

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
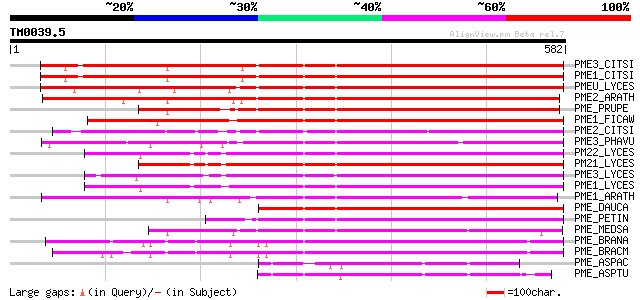
Query= TM0039.5
(582 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
PME3_CITSI (P83948) Pectinesterase 3 precursor (EC 3.1.1.11) (Pe... 462 e-129
PME1_CITSI (O04886) Pectinesterase 1 precursor (EC 3.1.1.11) (Pe... 461 e-129
PMEU_LYCES (Q43143) Pectinesterase U1 precursor (EC 3.1.1.11) (P... 454 e-127
PME2_ARATH (Q42534) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 446 e-124
PME_PRUPE (Q43062) Pectinesterase PPE8B precursor (EC 3.1.1.11) ... 443 e-124
PME1_FICAW (P83947) Pectinesterase precursor (EC 3.1.1.11) (Pect... 432 e-120
PME2_CITSI (O04887) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 426 e-118
PME3_PHAVU (Q43111) Pectinesterase 3 precursor (EC 3.1.1.11) (Pe... 418 e-116
PM22_LYCES (Q96575) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 414 e-115
PM21_LYCES (P09607) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 413 e-115
PME3_LYCES (Q96576) Pectinesterase 3 precursor (EC 3.1.1.11) (Pe... 405 e-112
PME1_LYCES (P14280) Pectinesterase 1 precursor (EC 3.1.1.11) (Pe... 403 e-112
PME1_ARATH (Q43867) Pectinesterase 1 precursor (EC 3.1.1.11) (Pe... 394 e-109
PME_DAUCA (P83218) Pectinesterase (EC 3.1.1.11) (Pectin methyles... 357 6e-98
PME_PETIN (Q43043) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 302 1e-81
PME_MEDSA (Q42920) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 290 7e-78
PME_BRANA (P41510) Probable pectinesterase precursor (EC 3.1.1.1... 265 2e-70
PME_BRACM (Q42608) Pectinesterase (EC 3.1.1.11) (Pectin methyles... 256 1e-67
PME_ASPAC (Q12535) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 107 7e-23
PME_ASPTU (P17872) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 105 2e-22
>PME3_CITSI (P83948) Pectinesterase 3 precursor (EC 3.1.1.11)
(Pectin methylesterase 3) (PE 3)
Length = 584
Score = 462 bits (1189), Expect = e-129
Identities = 252/568 (44%), Positives = 354/568 (61%), Gaps = 28/568 (4%)
Query: 33 RKKKLIILVLLAVVLIVASAVVLTA---VRSRAGGSGASTLRAKPTQAISNTCSKTRFPS 89
+KKK + L L A +L+VA+ + + A R +G +G A + ++CS TR+P
Sbjct: 25 KKKKKLFLALFATLLVVAAVIGIVAGVNSRKNSGDNGNEPHHA----ILKSSCSSTRYPD 80
Query: 90 LCVKSLLDFPGST--AASERDLVHISLNMTFQHLAKALYSSAAI--SASMDPRVRAAYDD 145
LC ++ P ++ S++D++ +SLN+T + + + ++ R + A D
Sbjct: 81 LCFSAIAAVPEASKKVTSQKDVIEMSLNITTTAVEHNYFGIQKLLKRTNLTKREKVALHD 140
Query: 146 CLELLDDSVDALGRSLSSF-------AAGSSSSDVMTWLSAALTNQDTCADGFA--DTSG 196
CLE +D+++D L +++ + + D+ T +SAA+TNQ TC DGF+ D +
Sbjct: 141 CLETIDETLDELHKAVEDLEEYPNKKSLSQHADDLKTLMSAAMTNQGTCLDGFSHDDANK 200
Query: 197 DVKDQMAGNLKDLSELVSNCLAIFAGAGD-DFSGVPIQNRRLLAMRD---DNFPRWLNRR 252
V+D ++ + ++ SN LA+ D D + N R L D +P WL+
Sbjct: 201 HVRDALSDGQVHVEKMCSNALAMIKNMTDTDMMIMRTSNNRKLIEETSTVDGWPAWLSTG 260
Query: 253 DRRLLSLPVSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEENNLKI 312
DRRLL S++ +VVV+ DG+G KT+A ++ P+ G++R+II ++AG Y EN +++
Sbjct: 261 DRRLLQS--SSVTPNVVVAADGSGNFKTVAASVAAAPQGGTKRYIIRIKAGVYREN-VEV 317
Query: 313 GRKRTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFENYAGPEK 372
+K N+M IGDG+ +T+ITG +NV+DG TTF +A+ A G GF+ARD+TF+N AGP K
Sbjct: 318 TKKHKNIMFIGDGRTRTIITGSRNVVDGS-TTFKSATVAVVGEGFLARDITFQNTAGPSK 376
Query: 373 HQAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVVFQNCS 432
HQAVALRVGAD + Y C+++ YQDT+YVHSNRQF+ C I GTVDFIFGNAA V QNC
Sbjct: 377 HQAVALRVGADLSAFYNCDMLAYQDTLYVHSNRQFFVNCLIAGTVDFIFGNAAAVLQNCD 436
Query: 433 LYARKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADLKASNESFETYLGRPWKLYS 492
++ARKP + QKN +TAQ R DPNQNTGI I R+ A +DLK SF TYLGRPWK YS
Sbjct: 437 IHARKPNSGQKNMVTAQGRADPNQNTGIVIQKSRIGATSDLKPVQGSFPTYLGRPWKEYS 496
Query: 493 RTVYMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGPGAAVGQRVNWPGYRVITSAL 552
RTV M S I D +H GW EW+ FAL+TL+YGE+ N G GA RV W G+RVITSA
Sbjct: 497 RTVIMQSSITDVIHPAGWHEWDGNFALNTLFYGEHQNAGAGAGTSGRVKWKGFRVITSAT 556
Query: 553 EASRFTVAQFILGATWLPSTGVAFLSGL 580
EA FT FI G++WL STG F GL
Sbjct: 557 EAQAFTPGSFIAGSSWLGSTGFPFSLGL 584
>PME1_CITSI (O04886) Pectinesterase 1 precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 584
Score = 461 bits (1187), Expect = e-129
Identities = 252/568 (44%), Positives = 353/568 (61%), Gaps = 28/568 (4%)
Query: 33 RKKKLIILVLLAVVLIVASAVVLTA---VRSRAGGSGASTLRAKPTQAISNTCSKTRFPS 89
+KKK + L L A +L+VA+ + + A R +G +G A + ++CS TR+P
Sbjct: 25 KKKKKLFLALFATLLVVAAVIGIVAGVNSRKNSGDNGNEPHHA----ILKSSCSSTRYPD 80
Query: 90 LCVKSLLDFPGST--AASERDLVHISLNMTFQHLAKALYSSAAI--SASMDPRVRAAYDD 145
LC ++ P ++ S++D++ +SLN+T + + + ++ R + A D
Sbjct: 81 LCFSAIAAVPEASKKVTSQKDVIEMSLNITTTAVEHNYFGIQKLLKRTNLTKREKVALHD 140
