

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
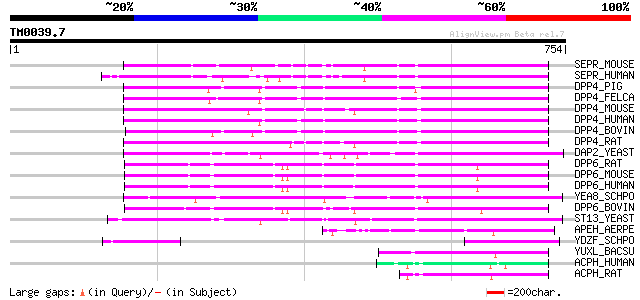
Query= TM0039.7
(754 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SEPR_MOUSE (P97321) Seprase (EC 3.4.21.-) (Fibroblast activation... 217 1e-55
SEPR_HUMAN (Q12884) Seprase (EC 3.4.21.-) (Fibroblast activation... 216 2e-55
DPP4_PIG (P22411) Dipeptidyl peptidase IV (EC 3.4.14.5) (DPP IV)... 197 9e-50
DPP4_FELCA (Q9N2I7) Dipeptidyl peptidase IV (EC 3.4.14.5) (DPP I... 196 2e-49
DPP4_MOUSE (P28843) Dipeptidyl peptidase IV (EC 3.4.14.5) (DPP I... 193 2e-48
DPP4_HUMAN (P27487) Dipeptidyl peptidase IV (EC 3.4.14.5) (DPP I... 191 6e-48
DPP4_BOVIN (P81425) Dipeptidyl peptidase IV (EC 3.4.14.5) (DPP I... 191 6e-48
DPP4_RAT (P14740) Dipeptidyl peptidase IV (EC 3.4.14.5) (DPP IV)... 190 1e-47
DAP2_YEAST (P18962) Dipeptidyl aminopeptidase B (EC 3.4.14.-) (D... 178 4e-44
DPP6_RAT (P46101) Dipeptidyl aminopeptidase-like protein 6 (Dipe... 165 5e-40
DPP6_MOUSE (Q9Z218) Dipeptidyl aminopeptidase-like protein 6 (Di... 164 6e-40
DPP6_HUMAN (P42658) Dipeptidyl aminopeptidase-like protein 6 (Di... 161 5e-39
YEA8_SCHPO (O14073) Putative dipeptidyl aminopeptidase C2E11.08 ... 161 7e-39
DPP6_BOVIN (P42659) Dipeptidyl aminopeptidase-like protein 6 (Di... 160 1e-38
ST13_YEAST (P33894) Dipeptidyl aminopeptidase A (EC 3.4.14.-) (D... 148 5e-35
APEH_AERPE (Q9YBQ2) Acylamino-acid-releasing enzyme (EC 3.4.19.1... 91 1e-17
YDZF_SCHPO (Q9P7E9) Putative dipeptidyl aminopeptidase C14C4.15c... 77 2e-13
YUXL_BACSU (P39839) Probable peptidase yuxL (EC 3.4.21.-) 69 5e-11
ACPH_HUMAN (P13798) Acylamino-acid-releasing enzyme (EC 3.4.19.1... 64 1e-09
ACPH_RAT (P13676) Acylamino-acid-releasing enzyme (EC 3.4.19.1) ... 62 7e-09
>SEPR_MOUSE (P97321) Seprase (EC 3.4.21.-) (Fibroblast activation
protein alpha) (Integral membrane serine protease)
Length = 761
Score = 217 bits (552), Expect = 1e-55
Identities = 171/596 (28%), Positives = 283/596 (46%), Gaps = 36/596 (6%)
Query: 155 SPDGSMLAYVRDHELHVLNLFTNESKQLTHGAKENGLTRGLAEYIAQEEM-DRKNGYWWS 213
SP GS LAYV + +++ + Q+T+ +EN + G+ +++ +EEM K WWS
Sbjct: 156 SPVGSKLAYVYQNNIYLKQRPGDPPFQITYTGRENRIFNGIPDWVYEEEMLATKYALWWS 215
Query: 214 LDSKYIAFTEVDSSEIPLFRIMHQGKSSVGLEAQEDHPYPFAGASNVKVRLGVVSAA-GS 272
D K++A+ E + S+IP+ + G + PYP AGA N VR+ +V
Sbjct: 216 PDGKFLAYVEFNDSDIPIIAYSYYGDGQYPRTI--NIPYPKAGAKNPVVRVFIVDTTYPH 273
Query: 273 SITWMDLLCGGKEQQANEEEYLARVNWMHGNTLTAQVLNRHHTKIKILKFDIRTG----- 327
+ M++ E A+ + Y + + W+ + Q L R + D R
Sbjct: 274 HVGPMEVPV--PEMIASSDYYFSWLTWVSSERVCLQWLKRVQNVSVLSICDFREDWHAWE 331
Query: 328 --QGKNILVEENSTWINIHDCFTP-LDKGITKFSGGFIWASEKTGFRHL-YLHDANGTCL 383
+ + + E + W TP + T + F S+K G++H+ Y+ D +
Sbjct: 332 CPKNQEHVEESRTGWAGGFFVSTPAFSQDATSYYKIF---SDKDGYKHIHYIKDTVENAI 388
Query: 384 GPITEGEWMVEQIAGVNEATGLLYFTGTLDG-PLESNLYCAKLYVDGTHPPEAPTRLTLN 442
IT G+W I V + + L Y + +G P N+Y + G PP +T +
Sbjct: 389 -QITSGKWEAIYIFRVTQDS-LFYSSNEFEGYPGRRNIYRISI---GNSPPSKKC-VTCH 442
Query: 443 KGKHIVVLDHHMQSFVDIYDSLGC-PPRVLLCSLEDGRV---IMPLFEQPFSVPRFKKLQ 498
K S+ Y +L C P + + +L DGR I L E + +Q
Sbjct: 443 LRKERCQYYTASFSYKAKYYALVCYGPGLPISTLHDGRTDQEIQVLEENKELENSLRNIQ 502
Query: 499 LEPPEIVEIQANDGTVLYGALYKPDASRFGPPPYKTMINVYGGPSVQLVSNSWLNTVDMR 558
L EI +++ T Y + P R Y +I VYGGP Q V + + V+
Sbjct: 503 LPKVEIKKLKDGGLTFWYKMILPPQFDR--SKKYPLLIQVYGGPCSQSVKSVF--AVNWI 558
Query: 559 AQYLRNQGILVWKLDNRGTARRGLKFESYLKHKLGQVDADDQLTGAEWLVKQGLAKAGHI 618
+GI++ +D RGTA +G KF + KLG + +DQLT ++ G I
Sbjct: 559 TYLASKEGIVIALVDGRGTAFQGDKFLHAVYRKLGVYEVEDQLTAVRKFIEMGFIDEERI 618
Query: 619 GLYGWSYGGYLSAMTLSRYPDFFKCAVAGAPVTSWDGYDTFYTEKYMGLPS--ENQSEYE 676
++GWSYGGY+S++ L+ FKC +A APV+SW+ Y + Y+E++MGLP+ +N Y+
Sbjct: 619 AIWGWSYGGYVSSLALASGTGLFKCGIAVAPVSSWEYYASIYSERFMGLPTKDDNLEHYK 678
Query: 677 SGSVMNQVHKLKG-RLLLVHGMIDENVHFRHTARLINALVAAGKPYELVLFPDERH 731
+ +VM + + LL+HG D+NVHF+++A++ ALV A ++ + + D+ H
Sbjct: 679 NSTVMARAEYFRNVDYLLIHGTADDNVHFQNSAQIAKALVNAQVDFQAMWYSDQNH 734
>SEPR_HUMAN (Q12884) Seprase (EC 3.4.21.-) (Fibroblast activation
protein alpha) (Integral membrane serine protease)
(170-kDa melanoma membrane-bound gelatinase)
Length = 760
Score = 216 bits (550), Expect = 2e-55
Identities = 180/642 (28%), Positives = 305/642 (47%), Gaps = 70/642 (10%)
Query: 125 AGIYIQDISQSKAELKLSSIPSSPIIDPHLSPDGSMLAYVRDHELHVLNLFTNESKQLTH 184
A YI D+S + ++ + +P PI SP GS LAYV + +++ + Q+T
Sbjct: 128 ATYYIYDLSNGEF-VRGNELPR-PIQYLCWSPVGSKLAYVYQNNIYLKQRPGDPPFQITF 185
Query: 185 GAKENGLTRGLAEYIAQEEM-DRKNGYWWSLDSKYIAFTEVDSSEIPLFRIMHQGKSSVG 243
+EN + G+ +++ +EEM K WWS + K++A+ E + +IP+ + G
Sbjct: 186 NGRENKIFNGIPDWVYEEEMLPTKYALWWSPNGKFLAYAEFNDKDIPVIAYSYYGDEQYP 245
Query: 244 LEAQEDHPYPFAGASNVKVRLGVVSAAGSSITWMDLLCGGKEQQ-----ANEEEYLARVN 298
+ PYP AGA N VR+ ++ T G +E A+ + Y + +
Sbjct: 246 RTI--NIPYPKAGAKNPVVRIFIID------TTYPAYVGPQEVPVPAMIASSDYYFSWLT 297
Query: 299 WMHGNTLTAQVLNRHHTKIKILKFDIRTGQGKNILVEENSTWINIHDCFTP---LDKGIT 355
W+ + Q L R + D R E+ TW DC +++ T
Sbjct: 298 WVTDERVCLQWLKRVQNVSVLSICDFR---------EDWQTW----DCPKTQEHIEESRT 344
Query: 356 KFSGGFIWA---------------SEKTGFRHL-YLHDANGTCLGPITEGEWMVEQIAGV 399
++GGF + S+K G++H+ Y+ D + IT G+W I V
Sbjct: 345 GWAGGFFVSRPVFSYDAISYYKIFSDKDGYKHIHYIKDTVENAI-QITSGKWEAINIFRV 403
