 215
Score = 140 bits (353), Expect = 3e-33
Identities = 73/191 (38%), Positives = 114/191 (59%), Gaps = 7/191 (3%)
Query: 65 AASATPSASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGV 124
A + P + + FL+I LKT +R GW+ + ESI+DHMYRM L ++ D V
Sbjct: 19 AILSKPHPNYQLAFLNIIQLLKTQRRTGWVDHGIDPCESISDHMYRMGLTTMLITD-KNV 77
Query: 125 DRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVL---GGGSRAKEV 181
DR+KC+++A+VHD AE++VGDITP D + KEEK+R E E + ++C+ + S ++E+
Sbjct: 78 DRNKCIRIALVHDFAESLVGDITPNDPMTKEEKHRREFETVKYLCESIIRPCSESASREI 137
Query: 182 AELWAEYEANSSPEAKFVKDLDKVEMILQALEYEHEQG--KDLDEFFQSTAGKFQTEIGK 239
+ W YE + E ++VKD+DK EM++Q EYE + KDL +F +T+ K
Sbjct: 138 LDDWLAYEKQTCLEGRYVKDIDKYEMLVQCFEYEQKYNGKKDLKQFL-GAINDIKTDEVK 196
Query: 240 AWASEIVSRRK 250
W ++ R+
Sbjct: 197 KWTQSLLEDRQ 207
>AGA1_YEAST (P32323) A-agglutinin attachment subunit precursor
Length = 725
Score = 43.1 bits (100), Expect = 7e-04
Identities = 27/73 (36%), Positives = 41/73 (55%)
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS 61
+T+T +S SSS +S SST S + SP S S T+ + S+ S S+T+S
Sbjct: 244 STSTSQSSTSTSSSSTSTSPSSTSTSSSSTSTSPSSKSTSASSTSTSSYSTSTSPSLTSS 303
Query: 62 PPSAASATPSASS 74
P+ AS +PS++S
Sbjct: 304 SPTLASTSPSSTS 316
Score = 42.4 bits (98), Expect = 0.001
Identities = 26/73 (35%), Positives = 41/73 (55%)
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS 61
+T+T +S SSS +S SST S + SP S S +T+ + SS+ S S T++
Sbjct: 195 STSTSPSSTSTSSSSTSTSSSSTSTSSSSTSTSPSSTSTSSSLTSTSSSSTSTSQSSTST 254
Query: 62 PPSAASATPSASS 74
S+ S +PS++S
Sbjct: 255 SSSSTSTSPSSTS 267
Score = 41.2 bits (95), Expect = 0.003
Identities = 26/74 (35%), Positives = 40/74 (53%)
Query: 1 MATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTT 60
++ T L S+ S+ +SLSST S + SP S S T+ + SS+ SSS T+
Sbjct: 166 ISPVTSTLSSTTSSNPTTTSLSSTSTSPSSTSTSPSSTSTSSSSTSTSSSSTSTSSSSTS 225
Query: 61 SPPSAASATPSASS 74
+ PS+ S + S +S
Sbjct: 226 TSPSSTSTSSSLTS 239
Score = 40.0 bits (92), Expect = 0.006
Identities = 26/74 (35%), Positives = 40/74 (53%)
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS 61
+T+T L+S SSS +S SST S + SP S S T+ + SS S+S T++
Sbjct: 230 STSTSSSLTSTSSSSTSTSQSSTSTSSSSTSTSPSSTSTSSSSTSTSPSSKSTSASSTST 289
Query: 62 PPSAASATPSASSA 75
+ S +PS +S+
Sbjct: 290 SSYSTSTSPSLTSS 303
Score = 35.0 bits (79), Expect = 0.18
Identities = 24/72 (33%), Positives = 38/72 (52%)
Query: 3 TATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSP 62
T T + +S SS +S SST S + S S S T+ + SS+ SSS+T++
Sbjct: 182 TTTSLSSTSTSPSSTSTSPSSTSTSSSSTSTSSSSTSTSSSSTSTSPSSTSTSSSLTSTS 241
Query: 63 PSAASATPSASS 74
S+ S + S++S
Sbjct: 242 SSSTSTSQSSTS 253
Score = 34.7 bits (78), Expect = 0.24
Identities = 24/77 (31%), Positives = 39/77 (50%)
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS 61
+T+T +S SS +S S T S + S S S T+ + SS+ SSS T++
Sbjct: 216 STSTSSSSTSTSPSSTSTSSSLTSTSSSSTSTSQSSTSTSSSSTSTSPSSTSTSSSSTST 275
Query: 62 PPSAASATPSASSAIDF 78
PS+ S + S++S +
Sbjct: 276 SPSSKSTSASSTSTSSY 292
Score = 34.3 bits (77), Expect = 0.31
Identities = 25/77 (32%), Positives = 39/77 (50%), Gaps = 6/77 (7%)
Query: 10 SSLFSSSIPSSLSS------TQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPP 63
S+ S + S+LSS T S + SP S S T+ + SS+ SSS T++
Sbjct: 162 SASIISPVTSTLSSTTSSNPTTTSLSSTSTSPSSTSTSPSSTSTSSSSTSTSSSSTSTSS 221
Query: 64 SAASATPSASSAIDFLS 80
S+ S +PS++S L+
Sbjct: 222 SSTSTSPSSTSTSSSLT 238
Score = 32.0 bits (71), Expect = 1.5
Identities = 24/79 (30%), Positives = 39/79 (48%)
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS 61
+T+T +S SS +S SST S + SP S + + + SS+ SS+ T S
Sbjct: 265 STSTSSSSTSTSPSSKSTSASSTSTSSYSTSTSPSLTSSSPTLASTSPSSTSISSTFTDS 324
Query: 62 PPSAASATPSASSAIDFLS 80
S S+ S+S+++ S
Sbjct: 325 TSSLGSSIASSSTSVSLYS 343
Score = 29.6 bits (65), Expect = 7.7
Identities = 21/62 (33%), Positives = 32/62 (50%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
+S S+S S+ +S L S TL+ SPS +T S+S SS+ +S S + +
Sbjct: 284 ASSTSTSSYSTSTSPSLTSSSPTLASTSPSSTSISSTFTDSTSSLGSSIASSSTSVSLYS 343
Query: 70 PS 71
PS
Sbjct: 344 PS 345
>NFL1_MOUSE (Q61985) Nuclear factor erythroid 2 related factor 1
(NF-E2 related factor 1) (NFE2-related factor 1)
(Nuclear factor, erythroid derived 2, like 1)
Length = 741
Score = 41.2 bits (95), Expect = 0.003
Identities = 33/109 (30%), Positives = 52/109 (47%), Gaps = 23/109 (21%)
Query: 34 SPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSASSAIDFLSICHRLKTTKRAG- 92
S HSPS ++++ GSSS SSS ++S S+AS++ S A+ + S L + G
Sbjct: 454 SSHSPSS---LSSSEGSSSSSSSSSSSSASSSASSSFSEEGAVGYSSDSETLDLEEAEGA 510
Query: 93 ---------WIRKDVQDPESIA----------DHMYRMSLMALIAADIP 122