Query: 146 CLELLDDSVDALGRSLSSF-------AAGSSSSDVMTWLSAALTNQDTCADGFA--DTSG 196
CLE +D+++D L +++ + + D+ T +SAA+TNQ TC DGF+ D +
Sbjct: 141 CLETIDETLDELHKAVEDLEEYPNKKSLSQHADDLKTLMSAAMTNQGTCLDGFSHDDANK 200
Query: 197 DVKDQMAGNLKDLSELVSNCLAIFAGAGD-DFSGVPIQNRRLLAMRD---DNFPRWLNRR 252
V+D ++ + ++ SN LA+ D D + N R L D +P WL+
Sbjct: 201 HVRDALSDGQVHVEKMCSNALAMIKNMTDTDMMIMRTSNNRKLTEETSTVDGWPAWLSPG 260
Query: 253 DRRLLSLPVSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEENNLKI 312
DRRLL S++ + VV+ DG+G KT+A A+ P+ G++R+II ++AG Y EN +++
Sbjct: 261 DRRLLQS--SSVTPNAVVAADGSGNFKTVAAAVAAAPQGGTKRYIIRIKAGVYREN-VEV 317
Query: 313 GRKRTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFENYAGPEK 372
+K N+M IGDG+ +T+ITG +NV+DG TTF +A+ A G GF+ARD+TF+N AGP K
Sbjct: 318 TKKHKNIMFIGDGRTRTIITGSRNVVDGS-TTFKSATAAVVGEGFLARDITFQNTAGPSK 376
Query: 373 HQAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVVFQNCS 432
HQAVALRVGAD + Y C+++ YQDT+YVHSNRQF+ C I GTVDFIFGNAA V QNC
Sbjct: 377 HQAVALRVGADLSAFYNCDMLAYQDTLYVHSNRQFFVNCLIAGTVDFIFGNAAAVLQNCD 436
Query: 433 LYARKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADLKASNESFETYLGRPWKLYS 492
++ARKP + QKN +TAQ R DPNQNTGI I R+ A +DLK SF TYLGRPWK YS
Sbjct: 437 IHARKPNSGQKNMVTAQGRTDPNQNTGIVIQKSRIGATSDLKPVQGSFPTYLGRPWKEYS 496
Query: 493 RTVYMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGPGAAVGQRVNWPGYRVITSAL 552
RTV M S I D +H GW EW+ FAL+TL+YGE+ N G GA RV W G+RVITSA
Sbjct: 497 RTVIMQSSITDLIHPAGWHEWDGNFALNTLFYGEHQNSGAGAGTSGRVKWKGFRVITSAT 556
Query: 553 EASRFTVAQFILGATWLPSTGVAFLSGL 580
EA FT FI G++WL STG F GL
Sbjct: 557 EAQAFTPGSFIAGSSWLGSTGFPFSLGL 584
>PMEU_LYCES (Q43143) Pectinesterase U1 precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 583
Score = 454 bits (1167), Expect = e-127
Identities = 244/574 (42%), Positives = 353/574 (60%), Gaps = 33/574 (5%)
Query: 33 RKKKLIILVLLAVVLIVASAVVLTAVRSRAGGSG----ASTLRAKPTQAISNTCSKTRFP 88
RKKK+ + ++ +V+L+ A V+ V+S + S + + + + CS T P
Sbjct: 17 RKKKIYLAIVASVLLVAAVIGVVAGVKSHSKNSDDHADIMAISSSAHAIVKSACSNTLHP 76
Query: 89 SLCVKSLLDFP--GSTAASERDLVHISLNMTFQHLAKALYSSAAISAS---MDPRVRAAY 143
LC ++++ S++D++ +SLN+T + + + Y+ + + + PR + A
Sbjct: 77 ELCYSAIVNVSDFSKKVTSQKDVIELSLNITVKAVRRNYYAVKELIKTRKGLTPREKVAL 136
Query: 144 DDCLELLDDSVDALGRSLSSFAAGSSSS-------DVMTWLSAALTNQDTCADGFADTSG 196
DCLE +D+++D L ++ + D+ T +S+A+TNQ+TC DGF+
Sbjct: 137 HDCLETMDETLDELHTAVEDLELYPNKKSLKEHVEDLKTLISSAITNQETCLDGFSHDEA 196
Query: 197 D--VKDQMAGNLKDLSELVSNCLAIFAGAGD-DFSG-----VPIQNRRLLAMRDDN--FP 246
D V+ + K + ++ SN LA+ D D + P NR+L+ +DN +P
Sbjct: 197 DKKVRKVLLKGQKHVEKMCSNALAMICNMTDTDIANEMKLSAPANNRKLV---EDNGEWP 253
Query: 247 RWLNRRDRRLLSLPVSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYE 306
WL+ DRRLL S + DVVV+ DG+G KT++EA++K PE S+R++I ++AG Y
Sbjct: 254 EWLSAGDRRLLQS--STVTPDVVVAADGSGDYKTVSEAVRKAPEKSSKRYVIRIKAGVYR 311
Query: 307 ENNLKIGRKRTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFEN 366
EN + + +K+TN+M +GDGK T+IT +NV DG TTFH+A+ +ARD+TF+N
Sbjct: 312 EN-VDVPKKKTNIMFMGDGKSNTIITASRNVQDGS-TTFHSATVVRVAGKVLARDITFQN 369
Query: 367 YAGPEKHQAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAV 426
AG KHQAVAL VG+D + YRC+++ YQDT+YVHSNRQF+ +C + GTVDFIFGN A
Sbjct: 370 TAGASKHQAVALCVGSDLSAFYRCDMLAYQDTLYVHSNRQFFVQCLVAGTVDFIFGNGAA 429
Query: 427 VFQNCSLYARKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADLKASNESFETYLGR 486
VFQ+C ++AR+P + QKN +TAQ R DPNQNTGI I CR+ A +DL+ +SF TYLGR
Sbjct: 430 VFQDCDIHARRPGSGQKNMVTAQGRTDPNQNTGIVIQKCRIGATSDLRPVQKSFPTYLGR 489
Query: 487 PWKLYSRTVYMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGPGAAVGQRVNWPGYR 546
PWK YSRTV M S I D + GW EWN FALDTL+YGEY N G GA RV W G++
Sbjct: 490 PWKEYSRTVIMQSSITDVIQPAGWHEWNGNFALDTLFYGEYANTGAGAPTSGRVKWKGHK 549
Query: 547 VITSALEASRFTVAQFILGATWLPSTGVAFLSGL 580
VITS+ EA +T +FI G +WL STG F GL
Sbjct: 550 VITSSTEAQAYTPGRFIAGGSWLSSTGFPFSLGL 583
>PME2_ARATH (Q42534) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase 2) (PE 2)
Length = 587
Score = 446 bits (1146), Expect = e-124
Identities = 246/570 (43%), Positives = 344/570 (60%), Gaps = 32/570 (5%)
Query: 35 KKLIILVLLAVVLIVASAVVLTAVRSRAGGSGASTLRAKPTQAI-SNTCSKTRFPSLCVK 93
KKLI+ +L++AS V + A + + T + + AI + CS T +P LC
Sbjct: 18 KKLILSSAAIALLLLASIVGIAATTTNQNKNQKITTLSSTSHAILKSVCSSTLYPELCFS 77
Query: 94 SLLDFPGSTAASERDLVHISLNMT---FQHLAKALYSSAAISASMDPRVRAAYDDCLELL 150
++ G S+++++ SLN+T +H A+ A + PR A DCLE +
Sbjct: 78 AVAATGGKELTSQKEVIEASLNLTTKAVKHNYFAVKKLIAKRKGLTPREVTALHDCLETI 137
Query: 151 DDSVDALGRSLSSF-------AAGSSSSDVMTWLSAALTNQDTCADGFA--DTSGDVKDQ 201
D+++D L ++ + + D+ T +S+A+TNQ TC DGF+ D V+