Query: 400 NEATGLLYFTGTLDG-PLESNLYCAKLYVDGTHPPEAPTRLTLNKGKHIVVLDHHMQSFV 458
+ + L Y + + P N+Y + G++PP +T + K ++ SF
Sbjct: 404 TQDS-LFYSSNEFEEYPGRRNIYRISI---GSYPPSKKC-VTCHLRKERC--QYYTASFS 456
Query: 459 DI--YDSLGC-PPRVLLCSLEDGRV---IMPLFEQPFSVPRFKKLQLEPPEIVEIQANDG 512
D Y +L C P + + +L DGR I L E K +QL EI +++ ++
Sbjct: 457 DYAKYYALVCYGPGIPISTLHDGRTDQEIKILEENKELENALKNIQLPKEEIKKLEVDEI 516
Query: 513 TVLYGALYKPDASRFGPPPYKTMINVYGGPSVQLVSNSWLNTVDMRAQYLRNQGILVWKL 572
T+ Y + P R Y +I VYGGP Q V + + V+ + +G+++ +
Sbjct: 517 TLWYKMILPPQFDR--SKKYPLLIQVYGGPCSQSVRSVF--AVNWISYLASKEGMVIALV 572
Query: 573 DNRGTARRGLKFESYLKHKLGQVDADDQLTGAEWLVKQGLAKAGHIGLYGWSYGGYLSAM 632
D RGTA +G K + KLG + +DQ+T ++ G I ++GWSYGGY+S++
Sbjct: 573 DGRGTAFQGDKLLYAVYRKLGVYEVEDQITAVRKFIEMGFIDEKRIAIWGWSYGGYVSSL 632
Query: 633 TLSRYPDFFKCAVAGAPVTSWDGYDTFYTEKYMGLPS--ENQSEYESGSVMNQVHKLKG- 689
L+ FKC +A APV+SW+ Y + YTE++MGLP+ +N Y++ +VM + +
Sbjct: 633 ALASGTGLFKCGIAVAPVSSWEYYASVYTERFMGLPTKDDNLEHYKNSTVMARAEYFRNV 692
Query: 690 RLLLVHGMIDENVHFRHTARLINALVAAGKPYELVLFPDERH 731
LL+HG D+NVHF+++A++ ALV A ++ + + D+ H
Sbjct: 693 DYLLIHGTADDNVHFQNSAQIAKALVNAQVDFQAMWYSDQNH 734
>DPP4_PIG (P22411) Dipeptidyl peptidase IV (EC 3.4.14.5) (DPP IV)
(T-cell activation antigen CD26)
Length = 766
Score = 197 bits (501), Expect = 9e-50
Identities = 159/601 (26%), Positives = 272/601 (44%), Gaps = 42/601 (6%)
Query: 155 SPDGSMLAYVRDHELHVLNLFTNESKQLTHGAKENGLTRGLAEYIAQEEM-DRKNGYWWS 213
SP G LAYV +++++V N S+++T KEN + G+ +++ +EE+ + WWS
Sbjct: 158 SPVGHKLAYVWNNDIYVKNEPNLSSQRITWTGKENVIYNGVTDWVYEEEVFSAYSALWWS 217
Query: 214 LDSKYIAFTEVDSSEIPLFRIMHQGKSSVGLEAQEDHPYPFAGASNVKVRLGVVS----A 269
+ ++A+ + + +E+PL S+ PYP AGA N V+ VV +
Sbjct: 218 PNGTFLAYAQFNDTEVPLIEYSFYSDESLQYPKTVRIPYPKAGAENPTVKFFVVDTRTLS 277
Query: 270 AGSSITWMDLLCGGKEQQANEEEYLARVNWMHGNTLTAQVLNR--HHTKIKILKFDIRTG 327
+S+T ++ + YL V W+ ++ Q + R +++ I I +D TG
Sbjct: 278 PNASVTSYQIVPPASVLIG--DHYLCGVTWVTEERISLQWIRRAQNYSIIDICDYDESTG 335
Query: 328 QGKNILVEEN-----STWINIHDCFTPLDKGITKFSGGFIWA-SEKTGFRHLYLHDANGT 381
+ + + ++ + W+ F P + T F S + G++H+ + +
Sbjct: 336 RWISSVARQHIEISTTGWVGR---FRPAEPHFTSDGNSFYKIISNEEGYKHICHFQTDKS 392
Query: 382 CLGPITEGEWMVEQIAGVNEATG--LLYFTGTLDG-PLESNLYCAKLYVDGTHPPEAPTR 438
IT+G W ++ G+ T L Y + G P NLY +L D T
Sbjct: 393 NCTFITKGAW---EVIGIEALTSDYLYYISNEHKGMPGGRNLYRIQLN-DYTKVTCLSCE 448
Query: 439 LTLNKGKHIVVLDHHMQSFVDIYDSLGCPPRVLLCSLEDGRVIMPLFEQPFSVPRFKKLQ 498
L + ++ + + + P L S + + L + + +Q
Sbjct: 449 LNPERCQYYSASFSNKAKYYQLRCFGPGLPLYTLHSSSSDKELRVLEDNSALDKMLQDVQ 508
Query: 499 LEPPEIVEIQANDGTVLYGALYKPDASRFGPPPYKTMINVYGGPSVQLVSN----SWLNT 554
+ ++ I + Y + P + Y +I VY GP Q V SW
Sbjct: 509 MPSKKLDVINLHGTKFWYQMILPPHFDK--SKKYPLLIEVYAGPCSQKVDTVFRLSW--- 563
Query: 555 VDMRAQYLRN-QGILVWKLDNRGTARRGLKFESYLKHKLGQVDADDQLTGAEWLVKQGLA 613
A YL + + I+V D RG+ +G K + +LG + +DQ+ K G
Sbjct: 564 ----ATYLASTENIIVASFDGRGSGYQGDKIMHAINRRLGTFEVEDQIEATRQFSKMGFV 619
Query: 614 KAGHIGLYGWSYGGYLSAMTLSRYPDFFKCAVAGAPVTSWDGYDTFYTEKYMGLPS--EN 671
I ++GWSYGGY+++M L FKC +A APV+ W+ YD+ YTE+YMGLP+ +N
Sbjct: 620 DDKRIAIWGWSYGGYVTSMVLGAGSGVFKCGIAVAPVSKWEYYDSVYTERYMGLPTPEDN 679
Query: 672 QSEYESGSVMNQVHKLKG-RLLLVHGMIDENVHFRHTARLINALVAAGKPYELVLFPDER 730
Y + +VM++ K LL+HG D+NVHF+ +A+L ALV AG ++ + + DE
Sbjct: 680 LDYYRNSTVMSRAENFKQVEYLLIHGTADDNVHFQQSAQLSKALVDAGVDFQTMWYTDED 739
Query: 731 H 731
H
Sbjct: 740 H 740
>DPP4_FELCA (Q9N2I7) Dipeptidyl peptidase IV (EC 3.4.14.5) (DPP IV)
(T-cell activation antigen CD26)
Length = 765
Score = 196 bits (498), Expect = 2e-49
Identities = 160/600 (26%), Positives = 279/600 (45%), Gaps = 40/600 (6%)
Query: 155 SPDGSMLAYVRDHELHVLNLFTNESKQLTHGAKENGLTRGLAEYIAQEEM-DRKNGYWWS 213
SP+G LAYV ++++V N + S ++T +EN + G+A+++ +EE+ + WWS
Sbjct: 157 SPEGHKLAYVWKNDVYVKNEPNSSSHRITWTGEENAIYNGIADWVYEEEIFSAYSALWWS 216
Query: 214 LDSKYIAFTEVDSSEIPLFRIMHQGKSSVGLEAQEDHPYPFAGASNVKVRLGVVSA---- 269
++A+ + + +++PL S+ PYP AGA+N V+L V+
Sbjct: 217 PKGTFLAYAQFNDTQVPLIEYSFYSDESLQYPMTMRIPYPKAGAANPTVKLFVIKTDNLN 276
Query: 270 AGSSITWMDLLCGGKEQQANEEEYLARVNWMHGNTLTAQVLNR--HHTKIKILKFDIRTG 327
++ T +++ + YL V W + ++ Q L R +++ + I ++ TG
Sbjct: 277 PNTNATSVEIT--PPAAMLTGDYYLCDVTWANEERISLQWLRRIQNYSVMDIRDYNNSTG 334
Query: 328 QGKNILVEEN-----STWINIHDCFTPLDKGITKFSGGFIWA-SEKTGFRHLYLHDANGT 381
+ + +E+ + W+ F P + T F S + G++H+ +
Sbjct: 335 KWISSAAQEHIEMSTTGWVGR---FRPAEPHFTSDGRNFYKIISNEDGYKHICRFQIDKK 391
Query: 382 CLGPITEGEWMVEQIAGVNEATG--LLYFTGTLDG-PLESNLYCAKLYVDGTHPPEAPTR 438
IT+G W ++ G+ T L Y + G P NLY +L D T
Sbjct: 392 DCTFITKGAW---EVIGIEALTTDYLYYISNEYKGMPGGRNLYKIQLN-DYTKVACLSCE 447
Query: 439 LTLNKGKHIVVLDHHMQSFVDIYDSLGCPPRVLLCSLEDGRVIMPLFEQPFSVPRFKKLQ 498
L + ++ V + + S P L + + L + +++Q
Sbjct: 448 LKPERCQYYSVSFSKEAKYYQLRCSGPGLPLYTLHRSSNDEELRVLEDNSALDKMLQEVQ 507
Query: 499 LEPPEIVEIQANDGTVLYGALYKP--DASRFGPPPYKTMINVYGGPSVQLVSNSW-LNTV 555
+ ++ I N+ Y + P D S+ Y +I+VY GP Q + LN
Sbjct: 508 MPSKKLDFIILNETKFWYQMILPPHFDTSK----KYPLLIDVYAGPCSQKADAIFRLNW- 562
Query: 556 DMRAQYLRN-QGILVWKLDNRGTARRGLKFESYLKHKLGQVDADDQLTGAEWLVKQGLAK 614
A YL + + I+V D RG+ +G K + +LG + +DQ+ A K G
Sbjct: 563 ---ATYLASTENIIVASFDGRGSGYQGDKIMHAVNRRLGTFEVEDQIEAARQFSKMGFVD 619
Query: 615 AGHIGLYGWSYGGYLSAMTLSRYPDFFKCAVAGAPVTSWDGYDTFYTEKYMGLPS--ENQ 672