+ R QDP ++ +H Y M+ AL +AD+P
Sbjct: 511 VGYQPEYSKFCRMSYQDPSQLSCLPYLEHVGHNHTYNMAPSALDSADLP 559
>DOT6_YEAST (P40059) Disrupter of telomere silencing protein 6
Length = 670
Score = 41.2 bits (95), Expect = 0.003
Identities = 36/96 (37%), Positives = 48/96 (49%), Gaps = 10/96 (10%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSS--PPSSSVTTSPPSAAS 67
+SL S+SI S T + S+T PHS S A SS P SSS +TS ++AS
Sbjct: 5 TSLNSASIHLSSMDTHPQLHSLTRQPHSSSTAMSKNEAQESSPSLPASSSSSTSASASAS 64
Query: 68 A-----TPSASSAIDFLSICHRLKTTKRAGWIRKDV 98
+ PS+ D L + H LK K+ GW KD+
Sbjct: 65 SKNSSKNPSSWDPQDDLLLRH-LKEVKKMGW--KDI 97
>WSC4_YEAST (P38739) Cell wall integrity and stress response
component 4 precursor
Length = 605
Score = 40.4 bits (93), Expect = 0.004
Identities = 29/76 (38%), Positives = 39/76 (51%), Gaps = 2/76 (2%)
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKS--VTLSPHSPSRVFHMTTAAGSSSPPSSSVT 59
+T T S+L S+S SS SST S ++ S S + TT+ SSS P+SS
Sbjct: 220 STTTSTTSSTLISTSTSSSSSSTPTTTSSAPISTSTTSSTSTSTSTTSPTSSSAPTSSSN 279
Query: 60 TSPPSAASATPSASSA 75
T+P S T S S+A
Sbjct: 280 TTPTSTTFTTTSPSTA 295
Score = 31.6 bits (70), Expect = 2.0
Identities = 20/64 (31%), Positives = 33/64 (51%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SS++ SS + SS+ + T+SP S T A +S+ PS+ +T++ P+ S
Sbjct: 127 SSVYVSSSSITSSSSTSIVDTTTISPTLTSTSTTPLTTASTSTTPSTDITSALPTTTSTK 186
Query: 70 PSAS 73
S S
Sbjct: 187 LSTS 190
Score = 31.6 bits (70), Expect = 2.0
Identities = 23/71 (32%), Positives = 36/71 (50%)
Query: 4 ATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPP 63
+T + S+ S+S +S SS+ SVT S + + T + S+S SSS T+
Sbjct: 188 STSIPTSTTSSTSTTTSTSSSTSTTVSVTSSTSTTTSTTSSTLISTSTSSSSSSTPTTTS 247
Query: 64 SAASATPSASS 74
SA +T + SS
Sbjct: 248 SAPISTSTTSS 258
>YL61_SCHPO (Q8TFG9) Hypothetical serine/threonine-rich protein
PB15E9.01c in chromosome I precursor
Length = 943
Score = 39.3 bits (90), Expect = 0.010
Identities = 26/74 (35%), Positives = 43/74 (57%), Gaps = 3/74 (4%)
Query: 3 TATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSP 62
T+ S+ SS+ PS +S+ L SV+ +P S + + TTA +SS P +SV ++
Sbjct: 506 TSANSTTSTSVSSTAPSYNTSSVLPTSSVSSTPLSSA---NSTTATSASSTPLTSVNSTT 562
Query: 63 PSAASATPSASSAI 76
++AS+TP +S I
Sbjct: 563 ATSASSTPFGNSTI 576
Score = 38.1 bits (87), Expect = 0.022
Identities = 26/67 (38%), Positives = 43/67 (63%), Gaps = 1/67 (1%)
Query: 10 SSLFSSSIPSS-LSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASA 68
SSL SSSI SS L+S+ + S+ S + S + +T + +S+ P+SS T+S S+ +A
Sbjct: 127 SSLASSSITSSSLASSSITSSSLASSSTTSSSLASSSTNSTTSATPTSSATSSSLSSTAA 186
Query: 69 TPSASSA 75
+ SA+S+
Sbjct: 187 SNSATSS 193
Score = 38.1 bits (87), Expect = 0.022
Identities = 29/89 (32%), Positives = 46/89 (51%), Gaps = 17/89 (19%)
Query: 2 ATATRVLLSSLFSSSIP------------SSLSSTQLRFKSVTLSPHS-----PSRVFHM 44
ATAT +SS SSS P +S +S+ ++ + +L P S P +
Sbjct: 238 ATATSSSISSTVSSSTPLTSSNSTTAATSASATSSSAQYNTSSLLPSSTPSSTPLSSANS 297
Query: 45 TTAAGSSSPPSSSVTTSPPSAASATPSAS 73
TTA +SS P +SV ++ ++AS+TP +S
Sbjct: 298 TTATSASSTPLTSVNSTTTTSASSTPLSS 326
Score = 36.2 bits (82), Expect = 0.082
Identities = 23/72 (31%), Positives = 37/72 (50%)
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS 61
+TA SS+ +S SS + + T + +P + TTA +SS P SSV ++
Sbjct: 404 STAPSYNTSSVLPTSSVSSTPLSSANSTTATSASSTPLSSVNSTTATSASSTPLSSVNST 463
Query: 62 PPSAASATPSAS 73
++AS+TP S
Sbjct: 464 TATSASSTPLTS 475
Score = 35.8 bits (81), Expect = 0.11
Identities = 21/62 (33%), Positives = 33/62 (52%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SSL SS PSS + + T + +P + TT +SS P SSV+++ + A++T
Sbjct: 279 SSLLPSSTPSSTPLSSANSTTATSASSTPLTSVNSTTTTSASSTPLSSVSSANSTTATST 338
Query: 70 PS 71
S
Sbjct: 339 SS 340
Score = 34.7 bits (78), Expect = 0.24
Identities = 23/77 (29%), Positives = 43/77 (54%), Gaps = 5/77 (6%)
Query: 3 TATRVLLSSLFSSSIPSSLSSTQLRFKSV-----TLSPHSPSRVFHMTTAAGSSSPPSSS 57
+A+ LSS+ S++ ++ S++ SV T + +P + TTA +SS P +S
Sbjct: 318 SASSTPLSSVSSANSTTATSTSSTPLSSVNSTTATSASSTPLTSVNSTTATSASSTPLTS 377
Query: 58 VTTSPPSAASATPSASS 74
V ++ ++AS+TP S+
Sbjct: 378 VNSTSATSASSTPLTSA 394
Score = 34.3 bits (77), Expect = 0.31
Identities = 24/74 (32%), Positives = 41/74 (54%), Gaps = 2/74 (2%)
Query: 3 TATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHS--PSRVFHMTTAAGSSSPPSSSVTT 60
+A+ LSS+ S++ S+ S+ S T + S P + TTA +SS P +SV +
Sbjct: 435 SASSTPLSSVNSTTATSASSTPLSSVNSTTATSASSTPLTSVNSTTATSASSTPLTSVNS 494
Query: 61 SPPSAASATPSASS 74
+ ++AS+TP S+
Sbjct: 495 TSATSASSTPLTSA 508
Score = 34.3 bits (77), Expect = 0.31
Identities = 29/80 (36%), Positives = 47/80 (58%), Gaps = 4/80 (5%)