Sbjct: 138 DETLDELHVAVEDLHQYPKQKSLRKHADDLKTLISSAITNQGTCLDGFSYDDADRKVRKA 197
Query: 202 MAGNLKDLSELVSNCLAIFAGAGD-DFSGVPIQ----------NRRLLAMR----DDNFP 246
+ + + SN LA+ + D + ++ NR+L + D +P
Sbjct: 198 LLKGQVHVEHMCSNALAMIKNMTETDIANFELRDKSSTFTNNNNRKLKEVTGDLDSDGWP 257
Query: 247 RWLNRRDRRLLSLPVSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYE 306
+WL+ DRRLL S ++AD V+ DG+G T+A A+ PE ++RF+I+++AG Y
Sbjct: 258 KWLSVGDRRLLQ--GSTIKADATVADDGSGDFTTVAAAVAAAPEKSNKRFVIHIKAGVYR 315
Query: 307 ENNLKIGRKRTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFEN 366
EN +++ +K+TN+M +GDG+GKT+ITG +NV+DG TTFH+A+ AA G F+ARD+TF+N
Sbjct: 316 EN-VEVTKKKTNIMFLGDGRGKTIITGSRNVVDGS-TTFHSATVAAVGERFLARDITFQN 373
Query: 367 YAGPEKHQAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAV 426
AGP KHQAVALRVG+D + Y+C++ YQDT+YVHSNRQF+ +C I GTVDFIFGNAA
Sbjct: 374 TAGPSKHQAVALRVGSDFSAFYQCDMFAYQDTLYVHSNRQFFVKCHITGTVDFIFGNAAA 433
Query: 427 VFQNCSLYARKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADLKASNESFETYLGR 486
V Q+C + AR+P + QKN +TAQ R DPNQNTGI I CR+ +DL A +F TYLGR
Sbjct: 434 VLQDCDINARRPNSGQKNMVTAQGRSDPNQNTGIVIQNCRIGGTSDLLAVKGTFPTYLGR 493
Query: 487 PWKLYSRTVYMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGPGAAVGQRVNWPGYR 546
PWK YSRTV M S I D + GW EW+ +FALDTL Y EY+N G GA RV W GY+
Sbjct: 494 PWKEYSRTVIMQSDISDVIRPEGWHEWSGSFALDTLTYREYLNRGGGAGTANRVKWKGYK 553
Query: 547 VITSALEASRFTVAQFILGATWLPSTGVAF 576
VITS EA FT QFI G WL STG F
Sbjct: 554 VITSDTEAQPFTAGQFIGGGGWLASTGFPF 583
>PME_PRUPE (Q43062) Pectinesterase PPE8B precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 522
Score = 443 bits (1139), Expect = e-124
Identities = 226/450 (50%), Positives = 302/450 (66%), Gaps = 25/450 (5%)
Query: 136 DPRVRAAYDDCLELLDDSVDALGRSLSSF--------AAGSSSSDVMTWLSAALTNQDTC 187
D R+ A DCL+LLD S D L SLS+ + G SSD+ TWLSAAL NQDTC
Sbjct: 83 DFRLANAISDCLDLLDFSADELNWSLSASQNQKGKNNSTGKLSSDLRTWLSAALVNQDTC 142
Query: 188 ADGFADTSGDVKDQMAGNLKDLSELVSNCLAIFAGAGDDFSGVPIQNRRLLAMRDDNFPR 247
++GF T+ V+ ++ L ++ LV L P N++ + P
Sbjct: 143 SNGFEGTNSIVQGLISAGLGQVTSLVQELLTQVH---------PNSNQQ---GPNGQIPS 190
Query: 248 WLNRRDRRLLSLPVSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEE 307
W+ +DR+LL + D +V++DG G + +A+ P+Y RR++IY++ G Y+E
Sbjct: 191 WVKTKDRKLLQ--ADGVSVDAIVAQDGTGNFTNVTDAVLAAPDYSMRRYVIYIKRGTYKE 248
Query: 308 NNLKIGRKRTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFENY 367
N ++I +K+ N+M+IGDG T+I+G ++ +DG TTF +A+FA SG GFIARD+TFEN
Sbjct: 249 N-VEIKKKKWNLMMIGDGMDATIISGNRSFVDGW-TTFRSATFAVSGRGFIARDITFENT 306
Query: 368 AGPEKHQAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVV 427
AGPEKHQAVALR +D +V YRCN+ GYQDT+Y H+ RQFYR+C I GTVDFIFG+A VV
Sbjct: 307 AGPEKHQAVALRSDSDLSVFYRCNIRGYQDTLYTHTMRQFYRDCKISGTVDFIFGDATVV 366
Query: 428 FQNCSLYARKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADLKASN-ESFETYLGR 486
FQNC + A+K + QKN+ITAQ RKDPN+ TGISI C + A +DL+A++ S TYLGR
Sbjct: 367 FQNCQILAKKGLPNQKNSITAQGRKDPNEPTGISIQFCNITADSDLEAASVNSTPTYLGR 426
Query: 487 PWKLYSRTVYMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGPGAAVGQRVNWPGYR 546
PWKLYSRTV M S++ + + GWLEWN FAL++L+YGEYMNYGPGA +G RV WPGY+
Sbjct: 427 PWKLYSRTVIMQSFLSNVIRPEGWLEWNGDFALNSLFYGEYMNYGPGAGLGSRVKWPGYQ 486
Query: 547 VITSALEASRFTVAQFILGATWLPSTGVAF 576
V + +A +TVAQFI G WLPSTGV +
Sbjct: 487 VFNESTQAKNYTVAQFIEGNLWLPSTGVKY 516
>PME1_FICAW (P83947) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 545
Score = 432 bits (1110), Expect = e-120
Identities = 236/505 (46%), Positives = 312/505 (61%), Gaps = 15/505 (2%)
Query: 82 CSKTRFPSLCVKSLLDFPGSTAASERDLVHISLNMTFQHLAKALYSSAAISASMD-PRVR 140
C ++ C+ + + G A R+L+ L T + KA ++ S ++ P+ R
Sbjct: 48 CDQSVNKESCLAMISEVTGLNMADHRNLLKSFLEKTTPRIQKAFETANDASRRINNPQER 107
Query: 141 AAYDDCLELLDDS----VDALGRSLSSFAAGSSSSDVMTWLSAALTNQDTCADGFADTSG 196
A DC EL+D S VD++ S D+ WLS LTN TC DG + S
Sbjct: 108 TALLDCAELMDLSKERVVDSISILFHQNLTTRSHEDLHVWLSGVLTNHVTCLDGLEEGST 167
Query: 197 D-VKDQMAGNLKDLSELVSNCLAIFAGAGDDFSGVPIQNRRLLAMRDDNFPRWLNRRDRR 255
D +K M +L +L LAIF S V + NFP W+ DRR
Sbjct: 168 DYIKTLMESHLNELILRARTSLAIFVTLFPAKSNV-------IEPVTGNFPTWVTAGDRR 220
Query: 256 LLSLPVSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEENNLKIGRK 315
LL ++ D+VV+KDG+G +T+ EA+ +P+ +R I+ VR G YEEN + G +
Sbjct: 221 LLQTLGKDIEPDIVVAKDGSGDYETLNEAVAAIPDNSKKRVIVLVRTGIYEEN-VDFGYQ 279
Query: 316 RTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFENYAGPEKHQA 375
+ NVM++G+G T+ITG +NV+DG TTF +A+ AA G GFIA+D+ F+N AGPEK+QA
Sbjct: 280 KKNVMLVGEGMDYTIITGSRNVVDGS-TTFDSATVAAVGDGFIAQDICFQNTAGPEKYQA 338
Query: 376 VALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVVFQNCSLYA 435
VALR+GAD V+ RC + YQDT+Y H+ RQFYR+ +I GTVDFIFGNAAVVFQNC+L