I ++GWSYGGY+++M L FKC +A APV+ W+ YD+ YTE+YMGLP+ +N
Sbjct: 620 DKRIAIWGWSYGGYVTSMVLGAGSGVFKCGIAVAPVSRWEYYDSVYTERYMGLPTPQDNL 679
Query: 673 SEYESGSVMNQVHKLKG-RLLLVHGMIDENVHFRHTARLINALVAAGKPYELVLFPDERH 731
Y++ +VM++ K LL+HG D+NVHF+ +A++ ALV AG ++ + + DE H
Sbjct: 680 DYYKNSTVMSRAENFKQVEYLLIHGTADDNVHFQQSAQISKALVDAGVDFQAMWYTDEDH 739
>DPP4_MOUSE (P28843) Dipeptidyl peptidase IV (EC 3.4.14.5) (DPP IV)
(T-cell activation antigen CD26) (Thymocyte-activating
molecule) (THAM)
Length = 760
Score = 193 bits (490), Expect = 2e-48
Identities = 162/599 (27%), Positives = 275/599 (45%), Gaps = 38/599 (6%)
Query: 155 SPDGSMLAYVRDHELHVLNLFTNESKQLTHGAKENGLTRGLAEYIAQEEM-DRKNGYWWS 213
SP+G LAYV ++++V S ++T +EN + G+ +++ +EE+ + WWS
Sbjct: 152 SPEGHKLAYVWKNDIYVKVEPHLPSHRITSTGEENVIYNGITDWVYEEEVFGAYSALWWS 211
Query: 214 LDSKYIAFTEVDSSEIPLFRIMHQGKSSVGLEAQEDHPYPFAGASNVKVRLGVVS--AAG 271
++ ++A+ + + + +PL S+ PYP AGA N V+ +V+ +
Sbjct: 212 PNNTFLAYAQFNDTGVPLIEYSFYSDESLQYPKTVWIPYPKAGAVNPTVKFFIVNIDSLS 271
Query: 272 SSITWMDLLCGGKEQQANEEEYLARVNWMHGNTLTAQVLNR--HHTKIKILKFD----IR 325
SS + + A + YL V W ++ Q L R +++ + I +D
Sbjct: 272 SSSSAAPIQIPAPASVARGDHYLCDVVWATEERISLQWLRRIQNYSVMAICDYDKINLTW 331
Query: 326 TGQGKNILVEENST-WINIHDCFTPLDKGITKFSGGFIWA-SEKTGFRHLYLHDANGTCL 383
+ VE ++T W+ F P + T F S+K G++H+ +
Sbjct: 332 NCPSEQQHVEMSTTGWVGR---FRPAEPHFTSDGSSFYKIISDKDGYKHICHFPKDKKDC 388
Query: 384 GPITEGEWMVEQIAGVNEATGLLYFTGTL--DGPLESNLYCAKLYVDGTHPPEAPTRLTL 441
IT+G W V I + + LY+ + P NLY +L D T+ L
Sbjct: 389 TFITKGAWEVISIEALT--SDYLYYISNQYKEMPGGRNLYKIQL-TDHTNVKCLSCDLNP 445
Query: 442 NKGKHIVVLDHHMQSFVDIYDSLGC----PPRVLLCSLEDGRVIMPLFEQPFSVPRFKKL 497
+ ++ V Y LGC P L D + + L + + +
Sbjct: 446 ERCQYYAVSFSKEAK----YYQLGCWGPGLPLYTLHRSTDHKELRVLEDNSALDRMLQDV 501
Query: 498 QLEPPEIVEIQANDGTVLYGALYKPDASRFGPPPYKTMINVYGGPSVQLVSNSW-LNTVD 556
Q+ ++ I N+ Y + P + Y +++VY GP Q S+ LN
Sbjct: 502 QMPSKKLDFIVLNETRFWYQMILPPHFDK--SKKYPLLLDVYAGPCSQKADASFRLNW-- 557
Query: 557 MRAQYLRN-QGILVWKLDNRGTARRGLKFESYLKHKLGQVDADDQLTGAEWLVKQGLAKA 615
A YL + + I+V D RG+ +G K + +LG ++ +DQ+ A VK G +
Sbjct: 558 --ATYLASTENIIVASFDGRGSGYQGDKIMHAINRRLGTLEVEDQIEAARQFVKMGFVDS 615
Query: 616 GHIGLYGWSYGGYLSAMTLSRYPDFFKCAVAGAPVTSWDGYDTFYTEKYMGL--PSENQS 673
+ ++GWSYGGY+++M L FKC +A APV+ W+ YD+ YTE+YMGL P +N
Sbjct: 616 KRVAIWGWSYGGYVTSMVLGSGSGVFKCGIAVAPVSRWEYYDSVYTERYMGLPIPEDNLD 675
Query: 674 EYESGSVMNQVHKLKG-RLLLVHGMIDENVHFRHTARLINALVAAGKPYELVLFPDERH 731
Y + +VM++ K LL+HG D+NVHF+ +A++ ALV AG ++ + + DE H
Sbjct: 676 HYRNSTVMSRAEHFKQVEYLLIHGTADDNVHFQQSAQISKALVDAGVDFQAMWYTDEDH 734
>DPP4_HUMAN (P27487) Dipeptidyl peptidase IV (EC 3.4.14.5) (DPP IV)
(T-cell activation antigen CD26) (TP103) (Adenosine
deaminase complexing protein-2) (ADABP)
Length = 766
Score = 191 bits (485), Expect = 6e-48
Identities = 158/596 (26%), Positives = 271/596 (44%), Gaps = 32/596 (5%)
Query: 155 SPDGSMLAYVRDHELHVLNLFTNESKQLTHGAKENGLTRGLAEYIAQEEM-DRKNGYWWS 213
SP G LAYV +++++V S ++T KE+ + G+ +++ +EE+ + WWS
Sbjct: 158 SPVGHKLAYVWNNDIYVKIEPNLPSYRITWTGKEDIIYNGITDWVYEEEVFSAYSALWWS 217
Query: 214 LDSKYIAFTEVDSSEIPLFRIMHQGKSSVGLEAQEDHPYPFAGASNVKVRLGVVSAAG-S 272
+ ++A+ + + +E+PL S+ PYP AGA N V+ VV+ S
Sbjct: 218 PNGTFLAYAQFNDTEVPLIEYSFYSDESLQYPKTVRVPYPKAGAVNPTVKFFVVNTDSLS 277
Query: 273 SIT-WMDLLCGGKEQQANEEEYLARVNWMHGNTLTAQVLNR--HHTKIKILKFDIRTGQG 329
S+T + + YL V W ++ Q L R +++ + I +D +G+
Sbjct: 278 SVTNATSIQITAPASMLIGDHYLCDVTWATQERISLQWLRRIQNYSVMDICDYDESSGRW 337
Query: 330 KNILVEEN-----STWINIHDCFTPLDKGITKFSGGFIWA-SEKTGFRHLYLHDANGTCL 383
++ ++ + W+ F P + T F S + G+RH+ +
Sbjct: 338 NCLVARQHIEMSTTGWVGR---FRPSEPHFTLDGNSFYKIISNEEGYRHICYFQIDKKDC 394
Query: 384 GPITEGEWMVEQIAGVNEATG--LLYFTGTLDG-PLESNLYCAKLYVDGTHPPEAPTRLT 440
IT+G W ++ G+ T L Y + G P NLY +L D T L
Sbjct: 395 TFITKGTW---EVIGIEALTSDYLYYISNEYKGMPGGRNLYKIQLS-DYTKVTCLSCELN 450
Query: 441 LNKGKHIVVLDHHMQSFVDIYDSLGCPPRVLLCSLEDGRVIMPLFEQPFSVPRFKKLQLE 500
+ ++ V + + S P L S + + + L + + +Q+
Sbjct: 451 PERCQYYSVSFSKEAKYYQLRCSGPGLPLYTLHSSVNDKGLRVLEDNSALDKMLQNVQMP 510
Query: 501 PPEIVEIQANDGTVLYGALYKPDASRFGPPPYKTMINVYGGPSVQLVSNSW-LNTVDMRA 559
++ I N+ Y + P + Y +++VY GP Q + LN A
Sbjct: 511 SKKLDFIILNETKFWYQMILPPHFDK--SKKYPLLLDVYAGPCSQKADTVFRLNW----A 564
Query: 560 QYLRN-QGILVWKLDNRGTARRGLKFESYLKHKLGQVDADDQLTGAEWLVKQGLAKAGHI 618
YL + + I+V D RG+ +G K + +LG + +DQ+ A K G I
Sbjct: 565 TYLASTENIIVASFDGRGSGYQGDKIMHAINRRLGTFEVEDQIEAARQFSKMGFVDNKRI 624
Query: 619 GLYGWSYGGYLSAMTLSRYPDFFKCAVAGAPVTSWDGYDTFYTEKYMGLPS--ENQSEYE 676
++GWSYGGY+++M L FKC +A APV+ W+ YD+ YTE+YMGLP+ +N Y
Sbjct: 625 AIWGWSYGGYVTSMVLGSGSGVFKCGIAVAPVSRWEYYDSVYTERYMGLPTPEDNLDHYR 684
Query: 677 SGSVMNQVHKLKG-RLLLVHGMIDENVHFRHTARLINALVAAGKPYELVLFPDERH 731
+ +VM++ K LL+HG D+NVHF+ +A++ ALV G ++ + + DE H
Sbjct: 685 NSTVMSRAENFKQVEYLLIHGTADDNVHFQQSAQISKALVDVGVDFQAMWYTDEDH 740
>DPP4_BOVIN (P81425) Dipeptidyl peptidase IV (EC 3.4.14.5) (DPP IV)
(T-cell activation antigen CD26) (Adenosine deaminase
complexing protein) (ADCP-I) (Activation molecule 3)
(ACT3) (WC10)
Length = 765
Score = 191 bits (485), Expect = 6e-48
Identities = 155/595 (26%), Positives = 272/595 (45%), Gaps = 36/595 (6%)
Query: 158 GSMLAYVRDHELHVLNLFTNESKQLTHGAKENGLTRGLAEYIAQEEM-DRKNGYWWSLDS 216
G LAYV +++++V N + S+++T K++ + G+ +++ +EE+ + WWS +S