Query: 3 TATRVLLSSLFSSSIPSS-LSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSV-TT 60
T++ + SS SSS+ SS +S+ L S+T S + S + +++ SSS SSS+ ++
Sbjct: 105 TSSSLTSSSATSSSLASSSTTSSSLASSSITSSSLASSSI--TSSSLASSSTTSSSLASS 162
Query: 61 SPPSAASATPSASSAIDFLS 80
S S SATP++S+ LS
Sbjct: 163 STNSTTSATPTSSATSSSLS 182
Score = 34.3 bits (77), Expect = 0.31
Identities = 31/101 (30%), Positives = 51/101 (49%), Gaps = 14/101 (13%)
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFK-SVTLSPHSPSRVFHMTTAAGSSSP------- 53
A + SSL SSS+ S+ S+T S T+S +P + TTAA S+S
Sbjct: 217 AASNSATSSSLASSSLNSTTSATATSSSISSTVSSSTPLTSSNSTTAATSASATSSSAQY 276
Query: 54 ------PSSSVTTSPPSAASATPSASSAIDFLSICHRLKTT 88
PSS+ +++P S+A++T + S++ L+ + TT
Sbjct: 277 NTSSLLPSSTPSSTPLSSANSTTATSASSTPLTSVNSTTTT 317
Score = 33.9 bits (76), Expect = 0.41
Identities = 23/72 (31%), Positives = 43/72 (58%), Gaps = 3/72 (4%)
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS 61
ATAT SSL S++ +S +S+ L S+ + + + +++ SS+P +SS +T+
Sbjct: 206 ATATS---SSLSSTAASNSATSSSLASSSLNSTTSATATSSSISSTVSSSTPLTSSNSTT 262
Query: 62 PPSAASATPSAS 73
++ASAT S++
Sbjct: 263 AATSASATSSSA 274
Score = 33.5 bits (75), Expect = 0.53
Identities = 25/74 (33%), Positives = 41/74 (54%), Gaps = 1/74 (1%)
Query: 3 TATRVLLSSLFSSSIPS-SLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS 61
T++ SSL SSS S SL+S+ + S+ S + S + +T + S + S++ TTS
Sbjct: 110 TSSSATSSSLASSSTTSSSLASSSITSSSLASSSITSSSLASSSTTSSSLASSSTNSTTS 169
Query: 62 PPSAASATPSASSA 75
+SAT S+ S+
Sbjct: 170 ATPTSSATSSSLSS 183
Score = 33.1 bits (74), Expect = 0.70
Identities = 29/72 (40%), Positives = 41/72 (56%), Gaps = 4/72 (5%)
Query: 14 SSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAG--SSSPPSSSVTTSPPSAASATPS 71
SS+ SSL+S+ S+T S S + TT+A SSS SSS T+S +++S T S
Sbjct: 68 SSASSSSLTSSSAASSSLTSSSSLASSSTNSTTSASPTSSSLTSSSATSSSLASSSTTSS 127
Query: 72 --ASSAIDFLSI 81
ASS+I S+
Sbjct: 128 SLASSSITSSSL 139
Score = 33.1 bits (74), Expect = 0.70
Identities = 31/89 (34%), Positives = 48/89 (53%), Gaps = 4/89 (4%)
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS 61
A+++ + SS SSS+ SS S S T + SP+ +++A SSS SSS T+S
Sbjct: 70 ASSSSLTSSSAASSSLTSSSSLASSSTNSTTSA--SPTSSSLTSSSATSSSLASSSTTSS 127
Query: 62 PPSAASATPS--ASSAIDFLSICHRLKTT 88
+++S T S ASS+I S+ T+
Sbjct: 128 SLASSSITSSSLASSSITSSSLASSSTTS 156
Score = 32.7 bits (73), Expect = 0.91
Identities = 26/73 (35%), Positives = 45/73 (61%), Gaps = 2/73 (2%)
Query: 4 ATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPP 63
A+ L S+ +++ SSLSST S T S + S + T+A +SS SS+V++S P
Sbjct: 196 ASSSLNSTTSATATSSSLSSTAAS-NSATSSSLASSSLNSTTSATATSSSISSTVSSSTP 254
Query: 64 -SAASATPSASSA 75
+++++T +A+SA
Sbjct: 255 LTSSNSTTAATSA 267
Score = 32.7 bits (73), Expect = 0.91
Identities = 24/71 (33%), Positives = 36/71 (49%), Gaps = 6/71 (8%)
Query: 3 TATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSP 62
+ T SS SS+ S+ S+T S LS + TTA +SS P +SV ++
Sbjct: 313 STTTTSASSTPLSSVSSANSTTATSTSSTPLSS------VNSTTATSASSTPLTSVNSTT 366
Query: 63 PSAASATPSAS 73
++AS+TP S
Sbjct: 367 ATSASSTPLTS 377
Score = 31.6 bits (70), Expect = 2.0
Identities = 23/72 (31%), Positives = 40/72 (54%), Gaps = 1/72 (1%)
Query: 4 ATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSP- 62
++ + SSL SSS SS ++ + + +P S + +++ A S+S SSS+ +S
Sbjct: 141 SSSITSSSLASSSTTSSSLASSSTNSTTSATPTSSATSSSLSSTAASNSATSSSLASSSL 200
Query: 63 PSAASATPSASS 74
S SAT ++SS
Sbjct: 201 NSTTSATATSSS 212
Score = 31.2 bits (69), Expect = 2.6
Identities = 26/86 (30%), Positives = 45/86 (52%), Gaps = 1/86 (1%)
Query: 4 ATRVLLSSLFSSSIPS-SLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSP 62
++ + SSL SSSI S SL+S+ S+ S + + T++A SSS S++ + S
Sbjct: 131 SSSITSSSLASSSITSSSLASSSTTSSSLASSSTNSTTSATPTSSATSSSLSSTAASNSA 190
Query: 63 PSAASATPSASSAIDFLSICHRLKTT 88
S++ A+ S +S + L +T
Sbjct: 191 TSSSLASSSLNSTTSATATSSSLSST 216
Score = 31.2 bits (69), Expect = 2.6
Identities = 24/68 (35%), Positives = 34/68 (49%), Gaps = 7/68 (10%)
Query: 14 SSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAA-------GSSSPPSSSVTTSPPSAA 66
SS+ PSS SST R S S P+ F T+ + GSSS S TTS S+
Sbjct: 643 SSAYPSSGSSTFSRLSSTLTSSIIPTETFGSTSGSATGTRPTGSSSQGSVVPTTSTGSSV 702
Query: 67 SATPSASS 74
++T + ++
Sbjct: 703 TSTGTGTT 710
Score = 30.0 bits (66), Expect = 5.9
Identities = 24/73 (32%), Positives = 40/73 (53%), Gaps = 3/73 (4%)
Query: 3 TATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGS---SSPPSSSVT 59
T++ L SS +S+ +S +S+ L S T S + S + A+ S SS SSS+T
Sbjct: 86 TSSSSLASSSTNSTTSASPTSSSLTSSSATSSSLASSSTTSSSLASSSITSSSLASSSIT 145
Query: 60 TSPPSAASATPSA 72