Sbjct: 339 VALRIGADETVINRCRIDAYQDTLYPHNYRQFYRDRNITGTVDFIFGNAAVVFQNCNLIP 398
Query: 436 RKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADLKASNESFETYLGRPWKLYSRTV 495
RK M Q+NTITAQ R DPNQNTG SI C + A+ADL+ ++F++YLGRPWK YSRTV
Sbjct: 399 RKQMKGQENTITAQGRTDPNQNTGTSIQNCEIFASADLEPVEDTFKSYLGRPWKEYSRTV 458
Query: 496 YMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGPGAAVGQRVNWPGYRVITSALEAS 555
M SYI D + GWLEW+ FAL TL+YGEY N GPG+ +RV WPGY VITS A
Sbjct: 459 VMESYISDVIDPAGWLEWDRDFALKTLFYGEYRNGGPGSGTSERVKWPGYHVITSPEVAE 518
Query: 556 RFTVAQFILGATWLPSTGVAFLSGL 580
+FTVA+ I G +WL STGV + +GL
Sbjct: 519 QFTVAELIQGGSWLGSTGVDYTAGL 543
>PME2_CITSI (O04887) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 510
Score = 426 bits (1094), Expect = e-118
Identities = 241/538 (44%), Positives = 326/538 (59%), Gaps = 35/538 (6%)
Query: 46 VLIVASAVVLTAVRSRAGGSGASTLRAKPTQAISNTCSKTRFPSLCVKSLLDFPGSTAAS 105
+LI S V+ + + G S + + + C KT P C L T+
Sbjct: 5 ILITVSLVLFSLSHTSFGYS---------PEEVKSWCGKTPNPQPCEYFLTQKTDVTSIK 55
Query: 106 E-RDLVHISLNMTFQHLAKALYSSAAI-SASMDPRVRAAYDDCLELLDDSVDALGRSLSS 163
+ D ISL + + A + + S + R +AA++DC EL + +V L ++ +S
Sbjct: 56 QDTDFYKISLQLALERATTAQSRTYTLGSKCRNEREKAAWEDCRELYELTVLKLNQTSNS 115
Query: 164 FAAGSSSSDVMTWLSAALTNQDTCADGFADTSGDVKDQMAGNLKD-LSELVSNCLAIFAG 222
+ G + D TWLS ALTN +TC D V + + L + +++L+SN L++
Sbjct: 116 -SPGCTKVDKQTWLSTALTNLETCRASLEDLG--VPEYVLPLLSNNVTKLISNTLSL--- 169
Query: 223 AGDDFSGVPIQNRRLLAMRDDNFPRWLNRRDRRLLSLPVSAMQADVVVSKDGNGTAKTIA 282
+ VP D FP W+ DR+LL + +A++VV++DG+G KTI
Sbjct: 170 -----NKVPYNEPSY----KDGFPTWVKPGDRKLLQ---TTPRANIVVAQDGSGNVKTIQ 217
Query: 283 EAIKKVPEYGSRRFIIYVRAGRYEENNLKIGRKRTNVMIIGDGKGKTVITGGKNVMDGLL 342
EA+ G R++IY++AG Y EN I K N+M +GDG GKT+ITG K+V G
Sbjct: 218 EAVAAASRAGGSRYVIYIKAGTYNEN---IEVKLKNIMFVGDGIGKTIITGSKSVGGGA- 273
Query: 343 TTFHTASFAASGAGFIARDMTFENYAGPEKHQAVALRVGADHAVVYRCNVIGYQDTMYVH 402
TTF +A+ A G FIARD+T N AGP HQAVALR G+D +V YRC+ GYQDT+YVH
Sbjct: 274 TTFKSATVAVVGDNFIARDITIRNTAGPNNHQAVALRSGSDLSVFYRCSFEGYQDTLYVH 333
Query: 403 SNRQFYRECDIYGTVDFIFGNAAVVFQNCSLYARKPMAQQKNTITAQNRKDPNQNTGISI 462
S RQFYRECDIYGTVDFIFGNAAVV QNC+++ARKP + NT+TAQ R DPNQ+TGI I
Sbjct: 334 SQRQFYRECDIYGTVDFIFGNAAVVLQNCNIFARKP-PNRTNTLTAQGRTDPNQSTGIII 392
Query: 463 HACRVLAAADLKASNESFETYLGRPWKLYSRTVYMMSYIGDHVHQRGWLEWNTTFALDTL 522
H CRV AA+DLK S +T+LGRPWK YSRTVY+ +++ ++ GW+EW+ FAL+TL
Sbjct: 393 HNCRVTAASDLKPVQSSVKTFLGRPWKQYSRTVYIKTFLDSLINPAGWMEWSGDFALNTL 452
Query: 523 YYGEYMNYGPGAAVGQRVNWPGYRVITSALEASRFTVAQFILGATWLPSTGVAFLSGL 580
YY EYMN GPG++ RV W GY V+TS + S+FTV FI G +WLP+T V F SGL
Sbjct: 453 YYAEYMNTGPGSSTANRVKWRGYHVLTSPSQVSQFTVGNFIAGNSWLPATNVPFTSGL 510
>PME3_PHAVU (Q43111) Pectinesterase 3 precursor (EC 3.1.1.11)
(Pectin methylesterase 3) (PE 3)
Length = 581
Score = 418 bits (1075), Expect = e-116
Identities = 246/569 (43%), Positives = 325/569 (56%), Gaps = 35/569 (6%)
Query: 34 KKKLIIL-----VLLAVVLIVASAVVLTAVRSRAGGSGASTLRAK--PTQAISNTCSKTR 86
+K+LII+ VL+AV++ + VV+ S + S S + + P ++ C TR
Sbjct: 26 RKRLIIIAVSSIVLIAVIIAAVAGVVIHNRNSESSPSSDSVPQTELSPAASLKAVCDTTR 85
Query: 87 FPSLCVKSLLDFPGSTAASERDLVHISLNMTFQHLAKALYSSAAISASMDPRVRAAYDDC 146
+PS C S+ P S L +SL + L+ + S +A D R++ A D C
Sbjct: 86 YPSSCFSSISSLPESNTTDPELLFKLSLRVAIDELS-SFPSKLRANAEQDARLQKAIDVC 144
Query: 147 -------LELLDDSVDALGRSLSSFAAGSSSSDVMTWLSAALTNQDTCADGFADTSGDVK 199
L+ L+DS+ ALG A+ +S S+V TWLSAALT+QDTC D + +
Sbjct: 145 SSVFGDALDRLNDSISALGTVAGRIASSASVSNVETWLSAALTDQDTCLDAVGELNSTAA 204
Query: 200 ----DQMAGNLKDLSELVSNCLAIFA---GAGDDFSGVPIQNRRLLAMRDDNFPRWLNRR 252
++ +++ +E SN LAI G F PI +RRLL FP WL
Sbjct: 205 RGALQEIETAMRNSTEFASNSLAIVTKILGLLSRFE-TPIHHRRLLG-----FPEWLGAA 258
Query: 253 DRRLLSLPVSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEENNLKI 312
+RRLL + D VV+KDG+G KTI EA+K V + RF +YV+ GRY EN + +
Sbjct: 259 ERRLLEEKNNDSTPDAVVAKDGSGQFKTIGEALKLVKKKSEERFSVYVKEGRYVEN-IDL 317
Query: 313 GRKRTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFENYAGPEK 372
+ NVMI GDGK KT + G +N MDG TF TA+FA G GFIA+D+ F N AG K
Sbjct: 318 DKNTWNVMIYGDGKDKTFVVGSRNFMDGT-PTFETATFAVKGKGFIAKDIGFVNNAGASK 376
Query: 373 HQAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVVFQNCS 432
HQAVALR G+D +V +RC+ G+QDT+Y HSNRQFYR+CDI GT+DFIFGNAAVVFQ+C
Sbjct: 377 HQAVALRSGSDRSVFFRCSFDGFQDTLYAHSNRQFYRDCDITGTIDFIFGNAAVVFQSCK 436
Query: 433 LYARKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADLKASNESFETYLGRPWKLYS 492
+ R+P+ Q NTITAQ +KDPNQNTGI I + +N + TYLGRPWK +S
Sbjct: 437 IMPRQPLPNQFNTITAQGKKDPNQNTGIIIQKSTITPF----GNNLTAPTYLGRPWKDFS 492
Query: 493 RTVYMMSYIGDHVHQRGWLEW-NTTFALDTLYYGEYMNYGPGAAVGQRVNWPGYRVITSA 551