Sbjct: 160 GHKLAYVWNNDIYVKNEPNSPSQRITWTGKKDVIYNGITDWVYEEEVFSAYSALWWSPNS 219
Query: 217 KYIAFTEVDSSEIPLFRIMHQGKSSVGLEAQEDHPYPFAGASNVKVRLGVVSAAGSS--- 273
++A+ + + +E+PL S+ PYP AGA N ++ VV+ + S
Sbjct: 220 TFLAYAQFNDTEVPLIEYSFYSDESLQYPKTVKIPYPKAGAVNPTIKFFVVNISSLSPNI 279
Query: 274 -ITWMDLLCGGKEQQANEEEYLARVNWMHGNTLTAQVLNR--HHTKIKILKFDIRTGQ-- 328
T ++ G + YL V W+ ++ Q L R +++ + I +D TG+
Sbjct: 280 NATSQQIVPPGSVLIG--DHYLCDVTWVTEERISLQWLRRIQNYSIMDICDYDRSTGRWI 337
Query: 329 ---GKNILVEENSTWINIHDCFTPLDKGITKFSGGFIWA-SEKTGFRHLYLHDANGTCLG 384
G+ + + W+ F P + T F S + G++H+ +
Sbjct: 338 SSVGRQHIEISTTGWVGR---FRPAEPHFTSDGNSFYKIISNEEGYKHICHFQTDKRNCT 394
Query: 385 PITEGEWMVEQIAGVNEATG--LLYFTGTLDG-PLESNLYCAKLYVDGTHPPEAPTRLTL 441
IT+G W ++ G+ T L Y + G P NLY +L D T L
Sbjct: 395 FITKGAW---EVIGIEALTSDYLYYISNEYKGMPGARNLYKIQLN-DYTKVTCLSCELNP 450
Query: 442 NKGKHIVVLDHHMQSFVDIYDSLGCPPRVLLCSLEDGRVIMPLFEQPFSVPRFKKLQLEP 501
++ ++ V + + S P L + + + + L + +Q+
Sbjct: 451 DRCQYYSVSFSQEAKYYQLRCSGPGLPLYTLHNSNNDKELRVLENNSDLDQVLQDVQMPS 510
Query: 502 PEIVEIQANDGTVLYGALYKPDASRFGPPPYKTMINVYGGPSVQLVSNSW-LNTVDMRAQ 560
++ I + Y + P + Y ++ VY GP Q + LN A
Sbjct: 511 KKLDFIHLHGTKFWYQMILPPHFDK--SKKYPLLLEVYAGPCSQKADAIFRLNW----AT 564
Query: 561 YLRN-QGILVWKLDNRGTARRGLKFESYLKHKLGQVDADDQLTGAEWLVKQGLAKAGHIG 619
YL + + I+V D RG+ +G K + +LG + +DQ+ K G I
Sbjct: 565 YLASTENIIVASFDGRGSGYQGDKIMHAINRRLGTFEVEDQIEATRQFSKMGFVDDKRIA 624
Query: 620 LYGWSYGGYLSAMTLSRYPDFFKCAVAGAPVTSWDGYDTFYTEKYMGLPS--ENQSEYES 677
++GWSYGGY+++M L FKC +A APV+ W+ YD+ YTE+YMGLP+ +N Y +
Sbjct: 625 IWGWSYGGYVTSMVLGAGSGVFKCGIAVAPVSKWEYYDSVYTERYMGLPTPEDNLDSYRN 684
Query: 678 GSVMNQVHKLKG-RLLLVHGMIDENVHFRHTARLINALVAAGKPYELVLFPDERH 731
+VM++ K LL+HG D+NVHF+ +A++ ALV AG ++ + + DE H
Sbjct: 685 STVMSRAENFKQVEYLLIHGTADDNVHFQQSAQISKALVDAGVDFQSMWYTDEDH 739
>DPP4_RAT (P14740) Dipeptidyl peptidase IV (EC 3.4.14.5) (DPP IV)
(T-cell activation antigen CD26) (GP110 glycoprotein)
(Bile canaliculus domain-specific membrane glycoprotein)
Length = 767
Score = 190 bits (483), Expect = 1e-47
Identities = 161/600 (26%), Positives = 275/600 (45%), Gaps = 37/600 (6%)
Query: 155 SPDGSMLAYVRDHELHVLNLFTNESKQLTHGAKENGLTRGLAEYIAQEEM-DRKNGYWWS 213
S +G LAYV ++++V S ++T KEN + G+ +++ +EE+ + WWS
Sbjct: 156 SQEGHKLAYVWKNDIYVKIEPHLPSHRITSTGKENVIFNGINDWVYEEEIFGAYSALWWS 215
Query: 214 LDSKYIAFTEVDSSEIPLFRIMHQGKSSVGLEAQEDHPYPFAGASNVKVRLGVVS--AAG 271
+ ++A+ + + + +PL S+ PYP AGA N V+ +V+ +
Sbjct: 216 PNGTFLAYAQFNDTGVPLIEYSFYSDESLQYPKTVWIPYPKAGAVNPTVKFFIVNTDSLS 275
Query: 272 SSITWMDLLCGGKEQQANEEEYLARVNWMHGNTLTAQVLNR--HHTKIKILKFDIRTGQG 329
S+ T + + + YL V W+ + ++ Q L R +++ + I +D T
Sbjct: 276 STTTTIPMQITAPASVTTGDHYLCDVAWVSEDRISLQWLRRIQNYSVMAICDYDKTTLVW 335
Query: 330 KNILVEENSTWINIHDC--FTPLDKGITKFSGGFIW-ASEKTGFRHLYLHDANG----TC 382
+E+ C F P + T F S+K G++H+ + C
Sbjct: 336 NCPTTQEHIETSATGWCGRFRPAEPHFTSDGSSFYKIVSDKDGYKHICQFQKDRKPEQVC 395
Query: 383 LGPITEGEWMVEQIAGVNEATGLLYFTGT--LDGPLESNLYCAKLYVDGTHPPEAPTRLT 440
IT+G W V I + + LY+ + P NLY +L D T+ L
Sbjct: 396 TF-ITKGAWEVISIEALT--SDYLYYISNEYKEMPGGRNLYKIQL-TDHTNKKCLSCDLN 451
Query: 441 LNKGKHIVVLDHHMQSFVDIYDSLGCP----PRVLLCSLEDGRVIMPLFEQPFSVPRFKK 496
+ ++ V S Y LGC P L D + + L + +
Sbjct: 452 PERCQYYSV----SLSKEAKYYQLGCRGPGLPLYTLHRSTDQKELRVLEDNSALDKMLQD 507
Query: 497 LQLEPPEIVEIQANDGTVLYGALYKPDASRFGPPPYKTMINVYGGPSVQLVSNSW-LNTV 555
+Q+ ++ I N+ Y + P + Y +I+VY GP Q ++ LN
Sbjct: 508 VQMPSKKLDFIVLNETRFWYQMILPPHFDK--SKKYPLLIDVYAGPCSQKADAAFRLNW- 564
Query: 556 DMRAQYLRN-QGILVWKLDNRGTARRGLKFESYLKHKLGQVDADDQLTGAEWLVKQGLAK 614
A YL + + I+V D RG+ +G K + +LG ++ +DQ+ A +K G
Sbjct: 565 ---ATYLASTENIIVASFDGRGSGYQGDKIMHAINKRLGTLEVEDQIEAARQFLKMGFVD 621
Query: 615 AGHIGLYGWSYGGYLSAMTLSRYPDFFKCAVAGAPVTSWDGYDTFYTEKYMGLPS--ENQ 672
+ + ++GWSYGGY+++M L FKC +A APV+ W+ YD+ YTE+YMGLP+ +N
Sbjct: 622 SKRVAIWGWSYGGYVTSMVLGSGSGVFKCGIAVAPVSRWEYYDSVYTERYMGLPTPEDNL 681
Query: 673 SEYESGSVMNQVHKLKG-RLLLVHGMIDENVHFRHTARLINALVAAGKPYELVLFPDERH 731
Y + +VM++ K LL+HG D+NVHF+ +A++ ALV AG ++ + + DE H
Sbjct: 682 DHYRNSTVMSRAENFKQVEYLLIHGTADDNVHFQQSAQISKALVDAGVDFQAMWYTDEDH 741
>DAP2_YEAST (P18962) Dipeptidyl aminopeptidase B (EC 3.4.14.-) (DPAP
B) (YSCV)
Length = 818
Score = 178 bits (452), Expect = 4e-44
Identities = 162/633 (25%), Positives = 280/633 (43%), Gaps = 52/633 (8%)
Query: 155 SPDGSMLAYVRDHELHVLNLFTNESKQLTHGAKENGLTRGLAEYIAQEEM-DRKNGYWWS 213
SP+ + +AYV+D+ +++ + + ++ + + L G +++ +EE+ + WWS
Sbjct: 195 SPNSNDIAYVQDNNIYIYSAISKKTIRAVTNDGSSFLFNGKPDWVYEEEVFEDDKAAWWS 254
Query: 214 LDSKYIAFTEVDSSEIPLFRIMHQGKSSVGLEAQ-EDHPYPFAGASNVKVRLGVVSAAGS 272
Y+AF ++D SE+ F I + + + + YP +G N L V S
Sbjct: 255 PTGDYLAFLKIDESEVGEFIIPYYVQDEKDIYPEMRSIKYPKSGTPNPHAELWVYSMKDG 314
Query: 273 SITWMDLLCGGKEQQANEEEYLARVNWMHGNTLTAQVLNRHHTKIKILKFDIRTGQGKNI 332
+ ++ + G K+ + + V W+ + + +R + + D + N+
Sbjct: 315 T-SFHPRISGNKKDGSL---LITEVTWVGNGNVLVKTTDRSSDILTVFLIDT-IAKTSNV 369
Query: 333 LVEENST---WINIHDC-FTPLDKGITKFSGGFIWASEKTGFRHL-YLHDANGTCLGPIT 387
+ E+S W H+ F P ++ + G++ G+ HL Y ++N + +T
Sbjct: 370 VRNESSNGGWWEITHNTLFIPANETFDRPHNGYVDILPIGGYNHLAYFENSNSSHYKTLT 429
Query: 388 EGEW-MVEQIAGVNEATGLLYFTGTLDGPLESNLYCAKLYVDGTHPPE----------AP 436
EG+W +V + LYF T E ++Y Y+D P E
Sbjct: 430 EGKWEVVNGPLAFDSMENRLYFISTRKSSTERHVY----YIDLRSPNEIIEVTDTSEDGV 485