+S +++S T S+
Sbjct: 146 SSSLASSSTTSSS 158
>YG46_YEAST (P53301) Hypothetical 52.8 kDa protein in BUB1-HIP1
intergenic region
Length = 507
Score = 39.3 bits (90), Expect = 0.010
Identities = 28/69 (40%), Positives = 41/69 (58%), Gaps = 1/69 (1%)
Query: 9 LSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSS-PPSSSVTTSPPSAAS 67
+SS SS++ SS+SST S T+S S V ++ + SSS PSSS TS + AS
Sbjct: 312 VSSSASSTVSSSVSSTVSSSASSTVSSSVSSTVSSSSSVSSSSSTSPSSSTATSSKTLAS 371
Query: 68 ATPSASSAI 76
++ + SS+I
Sbjct: 372 SSVTTSSSI 380
Score = 37.4 bits (85), Expect = 0.037
Identities = 26/75 (34%), Positives = 41/75 (54%)
Query: 1 MATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTT 60
+A + SS SS++ SS SST S T+S + S V ++ SSS SS ++
Sbjct: 296 LANGGSISSSSTSSSTVSSSASSTVSSSVSSTVSSSASSTVSSSVSSTVSSSSSVSSSSS 355
Query: 61 SPPSAASATPSASSA 75
+ PS+++AT S + A
Sbjct: 356 TSPSSSTATSSKTLA 370
Score = 33.5 bits (75), Expect = 0.53
Identities = 28/68 (41%), Positives = 39/68 (57%), Gaps = 5/68 (7%)
Query: 9 LSSLFSSSIPSSLSSTQLRFKSVTLSPH-SPSRVFHMTTAAGSSSPPSSSVTTSPPSAAS 67
+SS SS++ SS+SST SV+ S SPS +TA S + SSSVTTS ++
Sbjct: 328 VSSSASSTVSSSVSSTVSSSSSVSSSSSTSPSS----STATSSKTLASSSVTTSSSISSF 383
Query: 68 ATPSASSA 75
S+SS+
Sbjct: 384 EKQSSSSS 391
>SP96_DICDI (P14328) Spore coat protein SP96
Length = 600
Score = 39.3 bits (90), Expect = 0.010
Identities = 23/62 (37%), Positives = 40/62 (64%)
Query: 14 SSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSAS 73
S+S S++SS+ + + SP S + +++A SSSP SS+ ++SP S+AS++ S S
Sbjct: 439 SASGSSAVSSSASGSSAASSSPSSSAASSSPSSSAASSSPSSSAASSSPSSSASSSSSPS 498
Query: 74 SA 75
S+
Sbjct: 499 SS 500
Score = 37.7 bits (86), Expect = 0.028
Identities = 22/66 (33%), Positives = 42/66 (63%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
S++ SS+ SS +S+ + + SP S + +++A SSSP SS+ ++S PS+++++
Sbjct: 444 SAVSSSASGSSAASSSPSSSAASSSPSSSAASSSPSSSAASSSPSSSASSSSSPSSSASS 503
Query: 70 PSASSA 75
SA S+
Sbjct: 504 SSAPSS 509
Score = 37.4 bits (85), Expect = 0.037
Identities = 27/66 (40%), Positives = 40/66 (59%), Gaps = 2/66 (3%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SS SSS SS +S+ + + SP S + +++A SSS PSSS ++S SA S++
Sbjct: 453 SSAASSSPSSSAASSSPSSSAASSSPSSSAASSSPSSSASSSSSPSSSASSS--SAPSSS 510
Query: 70 PSASSA 75
S+SSA
Sbjct: 511 ASSSSA 516
Score = 35.4 bits (80), Expect = 0.14
Identities = 28/80 (35%), Positives = 43/80 (53%), Gaps = 1/80 (1%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSP-SRVFHMTTAAGSSSPPSSSVTTSPPSAASA 68
SS SSS SS +S+ + + SP S S +++A SSS PSSS ++S ++SA
Sbjct: 462 SSAASSSPSSSAASSSPSSSAASSSPSSSASSSSSPSSSASSSSAPSSSASSSSAPSSSA 521
Query: 69 TPSASSAIDFLSICHRLKTT 88
+ S++S+ S TT
Sbjct: 522 SSSSASSSSASSAATTAATT 541
Score = 34.3 bits (77), Expect = 0.31
Identities = 20/62 (32%), Positives = 36/62 (57%)
Query: 14 SSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSAS 73
SS++ S+ S+ +V+ S S +++ +SS PSSS +S PS+++A+ S S
Sbjct: 429 SSALGSTSESSASGSSAVSSSASGSSAASSSPSSSAASSSPSSSAASSSPSSSAASSSPS 488
Query: 74 SA 75
S+
Sbjct: 489 SS 490
Score = 32.3 bits (72), Expect = 1.2
Identities = 25/63 (39%), Positives = 39/63 (61%), Gaps = 5/63 (7%)
Query: 14 SSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSP-PSAASATPSA 72
S+S S+L ST S + + S + ++A SSSP SS+ ++SP SAAS++PS+
Sbjct: 425 STSDSSALGSTSESSASGSSAVSSSA----SGSSAASSSPSSSAASSSPSSSAASSSPSS 480
Query: 73 SSA 75
S+A
Sbjct: 481 SAA 483
Score = 30.8 bits (68), Expect = 3.5
Identities = 24/66 (36%), Positives = 37/66 (55%), Gaps = 1/66 (1%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SS +SS PSS +S+ S S +PS +++A SSS SSS ++S S+A+ T
Sbjct: 479 SSSAASSSPSSSASSSSSPSSSASSSSAPSSSAS-SSSAPSSSASSSSASSSSASSAATT 537
Query: 70 PSASSA 75
+ + A
Sbjct: 538 AATTIA 543
Score = 30.8 bits (68), Expect = 3.5
Identities = 24/74 (32%), Positives = 38/74 (50%)
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS 61
A+ + + SS SS SS S+ S + S S S +++ SSS SSS +S
Sbjct: 440 ASGSSAVSSSASGSSAASSSPSSSAASSSPSSSAASSSPSSSAASSSPSSSASSSSSPSS 499
Query: 62 PPSAASATPSASSA 75
S++SA S++S+
Sbjct: 500 SASSSSAPSSSASS 513
Score = 30.8 bits (68), Expect = 3.5
Identities = 22/66 (33%), Positives = 35/66 (52%), Gaps = 1/66 (1%)
Query: 10 SSLFSSSIP-SSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASA 68
SS SSS P SS SS+ S + S S S T A ++ +++ TT+ +A +A
Sbjct: 499 SSASSSSAPSSSASSSSAPSSSASSSSASSSSASSAATTAATTIATTAATTTATTTATTA 558
Query: 69 TPSASS 74
T +A++
Sbjct: 559 TTTATT 564
Score = 29.6 bits (65), Expect = 7.7
Identities = 25/67 (37%), Positives = 38/67 (56%), Gaps = 8/67 (11%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAAS-- 67