TV M S IG ++ GW+ W T++Y EY N GPGA V QRV W GY+ +
Sbjct: 493 TTVIMQSDIGALLNPVGWMSWVPNVEPPTTIFYAEYQNSGPGADVSQRVKWAGYKPTITD 552
Query: 552 LEASRFTVAQFILGATWLPSTGVAFLSGL 580
A FTV FI G WLP+ V F S L
Sbjct: 553 RNAEEFTVQSFIQGPEWLPNAAVQFDSTL 581
>PM22_LYCES (Q96575) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase 2) (PE 2)
Length = 550
Score = 414 bits (1065), Expect = e-115
Identities = 225/509 (44%), Positives = 305/509 (59%), Gaps = 18/509 (3%)
Query: 79 SNTCSKTRFPSLCVKSLLDFPGST-AASERDLVHISLNMTFQHLAKALYSSAAISASM-- 135
SN C + LC+ + D + ++ D + I + ++ + + +S
Sbjct: 51 SNLCKTAQDSQLCLSYVSDLMSNEIVTTDSDGLSILMKFLVNYVHQMNNAIPVVSKMKNQ 110
Query: 136 --DPRVRAAYDDCLELLDDSVDALGRSLSSF--AAGSSSSDVMTWLSAALTNQDTCADGF 191
D R A DCLELLD SVD + S+++ S ++ +WLS LTN TC D
Sbjct: 111 INDIRQEGALTDCLELLDQSVDLVSDSIAAIDKRTHSEHANAQSWLSGVLTNHVTCLD-- 168
Query: 192 ADTSGDVKDQMAGNLKDLSELVSNCLAIFAGAGDDFSGVPIQNRRLLAMRDDNFPRWLNR 251
+ K + G +L EL+S A + V N +L P W++
Sbjct: 169 -ELDSFTKAMINGT--NLDELISRAKVALAM----LASVTTPNDDVLRPGLGKMPSWVSS 221
Query: 252 RDRRLLSLPVSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEENNLK 311
RDR+L+ + A+ VV+KDG G +T+AEA+ P+ R++IYV+ G Y+EN ++
Sbjct: 222 RDRKLMESSGKDIGANAVVAKDGTGKYRTLAEAVAAAPDKSKTRYVIYVKRGIYKEN-VE 280
Query: 312 IGRKRTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFENYAGPE 371
+ ++ +MI+GDG T+ITG NV+DG TTFH+A+ AA G GFI +D+ +N AGP
Sbjct: 281 VSSRKMKLMIVGDGMHATIITGNLNVVDGS-TTFHSATLAAVGKGFILQDICIQNTAGPA 339
Query: 372 KHQAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVVFQNC 431
KHQAVALRVGAD +V+ RC + YQDT+Y HS RQFYR+ + GT+DFIFGNAAVVFQ C
Sbjct: 340 KHQAVALRVGADKSVINRCRIDAYQDTLYAHSQRQFYRDSYVTGTIDFIFGNAAVVFQKC 399
Query: 432 SLYARKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADLKASNESFETYLGRPWKLY 491
L ARKP Q+N +TAQ R DPNQ TG SI C ++A++DL+ + F TYLGRPWK Y
Sbjct: 400 KLVARKPGKYQQNMVTAQGRTDPNQATGTSIQFCNIIASSDLEPVLKEFPTYLGRPWKKY 459
Query: 492 SRTVYMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGPGAAVGQRVNWPGYRVITSA 551
SRTV M SY+G ++ GW EW+ FAL TLYYGE+MN GPGA +RV WPGY IT
Sbjct: 460 SRTVVMESYLGGLINPAGWAEWDGDFALKTLYYGEFMNNGPGAGTSKRVKWPGYHCITDP 519
Query: 552 LEASRFTVAQFILGATWLPSTGVAFLSGL 580
EA FTVA+ I G +WL STGVA++ GL
Sbjct: 520 AEAMPFTVAKLIQGGSWLRSTGVAYVDGL 548
>PM21_LYCES (P09607) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase 2) (PE 2)
Length = 550
Score = 413 bits (1062), Expect = e-115
Identities = 220/447 (49%), Positives = 284/447 (63%), Gaps = 13/447 (2%)
Query: 136 DPRVRAAYDDCLELLDDSVDALGRSLSSFAAGSSS--SDVMTWLSAALTNQDTCADGFAD 193
D R + A DCLELLD SVD + S+++ S S ++ +WLS LTN TC D +
Sbjct: 113 DIREQGALTDCLELLDLSVDLVCDSIAAIDKRSRSEHANAQSWLSGVLTNHVTCLD---E 169
Query: 194 TSGDVKDQMAGNLKDLSELVSNCLAIFAGAGDDFSGVPIQNRRLLAMRDDNFPRWLNRRD 253
K + G +L EL+S A + V N +L P W++ RD
Sbjct: 170 LDSFTKAMINGT--NLDELISRAKVALAM----LASVTTPNDEVLRPGLGKMPSWVSSRD 223
Query: 254 RRLLSLPVSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEENNLKIG 313
R+L+ + A+ VV+KDG G +T+AEA+ P+ R++IYV+ G Y+EN +++
Sbjct: 224 RKLMESSGKDIGANAVVAKDGTGKYRTLAEAVAAAPDKSKTRYVIYVKRGTYKEN-VEVS 282
Query: 314 RKRTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFENYAGPEKH 373
++ N+MIIGDG T+ITG NV+DG TTFH+A+ AA G GFI +D+ +N AGP KH
Sbjct: 283 SRKMNLMIIGDGMYATIITGSLNVVDGS-TTFHSATLAAVGKGFILQDICIQNTAGPAKH 341
Query: 374 QAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVVFQNCSL 433
QAVALRVGAD +V+ RC + YQDT+Y HS RQFYR+ + GT+DFIFGNAAVVFQ C L
Sbjct: 342 QAVALRVGADKSVINRCRIDAYQDTLYAHSQRQFYRDSYVTGTIDFIFGNAAVVFQKCQL 401
Query: 434 YARKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADLKASNESFETYLGRPWKLYSR 493
ARKP Q+N +TAQ R DPNQ TG SI C ++A+ DLK + F TYLGRPWK YSR
Sbjct: 402 VARKPGKYQQNMVTAQGRTDPNQATGTSIQFCDIIASPDLKPVVKEFPTYLGRPWKKYSR 461
Query: 494 TVYMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGPGAAVGQRVNWPGYRVITSALE 553
TV M SY+G + GW EW+ FAL TLYYGE+MN GPGA +RV WPGY VIT E
Sbjct: 462 TVVMESYLGGLIDPSGWAEWHGDFALKTLYYGEFMNNGPGAGTSKRVKWPGYHVITDPAE 521
Query: 554 ASRFTVAQFILGATWLPSTGVAFLSGL 580
A FTVA+ I G +WL ST VA++ GL
Sbjct: 522 AMSFTVAKLIQGGSWLRSTDVAYVDGL 548
>PME3_LYCES (Q96576) Pectinesterase 3 precursor (EC 3.1.1.11)
(Pectin methylesterase 3) (PE 3)
Length = 544
Score = 405 bits (1041), Expect = e-112
Identities = 222/508 (43%), Positives = 300/508 (58%), Gaps = 22/508 (4%)
Query: 79 SNTCSKTRFPSLCVKSLLDFPGSTAASERDLVHISLNMTFQHLAKALYSSAAI----SAS 134
SN C + LC L + +E D V + +++ + + + +
Sbjct: 51 SNLCKTAQDSQLC----LSYVSEIVTTESDGVTVLKKFLVKYVHQMNNAIPVVRKIKNQI 106
Query: 135 MDPRVRAAYDDCLELLDDSVDALGRSLSSFAAGSSS--SDVMTWLSAALTNQDTCADGFA 192
D R + A DCLELLD SVD + S+++ S S ++ +WLS LTN TC D