Query: 437 TRLTLNKGKHIVVLDHH-----MQSFVDIYDSLG--CPP-RVL---LCSLEDGRVIMPLF 485
++ + G+ +L + Q VD + C VL L LE V+ +
Sbjct: 486 YDVSFSSGRRFGLLTYKGPKVPYQKIVDFHSRKAEKCDKGNVLGKSLYHLEKNEVLTKIL 545
Query: 486 EQPFSVPR--FKKLQLEPPEI-VEIQANDGTVLYGALYKPDASRFGPPPYKTMINVYGGP 542
E ++VPR F++L L E +I N +L D Y YGGP
Sbjct: 546 ED-YAVPRKSFRELNLGKDEFGKDILVNSYEILPN-----DFDETLSDHYPVFFFAYGGP 599
Query: 543 SVQLVSNSWLNTVDMRAQYLRNQGILVWKLDNRGTARRGLKFESYLKHKLGQVDADDQLT 602
+ Q V ++ +V +V +D RGT +G F S ++ +LG +A DQ++
Sbjct: 600 NSQQVVKTF--SVGFNEVVASQLNAIVVVVDGRGTGFKGQDFRSLVRDRLGDYEARDQIS 657
Query: 603 GAEWLVKQGLAKAGHIGLYGWSYGGYLSAMTLSRYPD-FFKCAVAGAPVTSWDGYDTFYT 661
A I L+GWSYGGYL+ TL + FK ++ APVT W YD+ YT
Sbjct: 658 AASLYGSLTFVDPQKISLFGWSYGGYLTLKTLEKDGGRHFKYGMSVAPVTDWRFYDSVYT 717
Query: 662 EKYMGLPSENQSEYESGSVMNQVHKLK-GRLLLVHGMIDENVHFRHTARLINALVAAG-K 719
E+YM P EN Y SV N + R LL+HG D+NVHF+++ + ++ L G +
Sbjct: 718 ERYMHTPQENFDGYVESSVHNVTALAQANRFLLMHGTGDDNVHFQNSLKFLDLLDLNGVE 777
Query: 720 PYELVLFPDERHMPRRHSDRVYMEERMWEFIDR 752
Y++ +FPD H R H+ V + +++ ++ R
Sbjct: 778 NYDVHVFPDSDHSIRYHNANVIVFDKLLDWAKR 810
>DPP6_RAT (P46101) Dipeptidyl aminopeptidase-like protein 6
(Dipeptidylpeptidase VI) (Dipeptidylpeptidase 6)
(Dipeptidyl peptidase IV like protein) (Dipeptidyl
aminopeptidase-related protein) (DPPX)
Length = 859
Score = 165 bits (417), Expect = 5e-40
Identities = 146/589 (24%), Positives = 253/589 (42%), Gaps = 26/589 (4%)
Query: 156 PDGSMLAYVRDHELHVLNLFTNESKQLTHGAKENGLTRGLAEYIAQEEMDRKN-GYWWSL 214
P G L ++ ++ ++ ++ ++ KE + GL++++ +EE+ + + +WWS
Sbjct: 243 PKGQQLIFIFENNIYYCAHVGKQAIRVVSTGKEGVIYNGLSDWLYEEEILKSHIAHWWSP 302
Query: 215 DSKYIAFTEVDSSEIPLFRIMHQGKSSVGLEAQEDHPYPFAGASNVKVRLGVVSAAGSSI 274
D +A+ ++ S +PL + S + + YP AG+ N + L V+ G +
Sbjct: 303 DGTRLAYATINDSRVPLMELPTYTGSVY--PTVKPYHYPKAGSENPSISLHVIGLNGPT- 359
Query: 275 TWMDLLCGGKEQQANEEEYLARVNWMHGNTLTAQVLNRHHTKIKILKFDIRTGQGKNILV 334
DL + E Y+ V W + LNR + D TG
Sbjct: 360 --HDLEMMPPDDPRMREYYITMVKWATSTKVAVTWLNRAQNVSILTLCDATTGVCTKKHE 417
Query: 335 EENSTWINIHDCFTPLDKGITKFSGGFIWASEKTG---FRHLYLH----DANGTCLGPIT 387
+E+ W++ + K KF F+ A + G F H+ + +++ + IT
Sbjct: 418 DESEAWLHRQNEEPVFSKDGRKFF--FVRAIPQGGRGKFYHITVSSSQPNSSNDNIQSIT 475
Query: 388 EGEWMVEQIAGVNEATGLLYFTGTLDGPLESNLYCAKLYVDGTHPPEAPTRLTLNKGKHI 447
G+W V +I +E LYF T D P +LY A VD + L N
Sbjct: 476 SGDWDVTEILTYDEKRNKLYFLSTEDLPRRRHLYSANT-VDDFNRQCLSCDLVENCTYVS 534
Query: 448 VVLDHHMQSFVDIYDSLGCPPRVLLCSLEDGRVIMPLFEQPFSVPRFKKLQLEPPEIVEI 507
H+M F+ + G P V + + D R + L Q+ E +I
Sbjct: 535 ASFSHNMDFFLLKCEGPGVPT-VTVHNTTDKRRMFDLEANEQVQKAIYDRQMPKIEYRKI 593
Query: 508 QANDGTVLYGALYKPDASRFGPPPYKTMINVYGGPSVQLVSNSWLNTVDMRAQYLRNQGI 567
+ D ++ L KP A+ Y ++ V G P Q VS + V + + G
Sbjct: 594 EVEDYSLPMQIL-KP-ATFTDTAHYPLLLVVDGTPGSQSVSERF--EVTWETVLVSSHGA 649
Query: 568 LVWKLDNRGTARRGLKFESYLKHKLGQVDADDQLTGAEWLVKQGLAKAGHIGLYGWSYGG 627
+V K D RG+ +G K ++ +LG ++ DQ+ ++K+ + ++G YGG
Sbjct: 650 VVVKCDGRGSGFQGTKLLHEVRRRLGFLEEKDQMEAVRTMLKEQYIDKTRVAVFGKDYGG 709
Query: 628 YLSAMTL----SRYPDFFKCAVAGAPVTSWDGYDTFYTEKYMGLPSENQSEYESGSVMNQ 683
YLS L F C A +P+T + Y + ++E+Y+GL + YE + ++
Sbjct: 710 YLSTYILPAKGENQGQTFTCGSALSPITDFKLYASAFSERYLGLHGLDNRAYEMTKLAHR 769
Query: 684 VHKLKGR-LLLVHGMIDENVHFRHTARLINALVAAGKPYELVLFPDERH 731
V L+ + L++H DE +HF+HTA LI L+ Y L ++PDE H
Sbjct: 770 VSALEDQQFLIIHATADEKIHFQHTAELITQLIKGKANYSLQIYPDESH 818
>DPP6_MOUSE (Q9Z218) Dipeptidyl aminopeptidase-like protein 6
(Dipeptidylpeptidase VI) (Dipeptidylpeptidase 6)
(Dipeptidyl peptidase IV like protein) (Dipeptidyl
aminopeptidase-related protein) (DPPX)
Length = 804
Score = 164 bits (416), Expect = 6e-40
Identities = 144/589 (24%), Positives = 253/589 (42%), Gaps = 26/589 (4%)
Query: 156 PDGSMLAYVRDHELHVLNLFTNESKQLTHGAKENGLTRGLAEYIAQEEMDRKN-GYWWSL 214
P G L ++ ++ ++ ++ ++ KE + GL++++ +EE+ + + +WWS
Sbjct: 188 PKGQQLIFIFENNIYYCAHVGKQAIRVVSTGKEGVIYNGLSDWLYEEEILKSHIAHWWSP 247
Query: 215 DSKYIAFTEVDSSEIPLFRIMHQGKSSVGLEAQEDHPYPFAGASNVKVRLGVVSAAGSSI 274
D +A+ ++ S +PL + S + + YP AG+ N + L V+ G +
Sbjct: 248 DGTRLAYATINDSRVPLMELPTYTGSVY--PTVKPYHYPKAGSENPSISLHVIGLNGPT- 304
Query: 275 TWMDLLCGGKEQQANEEEYLARVNWMHGNTLTAQVLNRHHTKIKILKFDIRTGQGKNILV 334
DL + E Y+ V W + LNR + D TG
Sbjct: 305 --HDLEMMPPDDPRMREYYITMVKWATSTKVAVTWLNRAQNVSILTLCDATTGVCTKKHE 362
Query: 335 EENSTWINIHDCFTPLDKGITKFSGGFIWASEKTG---FRHLYLH----DANGTCLGPIT 387
+E+ W++ + K KF F+ A + G F H+ + +++ + IT
Sbjct: 363 DESEAWLHRQNEEPVFSKDGRKFF--FVRAIPQGGRGKFYHITVSSSQPNSSNDNIQSIT 420
Query: 388 EGEWMVEQIAGVNEATGLLYFTGTLDGPLESNLYCAKLYVDGTHPPEAPTRLTLNKGKHI 447
G+W V +I +E +YF T D P +LY A VD + L N
Sbjct: 421 SGDWDVTKILSYDEKRNKIYFLSTEDLPRRRHLYSANT-VDDFNRQCLSCDLVENCTYVS 479
Query: 448 VVLDHHMQSFVDIYDSLGCPPRVLLCSLEDGRVIMPLFEQPFSVPRFKKLQLEPPEIVEI 507
H+M F+ + G P V + + D R + L Q+ E +I
Sbjct: 480 ASFSHNMDFFLLKCEGPGVPT-VTVHNTTDKRRMFDLEANEEVQKAINDRQMPKIEYRKI 538
Query: 508 QANDGTVLYGALYKPDASRFGPPPYKTMINVYGGPSVQLVSNSWLNTVDMRAQYLRNQGI 567
+ D ++ L KP A+ Y ++ V G P Q V+ + V + + G
Sbjct: 539 EVEDYSLPMQIL-KP-ATFTDTAHYPLLLVVDGTPGSQSVTERF--EVTWETVLVSSHGA 594
Query: 568 LVWKLDNRGTARRGLKFESYLKHKLGQVDADDQLTGAEWLVKQGLAKAGHIGLYGWSYGG 627
+V K D RG+ +G K ++ +LG ++ DQ+ ++K+ + ++G YGG
Sbjct: 595 VVVKCDGRGSGFQGTKLLQEVRRRLGFLEEKDQMEAVRTMLKEQYIDKTRVAVFGKDYGG 654