SS SSS PSS +S+ S S +PS ++A SSS SSS +++ +AA+
Sbjct: 489 SSASSSSSPSSSASSSSAPSSSASSSSAPS------SSASSSSASSSSASSAATTAATTI 542
Query: 68 ATPSASS 74
AT +A++
Sbjct: 543 ATTAATT 549
>YS89_CAEEL (Q09624) Hypothetical protein ZK945.9 in chromosome II
Length = 3178
Score = 38.9 bits (89), Expect = 0.013
Identities = 20/73 (27%), Positives = 41/73 (55%)
Query: 3 TATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSP 62
T T +L+S + +S ++T++ S T++ P+ +TA+ S++ PS+S T+
Sbjct: 393 TFTTTMLTSTTTEEPSTSTTTTEVTSTSSTVTTTEPTTTLTTSTASTSTTEPSTSTVTTS 452
Query: 63 PSAASATPSASSA 75
PS + T + +S+
Sbjct: 453 PSTSPVTSTVTSS 465
Score = 36.2 bits (82), Expect = 0.082
Identities = 27/99 (27%), Positives = 45/99 (45%)
Query: 4 ATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPP 63
+T + S++ SSS S+ +T +S + SP S + ++ P SSS T S
Sbjct: 454 STSPVTSTVTSSSSSSTTVTTPTSTESTSTSPSSTVTTSTTAPSTSTTGPSSSSSTPSST 513
Query: 64 SAASATPSASSAIDFLSICHRLKTTKRAGWIRKDVQDPE 102
+++S + +ASS S TTK D +P+
Sbjct: 514 ASSSVSSTASSTQSSTSTQQSSTTTKSETTTSSDGTNPD 552
Score = 35.8 bits (81), Expect = 0.11
Identities = 26/77 (33%), Positives = 43/77 (55%), Gaps = 2/77 (2%)
Query: 1 MATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTT 60
+ T+T ++ S+S ++ ST +VT S S + V T+ +S+ PSS+VTT
Sbjct: 432 LTTSTASTSTTEPSTSTVTTSPSTSPVTSTVTSSSSSSTTVTTPTSTESTSTSPSSTVTT 491
Query: 61 S--PPSAASATPSASSA 75
S PS ++ PS+SS+
Sbjct: 492 STTAPSTSTTGPSSSSS 508
Score = 29.6 bits (65), Expect = 7.7
Identities = 26/72 (36%), Positives = 38/72 (52%), Gaps = 4/72 (5%)
Query: 3 TATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSP 62
++T + SS +SS PS+ SST L +VT P S TT+AGS++ S TTS
Sbjct: 664 SSTSAVASS--TSSTPSTPSST-LSTSTVT-EPSSTRSSDSTTTSAGSTTTLQESTTTSE 719
Query: 63 PSAASATPSASS 74
S ++ + S
Sbjct: 720 ESTTDSSTTTIS 731
>YB1E_SCHPO (P87179) Serine-rich protein C30B4.01c precursor
Length = 374
Score = 38.9 bits (89), Expect = 0.013
Identities = 26/71 (36%), Positives = 42/71 (58%)
Query: 5 TRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPS 64
T V SS+ S++ SS SS + T SP S S ++++ SSS SSS ++S S
Sbjct: 125 TTVSSSSVSSTTSSSSSSSPSSSSTTTTTSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 184
Query: 65 AASATPSASSA 75
++S++ S+SS+
Sbjct: 185 SSSSSSSSSSS 195
Score = 35.4 bits (80), Expect = 0.14
Identities = 25/66 (37%), Positives = 39/66 (58%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SS SSS SS SS+ S + S S S ++++ SSS P +S T+S S++S++
Sbjct: 165 SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSVPITSSTSSSHSSSSSS 224
Query: 70 PSASSA 75
S+SS+
Sbjct: 225 SSSSSS 230
Score = 35.4 bits (80), Expect = 0.14
Identities = 27/74 (36%), Positives = 39/74 (52%)
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS 61
+T T SS SSS SS SS+ S + S S S ++++ SSS SSS ++S
Sbjct: 148 STTTTTSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 207
Query: 62 PPSAASATPSASSA 75
P +S + S SS+
Sbjct: 208 VPITSSTSSSHSSS 221
Score = 34.7 bits (78), Expect = 0.24
Identities = 25/66 (37%), Positives = 37/66 (55%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SS SSS SS SS+ S + S S S + + +SS SSS ++S S++S+
Sbjct: 174 SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSVPITSSTSSSHSSSSSSSSSSSSSSR 233
Query: 70 PSASSA 75
PS+SS+
Sbjct: 234 PSSSSS 239
Score = 34.7 bits (78), Expect = 0.24
Identities = 30/73 (41%), Positives = 40/73 (54%), Gaps = 9/73 (12%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSS---VTTSPPSA- 65
SS SSS SS SS+ + S T S HS S ++++ SSS PSSS +TT S
Sbjct: 192 SSSSSSSSSSSSSSSSVPITSSTSSSHSSSS--SSSSSSSSSSRPSSSSSFITTMSSSTF 249
Query: 66 ---ASATPSASSA 75
+ TPS+SS+
Sbjct: 250 ISTVTVTPSSSSS 262
Score = 34.3 bits (77), Expect = 0.31
Identities = 23/55 (41%), Positives = 33/55 (59%), Gaps = 2/55 (3%)
Query: 23 STQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSS--VTTSPPSAASATPSASSA 75
S L V + S S V T+++ SSSP SSS TTSP S++S++ S+SS+
Sbjct: 114 SVYLTGNGVLQTTVSSSSVSSTTSSSSSSSPSSSSTTTTTSPSSSSSSSSSSSSS 168
Score = 33.9 bits (76), Expect = 0.41
Identities = 25/73 (34%), Positives = 41/73 (55%)
Query: 3 TATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSP 62
T + +SS SSS SS SS+ + S S S ++++ SSS SSS ++S
Sbjct: 126 TVSSSSVSSTTSSSSSSSPSSSSTTTTTSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 185
Query: 63 PSAASATPSASSA 75
S++S++ S+SS+
Sbjct: 186 SSSSSSSSSSSSS 198
Score = 33.5 bits (75), Expect = 0.53
Identities = 28/71 (39%), Positives = 38/71 (53%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SS SSS SS SS+ S + S S S T++ SSS SSS ++S S++S
Sbjct: 175 SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSVPITSSTSSSHSSSSSSSSSSSSSSRP 234