Sbjct: 107 NDIRQQGALTDCLELLDQSVDLVSDSIAAIDKRSRSEHANAQSWLSGVLTNHVTCLDELT 166
Query: 193 DTSGDVKDQMAGNLKDLSELVSNCLAIFAGAGDDFSGVPIQNRRLLAMRDDNFPRWLNRR 252
S K+ L EL++ A + V N +L P W++ R
Sbjct: 167 SFSLSTKNGTV-----LDELITRAKVALAM----LASVTTPNDEVLRQGLGKMPYWVSSR 217
Query: 253 DRRLLSLPVSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEENNLKI 312
DR+L+ + A+ VV++DG G +T+AEA+ P+ R++IYV+ G Y+EN + +
Sbjct: 218 DRKLMESSGKDIIANRVVAQDGTGDYQTLAEAVAAAPDKNKTRYVIYVKMGIYKEN-VVV 276
Query: 313 GRKRTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFENYAGPEK 372
+K+ N+MI+GDG T+ITG NV+DG +TF + + AA G GFI +D+ +N AGPEK
Sbjct: 277 TKKKMNLMIVGDGMNATIITGSLNVVDG--STFPSNTLAAVGQGFILQDICIQNTAGPEK 334
Query: 373 HQAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVVFQNCS 432
QAVALRVGAD +V+ RC + YQDT+Y HS RQFYR+ + GTVDFIFGNAAVVFQ C
Sbjct: 335 DQAVALRVGADMSVINRCRIDAYQDTLYAHSQRQFYRDSYVTGTVDFIFGNAAVVFQKCQ 394
Query: 433 LYARKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADLKASNESFETYLGRPWKLYS 492
+ ARKP +QKN +TAQ R DPNQ TG SI C ++A+ DL+ ++TYLGRPWK +S
Sbjct: 395 IVARKPNKRQKNMVTAQGRTDPNQATGTSIQFCDIIASPDLEPVMNEYKTYLGRPWKKHS 454
Query: 493 RTVYMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGPGAAVGQRVNWPGYRVITSAL 552
RTV M SY+ H+ GW EW FAL TLYYGE+MN GPGA +RV WPGY VIT
Sbjct: 455 RTVVMQSYLDGHIDPSGWFEWRGDFALKTLYYGEFMNNGPGAGTSKRVKWPGYHVITDPN 514
Query: 553 EASRFTVAQFILGATWLPSTGVAFLSGL 580
EA FTVA+ I G +WL ST VA++ GL
Sbjct: 515 EAMPFTVAELIQGGSWLNSTSVAYVEGL 542
>PME1_LYCES (P14280) Pectinesterase 1 precursor (EC 3.1.1.11)
(Pectin methylesterase 1) (PE 1)
Length = 546
Score = 403 bits (1036), Expect = e-112
Identities = 223/510 (43%), Positives = 300/510 (58%), Gaps = 20/510 (3%)
Query: 79 SNTCSKTRFPSLCVKSLLDFPGSTAASERDLVHISLNMTFQHLAKALYSSAAISASM--- 135
SN C + LC+ + D + + H L + + ++ + M
Sbjct: 47 SNLCKTAQDSQLCLSYVSDLISNEIVTTESDGHSILMKFLVNYVHQMNNAIPVVRKMKNQ 106
Query: 136 --DPRVRAAYDDCLELLDDSVDALGRSLSSFAAGSSS--SDVMTWLSAALTNQDTCADGF 191
D R A DCLELLD SVD S+++ S S ++ +WLS LTN TC D
Sbjct: 107 INDIRQHGALTDCLELLDQSVDFASDSIAAIDKRSRSEHANAQSWLSGVLTNHVTCLD-- 164
Query: 192 ADTSGDVKDQMAG-NLKDLSELVSNCLAIFAGAGDDFSGVPIQNRRLLAMRDDNFPRWLN 250
+ K + G NL++L LA+ A + Q+ + P W++
Sbjct: 165 -ELDSFTKAMINGTNLEELISRAKVALAMLAS-------LTTQDEDVFMTVLGKMPSWVS 216
Query: 251 RRDRRLLSLPVSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEENNL 310
DR+L+ + A+ VV++DG G +T+AEA+ P+ R++IYV+ G Y+EN +
Sbjct: 217 SMDRKLMESSGKDIIANAVVAQDGTGDYQTLAEAVAAAPDKSKTRYVIYVKRGTYKEN-V 275
Query: 311 KIGRKRTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFENYAGP 370
++ + N+MI+GDG T ITG NV+DG TTF +A+ AA G GFI +D+ +N AGP
Sbjct: 276 EVASNKMNLMIVGDGMYATTITGSLNVVDGS-TTFRSATLAAVGQGFILQDICIQNTAGP 334
Query: 371 EKHQAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVVFQN 430
K QAVALRVGAD +V+ RC + YQDT+Y HS RQFYR+ + GTVDFIFGNAAVVFQ
Sbjct: 335 AKDQAVALRVGADMSVINRCRIDAYQDTLYAHSQRQFYRDSYVTGTVDFIFGNAAVVFQK 394
Query: 431 CSLYARKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADLKASNESFETYLGRPWKL 490
C L ARKP Q+N +TAQ R DPNQ TG SI C ++A++DL+ + F TYLGRPWK
Sbjct: 395 CQLVARKPGKYQQNMVTAQGRTDPNQATGTSIQFCNIIASSDLEPVLKEFPTYLGRPWKE 454
Query: 491 YSRTVYMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGPGAAVGQRVNWPGYRVITS 550
YSRTV M SY+G ++ GW EW+ FAL TLYYGE+MN GPGA +RV WPGY VIT
Sbjct: 455 YSRTVVMESYLGGLINPAGWAEWDGDFALKTLYYGEFMNNGPGAGTSKRVKWPGYHVITD 514
Query: 551 ALEASRFTVAQFILGATWLPSTGVAFLSGL 580
+A FTVA+ I G +WL STGVA++ GL
Sbjct: 515 PAKAMPFTVAKLIQGGSWLRSTGVAYVDGL 544
>PME1_ARATH (Q43867) Pectinesterase 1 precursor (EC 3.1.1.11)
(Pectin methylesterase 1) (PE 1)
Length = 586
Score = 394 bits (1012), Expect = e-109
Identities = 229/568 (40%), Positives = 326/568 (57%), Gaps = 39/568 (6%)
Query: 34 KKKLIILVLLAVVLI--VASAVVLTAVRSRAGGSGASTL-RAKPTQAISNTCSKTRFPSL 90
+K+L++L + VVLI + +AVV T V S S P+ ++ CS TRFP
Sbjct: 26 RKRLLLLSISVVVLIAVIIAAVVATVVHKNKNESTPSPPPELTPSTSLKAICSVTRFPES 85
Query: 91 CVKSLLDFPGSTAASERDLVHISLNMTFQHLAKALYSSAAISASM-DPRVRAAYDDCLEL 149
C+ S+ P S L +SL + L +S D R+++A C +L
Sbjct: 86 CISSISKLPSSNTTDPETLFKLSLKVIIDELDSISDLPEKLSKETEDERIKSALRVCGDL 145
Query: 150 LDDSVDALGRSLSSF--------AAGSSSSDVMTWLSAALTNQDTCADGFADTSGD---- 197
++D++D L ++S+ + S D+ TWLSA +T+ +TC D + +
Sbjct: 146 IEDALDRLNDTVSAIDDEEKKKTLSSSKIEDLKTWLSATVTDHETCFDSLDELKQNKTEY 205
Query: 198 VKDQMAGNLKDL----SELVSNCLAIFAGAGDDFS--GVPIQNRRLLA----MRDDNFPR 247
+ NLK +E SN LAI + S G+PI RR L + +F +
Sbjct: 206 ANSTITQNLKSAMSRSTEFTSNSLAIVSKILSALSDLGIPIHRRRRLMSHHHQQSVDFEK 265
Query: 248 WLNRRDRRLLSLPVSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEE 307
W RR L + ++ DV V+ DG G T+ EA+ KVP+ + F+IYV++G Y E