Query: 628 YLSAMTL----SRYPDFFKCAVAGAPVTSWDGYDTFYTEKYMGLPSENQSEYESGSVMNQ 683
YLS L F C A +P+T + Y + ++E+Y+GL + YE + ++
Sbjct: 655 YLSTYILPAKGENQGQTFTCGSALSPITDFKLYASAFSERYLGLHGLDNRAYEMTKLAHR 714
Query: 684 VHKLKGR-LLLVHGMIDENVHFRHTARLINALVAAGKPYELVLFPDERH 731
V L+ + L++H DE +HF+HTA LI L+ Y L ++PDE H
Sbjct: 715 VSALEDQQFLIIHATADEKIHFQHTAELITQLIKGKANYSLQIYPDESH 763
>DPP6_HUMAN (P42658) Dipeptidyl aminopeptidase-like protein 6
(Dipeptidylpeptidase VI) (Dipeptidylpeptidase 6)
(Dipeptidyl peptidase IV like protein) (Dipeptidyl
aminopeptidase-related protein) (DPPX)
Length = 865
Score = 161 bits (408), Expect = 5e-39
Identities = 143/590 (24%), Positives = 252/590 (42%), Gaps = 28/590 (4%)
Query: 156 PDGSMLAYVRDHELHVLNLFTNESKQLTHGAKENGLTRGLAEYIAQEEMDRKN-GYWWSL 214
P G L ++ ++ ++ ++ ++ KE + GL++++ +EE+ + + +WWS
Sbjct: 249 PKGQQLIFIFENNIYYCAHVGKQAIRVVSTGKEGVIYNGLSDWLYEEEILKTHIAHWWSP 308
Query: 215 DSKYIAFTEVDSSEIPLFRIMHQGKSSVGLEAQEDHPYPFAGASNVKVRLGVVSAAGSSI 274
D +A+ ++ S +P+ + S + + YP AG+ N + L V+ G +
Sbjct: 309 DGTRLAYAAINDSRVPIMELPTYTGSIY--PTVKPYHYPKAGSENPSISLHVIGLNGPT- 365
Query: 275 TWMDLLCGGKEQQANEEEYLARVNWMHGNTLTAQVLNRHHTKIKILKFDIRTGQGKNILV 334
DL + E Y+ V W + LNR + D TG
Sbjct: 366 --HDLEMMPPDDPRMREYYITMVKWATSTKVAVTWLNRAQNVSILTLCDATTGVCTKKHE 423
Query: 335 EENSTWINIHDCFTPLDKGITKFSGGFIWASEKTG---FRHLYLH----DANGTCLGPIT 387
+E+ W++ + K KF FI A + G F H+ + +++ + IT
Sbjct: 424 DESEAWLHRQNEEPVFSKDGRKFF--FIRAIPQGGRGKFYHITVSSSQPNSSNDNIQSIT 481
Query: 388 EGEWMVEQIAGVNEATGLLYFTGTLDGPLESNLYCAKLYVDGTHPPEAPT-RLTLNKGKH 446
G+W V +I +E +YF T D P LY A +G + + L N
Sbjct: 482 SGDWDVTKILAYDEKGNKIYFLSTEDLPRRRQLYSAN--TEGNFNRQCLSCDLVENCTYF 539
Query: 447 IVVLDHHMQSFVDIYDSLGCPPRVLLCSLEDGRVIMPLFEQPFSVPRFKKLQLEPPEIVE 506
H M F+ + G P V + + D + + L Q+ E +
Sbjct: 540 SASFSHSMDFFLLKCEGPGVP-MVTVHNTTDKKKMFDLETNEHVKKAINDRQMPKVEYRD 598
Query: 507 IQANDGTVLYGALYKPDASRFGPPPYKTMINVYGGPSVQLVSNSWLNTVDMRAQYLRNQG 566
I+ +D + L KP A+ Y ++ V G P Q V+ + V + + G
Sbjct: 599 IEIDDYNLPMQIL-KP-ATFTDTTHYPLLLVVDGTPGSQSVAEKF--EVSWETVMVSSHG 654
Query: 567 ILVWKLDNRGTARRGLKFESYLKHKLGQVDADDQLTGAEWLVKQGLAKAGHIGLYGWSYG 626
+V K D RG+ +G K ++ +LG ++ DQ+ ++K+ + ++G YG
Sbjct: 655 AVVVKCDGRGSGFQGTKLLHEVRRRLGLLEEKDQMEAVRTMLKEQYIDRTRVAVFGKDYG 714
Query: 627 GYLSAMTL----SRYPDFFKCAVAGAPVTSWDGYDTFYTEKYMGLPSENQSEYESGSVMN 682
GYLS L F C A +P+T + Y + ++E+Y+GL + YE V +
Sbjct: 715 GYLSTYILPAKGENQGQTFTCGSALSPITDFKLYASAFSERYLGLHGLDNRAYEMTKVAH 774
Query: 683 QVHKLKGR-LLLVHGMIDENVHFRHTARLINALVAAGKPYELVLFPDERH 731
+V L+ + L++H DE +HF+HTA LI L+ Y L ++PDE H
Sbjct: 775 RVSALEEQQFLIIHPTADEKIHFQHTAELITQLIRGKANYSLQIYPDESH 824
>YEA8_SCHPO (O14073) Putative dipeptidyl aminopeptidase C2E11.08 (EC
3.4.14.-)
Length = 793
Score = 161 bits (407), Expect = 7e-39
Identities = 159/620 (25%), Positives = 261/620 (41%), Gaps = 46/620 (7%)
Query: 155 SPDGSMLAYVRDHELHVLNLFTNESKQLTHGAKENGLTRGLAEYIAQEE-MDRKNGYWWS 213
SP G L++V +++L+V N + G + + GL ++I +EE + + WWS
Sbjct: 177 SPTGHQLSFVYNNDLYVRKNDGNVQRLTYDGTVD--VFNGLTDWIYEEEVLSSPSTIWWS 234
Query: 214 LDSKYIAFTEVDSSEIPLFRI-MHQGKSSVGLEAQEDHP-----YPFAGASNVKVRLGVV 267
DS IAF +++ SEIP + ++ + L + + YP G N V L V
Sbjct: 235 PDSDKIAFLKLNESEIPTYHYPLYTAELDPSLPEFDYNKDMAIKYPKPGNPNPSVSLFVA 294
Query: 268 SAAGSSITWMDLLCGGKEQQANEEEYLARVNWMHGNTLTAQVLNRHHTKIKILKFDIRTG 327
++ + L + E + V W++ +++ Q NR+ T I D
Sbjct: 295 DLNSNASSNFSLW----HNEPLAEPVVQNVLWVNTSSVLVQFTNRNSTCITARLLDTELK 350
Query: 328 QGKNILVE-ENSTWINIHDC--FTPLDKGIT--KFSGGFIWASEKTGFRHLYLHDANGTC 382
+ E W + PL+ + +S G+ + HL NG+
Sbjct: 351 SIHTVKTECLEEGWYEVQQSAKMFPLNNSLVWENWSDGYFDILALDDYNHLAFIPFNGSS 410
Query: 383 LGPITEGEWMV-EQIAGVNEATGLLYFTGTLDGPLESNLYCAKLY------VDGTHPPEA 435
+T G W V + ++ G +YF TL E +LY L + E
Sbjct: 411 PIYLTSGAWDVTDGPIHIDGDFGNVYFLATLKDSTERHLYYVSLDTLEIYGITDNGEDEG 470
Query: 436 PTRLTLNKGKHIVVLDHHMQSFVDIYDSLGCPPRVLLCSLEDGRVIMPLFEQPFSVPRFK 495
+ + VL++H P L S +D + L +
Sbjct: 471 YYSTSFSPFGDFYVLNYHGPDV----------PWQELRSTKDKDYCLSLETNSRLKQQLS 520
Query: 496 KLQLEPPEIVEIQANDGTVLYGALYKPDASRFGPPPYKTMINVYGGPSVQLVSNSWLNTV 555
+ L E ++ ND T + + + + Y + YGGP Q V+ L V
Sbjct: 521 SITLPSVEYGKLTFNDTT--FNFMERRPRNFDVNKKYPVLFFAYGGPGSQQVAK--LFRV 576
Query: 556 DMRAQYLRNQG---ILVWKLDNRGTARRGLKFESYLKHKLGQVDADDQLTGAEWLVKQGL 612
D +A YL + +V LD RGT G F + LG+ ++ DQ ++
Sbjct: 577 DFQA-YLASHPDFEFIVVTLDGRGTGFNGNAFRYSVSRHLGEWESYDQGQAGKFWADLPF 635
Query: 613 AKAGHIGLYGWSYGGYLSAMTLSRYPDFFKCAVAGAPVTSWDGYDTFYTEKYMGLPSENQ 672
H+G++GWSYGGYL+ TL D F +A APVT W YD+ YTE+YM LP N+
Sbjct: 636 VDENHVGIWGWSYGGYLTLKTLET-QDVFSYGMAVAPVTDWRLYDSVYTERYMDLPQYNK 694
Query: 673 SEYESGSVMN-QVHKLKGRLLLVHGMIDENVHFRHTARLINAL-VAAGKPYELVLFPDER 730
Y++ + + + K R + HG D+NVHF+H+ L++ L +A Y++ +FPD
Sbjct: 695 EGYKNSQIHDYEKFKQLKRFFVAHGTGDDNVHFQHSMHLMDGLNLANCYNYDMAVFPDSA 754
Query: 731 HMPRRHSDRVYMEERMWEFI 750
H H+ + + R+ E+I
Sbjct: 755 HSISYHNASLSIYHRLSEWI 774
>DPP6_BOVIN (P42659) Dipeptidyl aminopeptidase-like protein 6
(Dipeptidylpeptidase VI) (Dipeptidylpeptidase 6)
(Dipeptidyl peptidase IV like protein) (Dipeptidyl
aminopeptidase-related protein) (DPPX)
Length = 863
Score = 160 bits (405), Expect = 1e-38
Identities = 143/593 (24%), Positives = 254/593 (42%), Gaps = 34/593 (5%)