Query: 70 PSASSAIDFLS 80
S+SS I +S
Sbjct: 235 SSSSSFITTMS 245
Score = 33.5 bits (75), Expect = 0.53
Identities = 25/72 (34%), Positives = 41/72 (56%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SS SSS SS ++T S + S S S ++++ SSS SSS ++S S++S++
Sbjct: 137 SSSSSSSPSSSSTTTTTSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 196
Query: 70 PSASSAIDFLSI 81
S+SS+ S+
Sbjct: 197 SSSSSSSSSSSV 208
Score = 33.5 bits (75), Expect = 0.53
Identities = 24/71 (33%), Positives = 42/71 (58%), Gaps = 5/71 (7%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SS +++ PSS SS+ S + S S S ++++ SSS SSS ++S S++S++
Sbjct: 147 SSTTTTTSPSSSSSSSSSSSSSSSSSSSSS-----SSSSSSSSSSSSSSSSSSSSSSSSS 201
Query: 70 PSASSAIDFLS 80
S+SS++ S
Sbjct: 202 SSSSSSVPITS 212
Score = 33.5 bits (75), Expect = 0.53
Identities = 25/66 (37%), Positives = 40/66 (59%), Gaps = 2/66 (3%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
+S SSS PSS S+T S + S S S ++++ SSS SSS ++S S++S++
Sbjct: 136 TSSSSSSSPSSSSTTTTTSPSSSSSSSSSSS--SSSSSSSSSSSSSSSSSSSSSSSSSSS 193
Query: 70 PSASSA 75
S+SS+
Sbjct: 194 SSSSSS 199
Score = 33.1 bits (74), Expect = 0.70
Identities = 24/66 (36%), Positives = 37/66 (55%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SS SSS SS SS+ S + S S S ++++ SSS S +T+S S+ S++
Sbjct: 162 SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSVPITSSTSSSHSSS 221
Query: 70 PSASSA 75
S+SS+
Sbjct: 222 SSSSSS 227
Score = 31.6 bits (70), Expect = 2.0
Identities = 25/68 (36%), Positives = 40/68 (58%), Gaps = 2/68 (2%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSS--PPSSSVTTSPPSAAS 67
SS SSS SS SS+ S + S S S ++++ SSS P +SS ++S S++S
Sbjct: 164 SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSVPITSSTSSSHSSSSS 223
Query: 68 ATPSASSA 75
++ S+SS+
Sbjct: 224 SSSSSSSS 231
Score = 31.2 bits (69), Expect = 2.6
Identities = 25/73 (34%), Positives = 39/73 (53%), Gaps = 5/73 (6%)
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS 61
+++T S SSS SS SS+ S + S S S ++++ SSS SSS ++S
Sbjct: 146 SSSTTTTTSPSSSSSSSSSSSSSSSSSSSSSSSSSSSS-----SSSSSSSSSSSSSSSSS 200
Query: 62 PPSAASATPSASS 74
S++S+ P SS
Sbjct: 201 SSSSSSSVPITSS 213
Score = 30.4 bits (67), Expect = 4.5
Identities = 24/74 (32%), Positives = 37/74 (49%)
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS 61
++ T S SSS SS SS+ S + S S S ++++ SSS SSS ++S
Sbjct: 147 SSTTTTTSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 206
Query: 62 PPSAASATPSASSA 75
S+T S+ S+
Sbjct: 207 SVPITSSTSSSHSS 220
Score = 30.0 bits (66), Expect = 5.9
Identities = 25/71 (35%), Positives = 39/71 (54%), Gaps = 3/71 (4%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SS SSS SS SS+ S + P + S ++ + SSS SSS ++S PS++S+
Sbjct: 184 SSSSSSSSSSSSSSSSSSSSSSSSVPITSST---SSSHSSSSSSSSSSSSSSRPSSSSSF 240
Query: 70 PSASSAIDFLS 80
+ S+ F+S
Sbjct: 241 ITTMSSSTFIS 251
>PRY3_YEAST (P47033) PRY3 protein (Pathogen related in Sc 3)
Length = 881
Score = 37.7 bits (86), Expect = 0.028
Identities = 24/75 (32%), Positives = 43/75 (57%)
Query: 1 MATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTT 60
+ ++T + SS SS+ SS++++ S +S + S ++ A SSS SS T+
Sbjct: 231 ITSSTESVGSSTVSSASSSSVTTSYATSSSTVVSSDATSSTTTTSSVATSSSTTSSDPTS 290
Query: 61 SPPSAASATPSASSA 75
S +A+S+ P++SSA
Sbjct: 291 STAAASSSDPASSSA 305
Score = 32.0 bits (71), Expect = 1.5
Identities = 25/76 (32%), Positives = 40/76 (51%), Gaps = 2/76 (2%)
Query: 2 ATATRVLLSSLFSSS--IPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVT 59
A+++ V S SSS + S +S+ SV S + S +TAA SSS P+SS
Sbjct: 246 ASSSSVTTSYATSSSTVVSSDATSSTTTTSSVATSSSTTSSDPTSSTAAASSSDPASSSA 305
Query: 60 TSPPSAASATPSASSA 75
+ SA++ ++SS+
Sbjct: 306 AASSSASTENAASSSS 321
Score = 32.0 bits (71), Expect = 1.5
Identities = 25/65 (38%), Positives = 31/65 (47%), Gaps = 2/65 (3%)
Query: 14 SSSIPSS--LSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPS 71
SS++ SS SST T S + S T AA SS P SSS S ++ S
Sbjct: 259 SSTVVSSDATSSTTTTSSVATSSSTTSSDPTSSTAAASSSDPASSSAAASSSASTENAAS 318
Query: 72 ASSAI 76
+SSAI
Sbjct: 319 SSSAI 323
>YEN1_SCHPO (O13695) Hypothetical serine-rich protein C11G7.01 in
chromosome I
Length = 536
Score = 37.0 bits (84), Expect = 0.048
Identities = 27/68 (39%), Positives = 41/68 (59%), Gaps = 4/68 (5%)
Query: 8 LLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAAS 67
+LSS +++I SS SS+ L S T SP+ +T+ SSS SSS + S S++S
Sbjct: 48 MLSSSSATAISSSSSSSPLSSSSFT----SPASSSFITSLVSSSSQQSSSSSASLTSSSS 103
Query: 68 ATPSASSA 75
AT ++SS+
Sbjct: 104 ATLTSSSS 111
Score = 33.1 bits (74), Expect = 0.70
Identities = 34/83 (40%), Positives = 38/83 (44%), Gaps = 16/83 (19%)