Sbjct: 266 WARRR-----LLQTAGLKPDVTVAGDGTGDVLTVNEAVAKVPKKSLKMFVIYVKSGTYVE 320
Query: 308 NNLKIGRKRTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFENY 367
N + + + + NVMI GDGKGKT+I+G KN +DG T+ TA+FA G GFI +D+ N
Sbjct: 321 N-VVMDKSKWNVMIYGDGKGKTIISGSKNFVDGT-PTYETATFAIQGKGFIMKDIGIINT 378
Query: 368 AGPEKHQAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVV 427
AG KHQAVA R G+D +V Y+C+ G+QDT+Y HSNRQFYR+CD+ GT+DFIFG+AAVV
Sbjct: 379 AGAAKHQAVAFRSGSDFSVYYQCSFDGFQDTLYPHSNRQFYRDCDVTGTIDFIFGSAAVV 438
Query: 428 FQNCSLYARKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADLKASNESFETYLGRP 487
FQ C + R+P++ Q NTITAQ +KDPNQ++G+SI C + A ++ A TYLGRP
Sbjct: 439 FQGCKIMPRQPLSNQFNTITAQGKKDPNQSSGMSIQRCTISANGNVIA-----PTYLGRP 493
Query: 488 WKLYSRTVYMMSYIGDHVHQRGWLEW-NTTFALDTLYYGEYMNYGPGAAVGQRVNWPGYR 546
WK +S TV M + IG V GW+ W + ++ YGEY N GPG+ V QRV W GY+
Sbjct: 494 WKEFSTTVIMETVIGAVVRPSGWMSWVSGVDPPASIVYGEYKNTGPGSDVTQRVKWAGYK 553
Query: 547 VITSALEASRFTVAQFILGATWLPSTGV 574
+ S EA++FTVA + GA W+P+TGV
Sbjct: 554 PVMSDAEAAKFTVATLLHGADWIPATGV 581
>PME_DAUCA (P83218) Pectinesterase (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 319
Score = 357 bits (915), Expect = 6e-98
Identities = 175/319 (54%), Positives = 226/319 (69%), Gaps = 2/319 (0%)
Query: 262 SAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEENNLKIGRKRTNVMI 321
S + +VVV+ DG+G KT++EA+ PE R++I ++AG Y EN + + +K+ N+M
Sbjct: 3 STVTPNVVVAADGSGDYKTVSEAVAAAPEDSKTRYVIRIKAGVYREN-VDVPKKKKNIMF 61
Query: 322 IGDGKGKTVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFENYAGPEKHQAVALRVG 381
+GDG+ T+IT KNV DG TTF++A+ AA GAGF+ARD+TF+N AG KHQAVALRVG
Sbjct: 62 LGDGRTSTIITASKNVQDGS-TTFNSATVAAVGAGFLARDITFQNTAGAAKHQAVALRVG 120
Query: 382 ADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVVFQNCSLYARKPMAQ 441
+D + YRC+++ YQD++YVHSNRQF+ C I GTVDFIFGNAAVV Q+C ++AR+P +
Sbjct: 121 SDLSAFYRCDILAYQDSLYVHSNRQFFINCFIAGTVDFIFGNAAVVLQDCDIHARRPGSG 180
Query: 442 QKNTITAQNRKDPNQNTGISIHACRVLAAADLKASNESFETYLGRPWKLYSRTVYMMSYI 501
QKN +TAQ R DPNQNTGI I R+ A +DL+ SF TYLGRPWK YSRTV M S I
Sbjct: 181 QKNMVTAQGRTDPNQNTGIVIQKSRIGATSDLQPVQSSFPTYLGRPWKEYSRTVVMQSSI 240
Query: 502 GDHVHQRGWLEWNTTFALDTLYYGEYMNYGPGAAVGQRVNWPGYRVITSALEASRFTVAQ 561
+ ++ GW W+ FALDTLYYGEY N G GAA RV W G++VITS+ EA FT
Sbjct: 241 TNVINPAGWFPWDGNFALDTLYYGEYQNTGAGAATSGRVTWKGFKVITSSTEAQGFTPGS 300
Query: 562 FILGATWLPSTGVAFLSGL 580
FI G +WL +T F GL
Sbjct: 301 FIAGGSWLKATTFPFSLGL 319
>PME_PETIN (Q43043) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 374
Score = 302 bits (774), Expect = 1e-81
Identities = 164/377 (43%), Positives = 221/377 (58%), Gaps = 12/377 (3%)
Query: 206 LKDLSELVSNCLAIFAGAGDDFSGVPIQNRRLLAMRDDNFPRWLNRRDRRLLSLPVSAMQ 265
L EL N L++ GD + NR+LL + RRLL +S +
Sbjct: 5 LNSTRELSINALSMLNSFGDMVAQATGLNRKLLTTDSSDATA------RRLLQ--ISNAK 56
Query: 266 ADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEENNLKIGRKRTNVMIIGDG 325
+ V+ DG+G KTI EA+ VP+ + FII+++AG Y+E + I + TNV++IG+G
Sbjct: 57 PNATVALDGSGQYKTIKEALDAVPKKNTEPFIIFIKAGVYKEY-IDIPKSMTNVVLIGEG 115
Query: 326 KGKTVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFENYAGPEKHQAVALRVGADHA 385
KT ITG K+V DG +TFHT + +GA F+A+++ FEN AGPEK QAVALRV AD A
Sbjct: 116 PTKTKITGNKSVKDGP-STFHTTTVGVNGANFVAKNIGFENTAGPEKEQAVALRVSADKA 174
Query: 386 VVYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVVFQNCSLYARKPMAQQKNT 445
++Y C + GYQDT+YVH+ RQFYR+C I GTVDFIFGN V QNC + RKP Q
Sbjct: 175 IIYNCQIDGYQDTLYVHTYRQFYRDCTITGTVDFIFGNGEAVLQNCKVIVRKPAQNQSCM 234
Query: 446 ITAQNRKDPNQNTGISIHACRVLAAADLKASNESFETYLGRPWKLYSRTVYMMSYIGDHV 505
+TAQ R +P Q I + C + D + + +TYLGRPWK YSRT+ M SYI +
Sbjct: 235 VTAQGRTEPIQKGAIVLQNCEIKPDTDYFSLSPPSKTYLGRPWKEYSRTIIMQSYIDKFI 294
Query: 506 HQRGWLEWN-TTFALDTLYYGEYMNYGPGAAVGQRVNWPGYRVITSALEASRFTVAQFIL 564
GW WN T F DT YY EY N GPGAA+ +R+ W G++ + A +FT +I
Sbjct: 295 EPEGWAPWNITNFGRDTSYYAEYQNRGPGAALDKRITWKGFQKGFTGEAAQKFTAGVYIN 354
Query: 565 G-ATWLPSTGVAFLSGL 580
WL V + +G+
Sbjct: 355 NDENWLQKANVPYEAGM 371
>PME_MEDSA (Q42920) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE) (P65)
Length = 447
Score = 290 bits (742), Expect = 7e-78
Identities = 171/449 (38%), Positives = 244/449 (54%), Gaps = 22/449 (4%)
Query: 146 CLELLDDSVDALGRSLSSF------AAGSSSSDVMTWLSAALTNQDTCADGFADTSGDVK 199
C E+LD +VD + +S+ + + D+ WL+ L++Q TC DGFA+T+
Sbjct: 4 CNEVLDYAVDGIHKSVGTLDQFDFHKLSEYAFDLKVWLTGTLSHQQTCLDGFANTTTKAG 63
Query: 200 DQMAGNLKDLSELVSNCLAIFAGAGDDFSGVPIQ----NRRLLAMRDDNFPRWLNRRDRR 255
+ M LK EL SN + + G +RRLL+ DD P W+N RR
Sbjct: 64 ETMTKVLKTSMELSSNAIDMMDAVSRILKGFDTSQYSVSRRLLS--DDGIPSWVNDGHRR 121
Query: 256 LLSLPVSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEENNLKIGRK 315