Query: 156 PDGSMLAYVRDHELHVLNLFTNESKQLTHGAKENGLTRGLAEYIAQEEMDRKN-GYWWSL 214
P G L ++ ++ ++ ++ ++ KE + GL++++ +EE+ + + +WWS
Sbjct: 247 PKGQQLIFIFENNIYYCAHVGKQAIRVVSTGKEGVIYNGLSDWLYEEEILKTHIAHWWSP 306
Query: 215 DSKYIAFTEVDSSEIPLFRIMHQGKSSVGLEAQEDHPYPFAGASNVKVRLGVVSAAGSSI 274
D +A+ ++ S +P+ + SV A+ H YP AG N + L V+ G +
Sbjct: 307 DGTRLAYATINDSRVPVMELPTY-TGSVYPTAKPYH-YPKAGCENPSISLHVIGLNGPT- 363
Query: 275 TWMDLLCGGKEQQANEEEYLARVNWMHGNTLTAQVLNRHHTKIKILKFDIRTGQGKNILV 334
DL + E Y+ V W + L+R + D TG
Sbjct: 364 --HDLEMTPPDDPRMREYYITMVKWATSTKVAVNWLSRAQNVSILTLCDATTGVCTKKHE 421
Query: 335 EENSTWINIHDCFTPLDKGITKFSGGFIWASEKTG---FRHLYLH----DANGTCLGPIT 387
+E+ W++ + K KF F+ A + G F H+ + +++ + IT
Sbjct: 422 DESEAWLHRQNEEPVFSKDGRKFF--FVRAIPQGGQGKFYHITVSSSQPNSSNDNIQSIT 479
Query: 388 EGEWMVEQIAGVNEATGLLYFTGTLDGPLESNLYCAKLYVDGTHPPEAPTRLTLNKGKHI 447
G+W V +I +E +YF T D P LY A G+ + L+ + +
Sbjct: 480 SGDWDVTKILSYDEKRSQIYFLSTEDLPRRRQLYSASTV--GSFNRQC---LSCDLVDNC 534
Query: 448 VVLDHHMQSFVDIYDSLGCP----PRVLLCSLEDGRVIMPLFEQPFSVPRFKKLQLEPPE 503
D + L C P V + + D + + L Q+ E
Sbjct: 535 TYFSASFSPGADFF-LLKCEGPGVPTVSVHNTTDKKKMFDLETNEHVQKAISDRQMPKVE 593
Query: 504 IVEIQANDGTVLYGALYKPDASRFGPPPYKTMINVYGGPSVQLVSNSWLNTVDMRAQYLR 563
+I+ +D + L KP A+ Y ++ V G P Q V+ + V +
Sbjct: 594 YRKIETDDYNLPIQIL-KP-ATFTDTAHYPLLLVVDGTPGSQSVAEKF--AVTWETVMVS 649
Query: 564 NQGILVWKLDNRGTARRGLKFESYLKHKLGQVDADDQLTGAEWLVKQGLAKAGHIGLYGW 623
+ G +V K D RG+ +G + ++ +LG ++ DQ+ ++K+ + ++G
Sbjct: 650 SHGAVVVKCDGRGSGFQGTRLLHEVRRRLGSLEEKDQMEAVRVMLKEPYIDKTRVAVFGK 709
Query: 624 SYGGYLSAMTLSRYPD----FFKCAVAGAPVTSWDGYDTFYTEKYMGLPSENQSEYESGS 679
YGGYLS L D F C A +P+T + Y + ++E+Y+GL + YE
Sbjct: 710 DYGGYLSTYLLPAKGDGQAPVFSCGSALSPITDFKLYASAFSERYLGLHGLDNRAYEMAK 769
Query: 680 VMNQVHKLKGR-LLLVHGMIDENVHFRHTARLINALVAAGKPYELVLFPDERH 731
V ++V L+G+ L++H DE +HF+HTA LI L+ Y L ++PDE H
Sbjct: 770 VAHRVSALEGQQFLVIHATADEKIHFQHTAELITQLIKGKANYSLQIYPDESH 822
>ST13_YEAST (P33894) Dipeptidyl aminopeptidase A (EC 3.4.14.-) (DPAP
A) (YSCIV)
Length = 931
Score = 148 bits (374), Expect = 5e-35
Identities = 151/647 (23%), Positives = 274/647 (42%), Gaps = 50/647 (7%)
Query: 133 SQSKAELKLSSIPSSPIIDPHLSPDGSMLAYVRDHELHVLNLFTNESKQLTHGAKENGLT 192
S EL LS + + H SP + + +V ++ L + + + +K++T ++ +
Sbjct: 290 SDDNYELGLSKLSYA-----HFSPAYNYIYFVYENNLFLQQVNSGVAKKVTEDGSKD-IF 343
Query: 193 RGLAEYIAQEE-MDRKNGYWWSLDSKYIAFTEVDSSEIPLFRIMHQGKSSVGLEAQEDHP 251
++I +EE + WW+ D F + + + R+ + +
Sbjct: 344 NAKPDWIYEEEVLASDQAIWWAPDDSKAVFARFNDTSVDDIRLNRYTNMNEAYLSDTKIK 403
Query: 252 YPFAGASNVKVRLGVVSAAGSSITWMDLLCGGKEQQANEEEYLARVNWMHGNTLTAQVLN 311
YP G N + L +V+ I + GG++ + L W+ +T ++ +
Sbjct: 404 YPKPGFQNPQFDLFLVNLQNGII--YSINTGGQK-----DSILYNGKWISPDTFRFEITD 456
Query: 312 RHHTKIKILKFDIRTGQGKNILVEENST---WIN-IHDCFT-PLDKGITKFSGGFIWA-S 365
R+ + + +DI + Q + ++ WI D + P + + G+I +
Sbjct: 457 RNSKILDVKVYDIPSSQMLTVRNTNSNLFNGWIEKTKDILSIPPKPELKRMDYGYIDIHA 516
Query: 366 EKTGFRHLYLHDANGTCLGPI--TEGEWMV--EQIAGVNEATGLLYFTGTLDGPLESNLY 421
+ GF HL+ + PI T+G W V I G T ++FT G + +LY
Sbjct: 517 DSRGFSHLFYYPTV-FAKEPIQLTKGNWEVTGNGIVGYEYETDTIFFTANEIGVMSQHLY 575
Query: 422 CAKLYVDGTHPPEAPTRLTLNKGKHIVVLDHHMQSFVDIYDSLGCPP-----------RV 470
L D T + + N D + S S P RV
Sbjct: 576 SISL-TDST--TQNTFQSLQNPSDKYDFYDFELSSSARYAISKKLGPDTPIKVAGPLTRV 632
Query: 471 L-LCSLEDGRVIMPLFEQPFSVPRFKKLQLEPPEIVEIQANDGTVLYGALYKPDASRFGP 529
L + + D ++ ++ F + K L + +DG + KP A+
Sbjct: 633 LNVAEIHDDSILQLTKDEKFK-EKIKNYDLPITSYKTMVLDDGVEINYIEIKP-ANLNPK 690
Query: 530 PPYKTMINVYGGPSVQLVSNSWLNTVDMRAQYLRNQGILVWKLDNRGTARRGLKFESYLK 589
Y ++N+YGGP Q + +++ + ++V +++ RGT +G F S+ +
Sbjct: 691 KKYPILVNIYGGPGSQTFTTK--SSLAFEQAVVSGLDVIVLQIEPRGTGGKGWSFRSWAR 748
Query: 590 HKLGQVDADDQLTGAEWLVKQGLAKAGH--IGLYGWSYGGYLSAMTLSR-YPDFFKCAVA 646
KLG + D + +++ I ++GWSYGG+ S T+ D FK A+A
Sbjct: 749 EKLGYWEPRDITEVTKKFIQRNSQHIDESKIAIWGWSYGGFTSLKTVELDNGDTFKYAMA 808
Query: 647 GAPVTSWDGYDTFYTEKYMGLPSENQSEYESGSVMNQVHKLKG--RLLLVHGMIDENVHF 704
APVT+W YD+ YTE+YM PSEN Y S + + RL +VHG D+NVH
Sbjct: 809 VAPVTNWTLYDSVYTERYMNQPSENHEGYFEVSTIQNFKSFESLKRLFIVHGTFDDNVHI 868
Query: 705 RHTARLINALVAAG-KPYELVLFPDERHMPRRHSDRVYMEERMWEFI 750
++T RL++ L G Y++ +FPD H R H+ + + ++++ ++
Sbjct: 869 QNTFRLVDQLNLLGLTNYDMHIFPDSDHSIRYHNAQRIVFQKLYYWL 915
>APEH_AERPE (Q9YBQ2) Acylamino-acid-releasing enzyme (EC 3.4.19.1)
(AARE) (Acyl-peptide hydrolase) (APH)
(Acylaminoacyl-peptidase)
Length = 582
Score = 90.5 bits (223), Expect = 1e-17
Identities = 91/324 (28%), Positives = 141/324 (43%), Gaps = 37/324 (11%)
Query: 425 LYVDGTHPPEAPT----RLTLNKGKHIVVLDHHMQSFVDIYDSLGCPPRVLLCSLEDGRV 480
+++DG EAP R+ L +GK V + SL PPR++ SL G
Sbjct: 271 VFIDGERV-EAPQGNHGRVVLWRGK-----------LVTSHTSLSTPPRIV--SLPSGE- 315
Query: 481 IMPLFEQPFSVPRFKKLQLEPPEIVEIQANDGTVLYGALYKPDASRFGPPPYKTMINVYG 540
PL E +P + + +V +++ DG+ + Y ++ R P P T++ V+G
Sbjct: 316 --PLLEG--GLPEDLRRSIAGSRLVWVESFDGSRV--PTYVLESGR-APTPGPTVVLVHG 368
Query: 541 GPSVQLVSNSWLNTVDMRAQYLRNQGILVWKLDNRGTARRGLKFESYLKHKLGQVDADDQ 600
GP + S+SW D A L G V + RG+ G ++ + + +D
Sbjct: 369 GPFAE-DSDSW----DTFAASLAAAGFHVVMPNYRGSTGYGEEWRLKIIGDPCGGELEDV 423
Query: 601 LTGAEWLVKQGLAKAGHIGLYGWSYGGYLSAMTLSRYPDFFKCAVAGAPVTSWDGY---- 656
A W + GLA +I G+SYGGY++ L+ P FK VAGA V W+
Sbjct: 424 SAAARWARESGLASELYI--MGYSYGGYMTLCALTMKPGLFKAGVAGASVVDWEEMYELS 481