Query: 9 LSSLFSSSIPSSLSS----TQLRFKSVTLSPHSPSRVFHMTTAAGSSS------------ 52
LSS SSSIP+S SS T L SV S S TA SSS
Sbjct: 14 LSSSASSSIPASSSSAAASTSLSSSSVIPSSSSSMLSSSSATAISSSSSSSPLSSSSFTS 73
Query: 53 PPSSSVTTSPPSAASATPSASSA 75
P SSS TS S++S S+SSA
Sbjct: 74 PASSSFITSLVSSSSQQSSSSSA 96
Score = 31.6 bits (70), Expect = 2.0
Identities = 27/84 (32%), Positives = 43/84 (51%), Gaps = 2/84 (2%)
Query: 1 MATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTT 60
+ +++ L+S S+S SS SS L S +L S S ++ + SSS PSSS +
Sbjct: 98 LTSSSSATLTSSSSASPTSSSSSHALSSSSSSLVASSSSSGMSSSSLSHSSSVPSSS--S 155
Query: 61 SPPSAASATPSASSAIDFLSICHR 84
S S++ T SS+ +S +R
Sbjct: 156 SYHSSSMTTSGLSSSASIVSSTYR 179
>MID2_YEAST (P36027) Mating process protein MID2 (Serine-rich
protein SMS1) (Protein kinase A interference protein)
Length = 376
Score = 37.0 bits (84), Expect = 0.048
Identities = 28/84 (33%), Positives = 43/84 (50%), Gaps = 10/84 (11%)
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTL----------SPHSPSRVFHMTTAAGSS 51
+T++R L+S SS+ +S+S T F S + S S S T+A S
Sbjct: 74 STSSRSLVSHTSSSTSIASISFTSFSFSSDSSTSSSSSASSDSSSSSSFSISSTSATSES 133
Query: 52 SPPSSSVTTSPPSAASATPSASSA 75
S S+ +TS S+ S+TPS+SS+
Sbjct: 134 STSSTQTSTSSSSSLSSTPSSSSS 157
Score = 32.7 bits (73), Expect = 0.91
Identities = 26/66 (39%), Positives = 34/66 (51%), Gaps = 1/66 (1%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSS-VTTSPPSAASA 68
SS SSS SS SS+ F + S S S T+ SSS SS+ ++S PS ++
Sbjct: 104 SSTSSSSSASSDSSSSSSFSISSTSATSESSTSSTQTSTSSSSSLSSTPSSSSSPSTITS 163
Query: 69 TPSASS 74
PS SS
Sbjct: 164 APSTSS 169
Score = 31.6 bits (70), Expect = 2.0
Identities = 23/73 (31%), Positives = 40/73 (54%), Gaps = 2/73 (2%)
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS 61
+T++ SS SSS S+SST +S T S + + ++ SSS S++T++
Sbjct: 105 STSSSSSASSDSSSSSSFSISSTSATSESSTSSTQTSTSSSSSLSSTPSSSSSPSTITSA 164
Query: 62 PPSAASATPSASS 74
P + S+TPS ++
Sbjct: 165 P--STSSTPSTTA 175
>WSC2_YEAST (P53832) Cell wall integrity and stress response
component 2 precursor
Length = 503
Score = 36.6 bits (83), Expect = 0.063
Identities = 23/73 (31%), Positives = 41/73 (55%)
Query: 3 TATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSP 62
T T S+ SSS S+ S+T ++ T S S S TT+ S++ ++S ++SP
Sbjct: 153 TKTSTSSSATHSSSSSSTTSTTTSSSETTTSSSSSSSSSSTSTTSTTSTTSSTTSTSSSP 212
Query: 63 PSAASATPSASSA 75
+ +S+T ++SS+
Sbjct: 213 STTSSSTSASSSS 225
Score = 35.4 bits (80), Expect = 0.14
Identities = 22/65 (33%), Positives = 35/65 (53%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SS SSS +S +ST S T + SPS T+A+ SS S+ T+S ++ S++
Sbjct: 184 SSSSSSSSSTSTTSTTSTTSSTTSTSSSPSTTSSSTSASSSSETSSTQATSSSTTSTSSS 243
Query: 70 PSASS 74
S ++
Sbjct: 244 TSTAT 248
Score = 33.5 bits (75), Expect = 0.53
Identities = 28/73 (38%), Positives = 39/73 (53%), Gaps = 8/73 (10%)
Query: 3 TATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSP 62
T+T++ + SSS S SS+ S T + S S TT++ SSS SS+ TTS
Sbjct: 147 TSTKLDTKTSTSSSATHSSSSS-----STTSTTTSSSET---TTSSSSSSSSSSTSTTST 198
Query: 63 PSAASATPSASSA 75
S S+T S SS+
Sbjct: 199 TSTTSSTTSTSSS 211
Score = 31.6 bits (70), Expect = 2.0
Identities = 24/87 (27%), Positives = 43/87 (48%)
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS 61
A +T SS +S+ +S SST + K+ T S T ++ SSS S++ ++S
Sbjct: 119 AASTADSTSSTATSTSTTSSSSTSVSSKTSTKLDTKTSTSSSATHSSSSSSTTSTTTSSS 178
Query: 62 PPSAASATPSASSAIDFLSICHRLKTT 88
+ +S++ S+SS+ S +T
Sbjct: 179 ETTTSSSSSSSSSSTSTTSTTSTTSST 205
Score = 31.6 bits (70), Expect = 2.0
Identities = 33/91 (36%), Positives = 46/91 (50%), Gaps = 18/91 (19%)
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSP--HSPSRVF---------HMTTAAGS 50
+TAT +S S+S+ SS +ST+L K+ T S HS S TT++ S
Sbjct: 128 STATSTSTTSSSSTSV-SSKTSTKLDTKTSTSSSATHSSSSSSTTSTTTSSSETTTSSSS 186
Query: 51 SSPPSSSVTTSPPSAASAT------PSASSA 75
SS SS+ TTS S S+T PS +S+
Sbjct: 187 SSSSSSTSTTSTTSTTSSTTSTSSSPSTTSS 217
Score = 31.2 bits (69), Expect = 2.6
Identities = 24/62 (38%), Positives = 34/62 (54%), Gaps = 1/62 (1%)
Query: 14 SSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSAS 73
SSS SS SST + T S + S +T + S+S SSS T+S + +S+T S S
Sbjct: 183 SSSSSSSSSSTSTTSTTSTTS-STTSTSSSPSTTSSSTSASSSSETSSTQATSSSTTSTS 241
Query: 74 SA 75
S+
Sbjct: 242 SS 243
>NFL1_HUMAN (Q14494) Nuclear factor erythroid 2 related factor 1
(NF-E2 related factor 1) (NFE2-related factor 1)
(Nuclear factor, erythroid derived 2, like 1)
(Transcription factor 11) (Transcription factor HBZ17)
(Transcription factor LCR-F1) (Locus con
Length = 772
Score = 36.2 bits (82), Expect = 0.082
Identities = 37/132 (28%), Positives = 55/132 (41%), Gaps = 41/132 (31%)
Query: 11 SLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATP 70
SL SS PSSLSS++ + SSS SSS ++S S+AS++
Sbjct: 480 SLDSSHSPSSLSSSE---------------------GSSSSSSSSSSSSSSASSSASSSF 518
Query: 71 SASSAIDFLSICHRLKTTKRAG----------WIRKDVQDPESIA----------DHMYR 110
S A+ + S L + G + R QDP ++ +H Y
Sbjct: 519 SEEGAVGYSSDSETLDLEEAEGAVGYQPEYSKFCRMSYQDPAQLSCLPYLEHVGHNHTYN 578
Query: 111 MSLMALIAADIP 122
M+ AL +AD+P
Sbjct: 579 MAPSALDSADLP 590
>HMCT_HELPY (Q59465) Cadmium, zinc and cobalt transporting ATPase
(EC 3.6.3.3) (EC 3.6.3.5)
Length = 686
Score = 35.8 bits (81), Expect = 0.11
Identities = 30/100 (30%), Positives = 44/100 (44%), Gaps = 15/100 (15%)
Query: 135 VHDIAEAIVGDITPTDGVPKEEKNRLEQEAL---------DHMCKVLGGGSRAKEVAELW 185
V+DI VG+ P DGV + ++ L++ AL KVLGG K V E+
Sbjct: 209 VNDIVVVKVGEKVPVDGVVVKGESLLDERALSGESMPVNVSENSKVLGGSLNLKAVLEIQ 268
Query: 186 AEYEANSSPEAKFVKDLDKVEMILQALEYEHEQGKDLDEF 225
E S AK V+++ QA + E K + +F
Sbjct: 269 VEKMYKDSSIAKV------VDLVQQATNEKSETEKFITKF 302
>GUNA_PSEFL (P10476) Endoglucanase A precursor (EC 3.2.1.4)
(Endo-1,4-beta-glucanase) (Cellulase) (EGA)
Length = 962
Score = 35.4 bits (80), Expect = 0.14
Identities = 26/84 (30%), Positives = 43/84 (50%), Gaps = 7/84 (8%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SS SSS+P S SS+ S + S PS ++ SS P SSS ++S +++S++
Sbjct: 612 SSSSSSSVPVSSSSSSSIIPSSSSSSIQPS-------SSSSSMPSSSSSSSSVVASSSSS 664
Query: 70 PSASSAIDFLSICHRLKTTKRAGW 93
S ++ + L T ++GW
Sbjct: 665 VSGGLRCNWYGTLYPLCVTTQSGW 688
>FLO5_YEAST (P38894) Flocculation protein FLO5 precursor (Flocculin
5)
Length = 1075
Score = 35.4 bits (80), Expect = 0.14
Identities = 27/80 (33%), Positives = 43/80 (53%), Gaps = 2/80 (2%)
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS 61
+ T V+ SS+ SSS+ SSL ++ S +S + + + ++ SS P+SS T+
Sbjct: 662 SNGTSVISSSVISSSVTSSLVTSSSFISSSVISSSTTTSTSIFSESSTSSVIPTSSSTSG 721
Query: 62 PPSAASATPSASSAIDFLSI 81
S+ S T SASS+ SI
Sbjct: 722 --SSESKTSSASSSSSSSSI 739
Score = 33.1 bits (74), Expect = 0.70
Identities = 23/73 (31%), Positives = 36/73 (48%), Gaps = 4/73 (5%)
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS 61
+T++ + SS S S S SS S ++S SP T + SS PP +S TT
Sbjct: 708 STSSVIPTSSSTSGSSESKTSSASSSSSSSSISSESPKS----PTNSSSSLPPVTSATTG 763
Query: 62 PPSAASATPSASS 74
+A+S P+ ++
Sbjct: 764 QETASSLPPATTT 776
Score = 32.7 bits (73), Expect = 0.91
Identities = 27/83 (32%), Positives = 43/83 (51%), Gaps = 10/83 (12%)
Query: 3 TATRVLLSSLFSSSIPSSLSSTQLRF------KSVTLSPHSPSRVFHMTTAAGSSSPPSS 56
T++ V SS SSS+ SS ++T SV + S S T++ SSS SS
Sbjct: 678 TSSLVTSSSFISSSVISSSTTTSTSIFSESSTSSVIPTSSSTSGSSESKTSSASSSSSSS 737
Query: 57 SVTT----SPPSAASATPSASSA 75
S+++ SP +++S+ P +SA
Sbjct: 738 SISSESPKSPTNSSSSLPPVTSA 760
>DP87_DICDI (Q04503) Prespore protein DP87 precursor
Length = 555
Score = 35.4 bits (80), Expect = 0.14
Identities = 23/66 (34%), Positives = 40/66 (59%), Gaps = 6/66 (9%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SS SS+ S+ SS+ + + + PS ++A SSS PSS+ +++P SA+S+
Sbjct: 476 SSAASSADSSAASSSPSSSAASSAASSEPS------SSAASSSAPSSASSSAPSSASSSA 529
Query: 70 PSASSA 75
PS+S++
Sbjct: 530 PSSSAS 535
Score = 34.3 bits (77), Expect = 0.31
Identities = 24/66 (36%), Positives = 38/66 (57%), Gaps = 2/66 (3%)
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SS SSS SS +S++ S S +PS ++ SSS PSSS ++S S+A+++
Sbjct: 488 SSSPSSSAASSAASSEP--SSSAASSSAPSSASSSAPSSASSSAPSSSASSSAASSAASS 545
Query: 70 PSASSA 75
S+ S+
Sbjct: 546 ESSESS 551
Score = 33.9 bits (76), Expect = 0.41
Identities = 25/74 (33%), Positives = 41/74 (54%), Gaps = 2/74 (2%)
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS 61
+T+ SS SS+ SS SS+ S S S S ++ SSS PSS+ +++
Sbjct: 472 STSDSSAASSADSSAASSSPSSSAA--SSAASSEPSSSAASSSAPSSASSSAPSSASSSA 529
Query: 62 PPSAASATPSASSA 75
P S+AS++ ++S+A
Sbjct: 530 PSSSASSSAASSAA 543
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.312 0.125 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,498,396
Number of Sequences: 164201
Number of extensions: 1122498
Number of successful extensions: 7137
Number of sequences better than 10.0: 154
Number of HSP's better than 10.0 without gapping: 67
Number of HSP's successfully gapped in prelim test: 92
Number of HSP's that attempted gapping in prelim test: 5892
Number of HSP's gapped (non-prelim): 930
length of query: 261
length of database: 59,974,054
effective HSP length: 108
effective length of query: 153
effective length of database: 42,240,346
effective search space: 6462772938
effective search space used: 6462772938
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0041.3