Query: 657 DTFYTEKYMGLPSENQSEYESGSVMNQVHKLKGRLLLVHGMIDENVHFRHTARLINALVA 716
D + L ++ S S +N V ++K L L+H D + RL+ L+A
Sbjct: 482 DAAFRNFIEQLTGGSREIMRSRSPINHVDRIKEPLALIHPQNDSRTPLKPLLRLMGELLA 541
Query: 717 AGKPYELVLFPDERHMPRRHSDRV 740
GK +E + PD H D V
Sbjct: 542 RGKTFEAHIIPDAGHAINTMEDAV 565
>YDZF_SCHPO (Q9P7E9) Putative dipeptidyl aminopeptidase C14C4.15c
(EC 3.4.14.-)
Length = 853
Score = 77.0 bits (188), Expect = 2e-13
Identities = 46/133 (34%), Positives = 71/133 (52%), Gaps = 6/133 (4%)
Query: 618 IGLYGWSYGGYLSAMTLSRYPDFFKCAVAGAPVTSWDGYDTFYTEKYMGLPSENQSE-YE 676
+G++GWS+GGYL+ L DF K APVT W YD +Y+E +G S+ + Y+
Sbjct: 713 VGIWGWSFGGYLTLKILEN-ADFIKTGAVVAPVTDWRYYDAYYSENLLGAYSKQTTAIYD 771
Query: 677 SGSV--MNQVHKLKGRLLLVHGMIDENVHFRHTARLINALVAAG-KPYELVLFPDERHMP 733
+V KL G LL++HG D+NVH +T +L A+V G Y + P+ H
Sbjct: 772 KTAVHYSENFRKLCG-LLVLHGTSDDNVHIENTMQLTKAMVEKGVYNYYPFIVPNANHEF 830
Query: 734 RRHSDRVYMEERM 746
+D ++ E++
Sbjct: 831 SDPTDYTFLREKL 843
Score = 45.1 bits (105), Expect = 7e-04
Identities = 26/106 (24%), Positives = 51/106 (47%), Gaps = 2/106 (1%)
Query: 127 IYIQDISQSKAELKLSSIPSSPIIDPHLSPDGSMLAYVRDHELHVLNLFTNESKQLTHGA 186
+Y+ + + + E L+S S I+ SP G L Y L + F+ +T +
Sbjct: 206 VYLVERATGRIE-HLASDQSKKIVVAEWSPIGHKLVYGLGSNLFIWESFSEPPVCITDQS 264
Query: 187 KENGLTRGLAEYIAQEE-MDRKNGYWWSLDSKYIAFTEVDSSEIPL 231
+GL G ++++ +EE + WWS D +++ +D S++P+
Sbjct: 265 DLDGLFNGNSDWVYEEEILQSSKAVWWSPDGNCLSYLSIDDSKVPV 310
>YUXL_BACSU (P39839) Probable peptidase yuxL (EC 3.4.21.-)
Length = 657
Score = 68.9 bits (167), Expect = 5e-11
Identities = 58/238 (24%), Positives = 108/238 (45%), Gaps = 15/238 (6%)
Query: 502 PEIVEIQANDGTVLYGALYKPDASRFGPPPYKTMINVYGGPSVQLVSNSWLNTVDMRAQY 561
PE ++ DG ++ G L +P A G Y ++N++GGP + + +T Q
Sbjct: 401 PEEIQYATEDGVMVNGWLMRP-AQMEGETTYPLILNIHGGPHMM-----YGHTYFHEFQV 454
Query: 562 LRNQGILVWKLDNRGTARRGLKFESYLKHKLGQVDADDQLTGAEWLVKQGL-AKAGHIGL 620
L +G V ++ RG+ G +F + ++ G D DD + + +K+ +G+
Sbjct: 455 LAAKGYAVVYINPRGSHGYGQEFVNAVRGDYGGKDYDDVMQAVDEAIKRDPHIDPKRLGV 514
Query: 621 YGWSYGGYLSAMTLSRYPDFFKCAVAGAPVTSWDGYDT------FYTEKYMGLPS-ENQS 673
G SYGG+++ + + + FK AV +++W + F+T+ + E+
Sbjct: 515 TGGSYGGFMTNWIVGQ-TNRFKAAVTQRSISNWISFHGVSDIGYFFTDWQLEHDMFEDTE 573
Query: 674 EYESGSVMNQVHKLKGRLLLVHGMIDENVHFRHTARLINALVAAGKPYELVLFPDERH 731
+ S + ++ LL++HG D+ +L AL GK +LV FP+ H
Sbjct: 574 KLWDRSPLKYAANVETPLLILHGERDDRCPIEQAEQLFIALKKMGKETKLVRFPNASH 631
>ACPH_HUMAN (P13798) Acylamino-acid-releasing enzyme (EC 3.4.19.1)
(AARE) (Acyl-peptide hydrolase) (APH)
(Acylaminoacyl-peptidase) (Oxidized protein hydrolase)
(OPH) (DNF15S2 protein)
Length = 732
Score = 63.9 bits (154), Expect = 1e-09
Identities = 66/252 (26%), Positives = 101/252 (39%), Gaps = 31/252 (12%)
Query: 499 LEPPEIVEIQANDGTVLYGALYKPDASRFGPPPYKTMINV----YGGPSVQLVSNSWLNT 554
L+PP E G L +P G PP KT + + +GGP V+ +W+
Sbjct: 468 LQPPPEQENVQYAGLDFEAILLQP-----GSPPDKTQVPMVVMPHGGPHSSFVT-AWMLF 521
Query: 555 VDMRAQYLRNQGILVWKLDNRGTARRGLKFESYLKHKLGQVDADDQLTGAEWLVKQGLAK 614
M L G V ++ RG+ G L +G D D E ++++
Sbjct: 522 PAM----LCKMGFAVLLVNYRGSTGFGQDSILSLPGNVGHQDVKDVQFAVEQVLQEEHFD 577
Query: 615 AGHIGLYGWSYGGYLSAMTLSRYPDFFKCAVAGAPVT-------SWDGYDTFYTEKYMGL 667
A H+ L G S+GG++S + +YP+ ++ VA PV S D D E G
Sbjct: 578 ASHVALMGGSHGGFISCHLIGQYPETYRACVARNPVINIASMLGSTDIPDWCVVE--AGF 635
Query: 668 PSENQ--------SEYESGSVMNQVHKLKGRLLLVHGMIDENVHFRHTARLINALVAAGK 719
P + +E S + + ++K LLL+ G D V F+ AL
Sbjct: 636 PFSSDCLPDLSVWAEMLDKSPIRYIPQVKTPLLLMLGQEDRRVPFKQGMEYYRALKTRNV 695
Query: 720 PYELVLFPDERH 731
P L+L+P H
Sbjct: 696 PVRLLLYPKSTH 707
>ACPH_RAT (P13676) Acylamino-acid-releasing enzyme (EC 3.4.19.1)
(AARE) (Acyl-peptide hydrolase) (APH)
(Acylaminoacyl-peptidase)
Length = 732
Score = 61.6 bits (148), Expect = 7e-09
Identities = 56/219 (25%), Positives = 90/219 (40%), Gaps = 22/219 (10%)
Query: 530 PPYKTMINV----YGGPSVQLVSNSWLNTVDMRAQYLRNQGILVWKLDNRGTARRGLKFE 585
PP KT + + +GGP V+ +W+ M L G V ++ RG+ G
Sbjct: 494 PPDKTQVPMVVMPHGGPHSSFVT-AWMLFPAM----LCKMGFAVLLVNYRGSTGFGQDSI 548
Query: 586 SYLKHKLGQVDADDQLTGAEWLVKQGLAKAGHIGLYGWSYGGYLSAMTLSRYPDFFKCAV 645
L +G D D E ++++ A + L G S+GG+LS + +YP+ + +
Sbjct: 549 LSLPGNVGHQDVKDVQFAVEQVLQEEHFDARRVALMGGSHGGFLSCHLIGQYPETYSACI 608
Query: 646 AGAPVTS------------WDGYDTFYTEKYMGLPSEN-QSEYESGSVMNQVHKLKGRLL 692
A PV + W +T + LP N E S + + ++K +L
Sbjct: 609 ARNPVINIASMMGSTDIPDWCMVETGFPYSNSCLPDLNVWEEMLDKSPIKYIPQVKTPVL 668
Query: 693 LVHGMIDENVHFRHTARLINALVAAGKPYELVLFPDERH 731
L+ G D V F+ AL A P L+L+P H
Sbjct: 669 LMLGQEDRRVPFKQGMEYYRALKARNVPVRLLLYPKSNH 707
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.137 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 98,809,379
Number of Sequences: 164201
Number of extensions: 4579906
Number of successful extensions: 9352
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 32
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 9213
Number of HSP's gapped (non-prelim): 91
length of query: 754
length of database: 59,974,054
effective HSP length: 118
effective length of query: 636
effective length of database: 40,598,336
effective search space: 25820541696
effective search space used: 25820541696
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0039.7