LL+ +Q + VV++DG+G KT+ +A+K VP + F+I+V+AG Y+E + + ++
Sbjct: 122 LLA--GGNVQPNAVVAQDGSGQFKTLTDALKTVPPKNAVPFVIHVKAGVYKET-VNVAKE 178
Query: 316 RTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFENYAGPEKHQA 375
V +IGDG KT TG N DG+ T++TA+F +GA F+A+D+ FEN AG KHQA
Sbjct: 179 MNYVTVIGDGPTKTKFTGSLNYADGI-NTYNTATFGVNGANFMAKDIGFENTAGTGKHQA 237
Query: 376 VALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVVFQNCSLYA 435
VALRV AD A+ Y C + G+QDT+YV S RQFYR+C I GT+DF+FG VFQNC L
Sbjct: 238 VALRVTADQAIFYNCQMDGFQDTLYVQSQRQFYRDCSISGTIDFVFGERFGVFQNCKLVC 297
Query: 436 RKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADLKASNESFETYLGRPWKLYSRTV 495
R P Q+ +TA R+ N + + + L + +YLGRPWK YS+ V
Sbjct: 298 RLPAKGQQCLVTAGGREKQNSASALVFQSSHFTGEPALTSVTPKV-SYLGRPWKQYSKVV 356
Query: 496 YMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGPGAAVGQRVNWPGYRVITSALEAS 555
M S I G++ W + +T Y EY N GPGA RV W G +V+TS + A
Sbjct: 357 IMDSTIDAIFVPEGYMPWMGSAFKETCTYYEYNNKGPGADTNLRVKWHGVKVLTSNVAAE 416
Query: 556 R-----FTVAQFILGATWLPSTGVAFLSG 579
F + TW+ +GV + G
Sbjct: 417 YYPGKFFEIVNATARDTWIVKSGVPYSLG 445
>PME_BRANA (P41510) Probable pectinesterase precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 584
Score = 265 bits (678), Expect = 2e-70
Identities = 184/589 (31%), Positives = 282/589 (47%), Gaps = 56/589 (9%)
Query: 38 IILVLLAVVLIVASAVVLTAVRSRAGGSGASTLRAKPTQAISNTCSKTRFPSLCVKSLLD 97
I++ + +++L+V A+ + ++ GG+G +A+ + C+ C K+L
Sbjct: 6 IVISVASMLLVVGVAIGVVTFVNKGGGAGGDKTLNSHQKAVESLCASATDKGSCAKTLDP 65
Query: 98 FPGSTAASERDLVHISLNMTFQHLAKALYSSAAISASMDPRV----RAAYDDC------- 146
+ L+ + T + K+ +A+ M + +A D C
Sbjct: 66 VKSDDPSK---LIKAFMLATKDAVTKSTNFTASTEEGMGKNINATSKAVLDYCKRVLMYA 122
Query: 147 LELLDDSVDALGRSLSSFAAGSSSSDVMTWLSAALTNQDTCADGFADTSGDVKDQMAGNL 206
LE L+ V+ +G L +GS + WL+ Q C D ++ +++ M +
Sbjct: 123 LEDLETIVEEMGEDLQQ--SGSKMDQLKQWLTGVFNYQTDCIDDIEES--ELRKVMGEGI 178
Query: 207 KDLSELVSNCLAIFAGAGDDFSGV---------------PIQNRRLLAMRDDN-FPRWLN 250
L SN + IF S + P +R LL D P+W +
Sbjct: 179 AHSKILSSNAIDIFHALTTAMSQMNVKVDDMKKGNLGETPAPDRDLLEDLDQKGLPKWHS 238
Query: 251 RRDRRLLSL---PVSAMQADV--------------VVSKDGNGTAKTIAEAIKKVPEYGS 293
+DR+L++ P + + VV+KDG+G KTI+EA+K PE
Sbjct: 239 DKDRKLMAQAGRPGAPADEGIGEGGGGGGKIKPTHVVAKDGSGQFKTISEAVKACPEKNP 298
Query: 294 RRFIIYVRAGRYEENNLKIGRKRTNVMIIGDGKGKTVITGGKNV-MDGLLTTFHTASFAA 352
R IIY++AG Y+E + I +K NV + GDG +T+IT ++V + TT + +
Sbjct: 299 GRCIIYIKAGVYKEQ-VTIPKKVNNVFMFGDGATQTIITFDRSVGLSPGTTTSLSGTVQV 357
Query: 353 SGAGFIARDMTFENYAGPEKHQAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECD 412
GF+A+ + F+N AGP HQAVA RV D AV++ C GYQDT+YV++ RQFYR
Sbjct: 358 ESEGFMAKWIGFQNTAGPLGHQAVAFRVNGDRAVIFNCRFDGYQDTLYVNNGRQFYRNIV 417
Query: 413 IYGTVDFIFGNAAVVFQNCSLYARKPMAQQKNTITAQ-NRKDPNQNTGISIHACRVLAAA 471
+ GTVDFIFG +A V QN + RK Q N +TA N K GI +H CR++A
Sbjct: 418 VSGTVDFIFGKSATVIQNSLILCRKGSPGQTNHVTADGNEKGKAVKIGIVLHNCRIMADK 477
Query: 472 DLKASNESFETYLGRPWKLYSRTVYMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYG 531
+L+A + ++YLGRPWK ++ T + + IGD + GW EW T Y E+ N G
Sbjct: 478 ELEADRLTVKSYLGRPWKPFATTAVIGTEIGDLIQPTGWNEWQGEKFHLTATYVEFNNRG 537
Query: 532 PGAAVGQRVNWPGYRVITSALEASRFTVAQFILGATWLPSTGVAFLSGL 580
PGA RV W ++ SA E RFTVA ++ A W+ V GL
Sbjct: 538 PGANTAARVPWA--KMAKSAAEVERFTVANWLTPANWIQEANVPVQLGL 584
>PME_BRACM (Q42608) Pectinesterase (EC 3.1.1.11) (Pectin
methylesterase) (PE) (Fragment)
Length = 571
Score = 256 bits (653), Expect = 1e-67
Identities = 183/585 (31%), Positives = 277/585 (47%), Gaps = 64/585 (10%)
Query: 46 VLIVASAVVLTAVRSRAGGSGASTLRAKPTQAISNTCSKTRFPSLCVKSL----LDFPGS 101
+L+V A+ + ++ GG+G +A+ + C+ C K+L D P
Sbjct: 1 LLVVGVAIGVVTFVNKGGGAGGDKTLNSHQKAVESLCASATDKGSCAKTLDPVKSDDPSK 60
Query: 102 TAAS----ERDLVHISLNMTFQHLAKALYSSAAISASMDPRVRAAYDDC-------LELL 150
+ +D V S N T + + +M+ +A D C LE L
Sbjct: 61 LIKAFMLATKDAVTKSTNFTAS-------TEEGMGKNMNATSKAVLDYCKRVLMYALEDL 113
Query: 151 DDSVDALGRSLSSFAAGSSSSDVMTWLSAALTNQDTCADGFADTSGDVKDQMAGNLKDLS 210
+ V+ +G L +GS + WL+ Q C D ++ +++ M ++
Sbjct: 114 ETIVEEMGEDLQQ--SGSKMDQLKQWLTGVFNYQTDCIDDIEES--ELRKVMGEGIRHSK 169
Query: 211 ELVSNCLAIFAGAGDDFSGV---------------PIQNRRLLAMRDDN-FPRWLNRRDR 254
L SN + IF S + P +R LL D P+W + +DR
Sbjct: 170 ILSSNAIDIFHALTTAMSQMNVKVDDMKKGNLGETPAPDRDLLEDLDQKGLPKWHSDKDR 229
Query: 255 RLLSL---PVSAMQADV--------------VVSKDGNGTAKTIAEAIKKVPEYGSRRFI 297
+L++ P + + VV+KDG+G KTI+EA+K PE R I
Sbjct: 230 KLMAQAGRPGAPADEGIGEGGGGGGKIKPTHVVAKDGSGQFKTISEAVKACPEKNPGRCI 289
Query: 298 IYVRAGRYEENNLKIGRKRTNVMIIGDGKGKTVITGGKNV-MDGLLTTFHTASFAASGAG 356
IY++AG Y+E + I +K NV + GDG +T+IT ++V + TT + + G
Sbjct: 290 IYIKAGVYKEQ-VTIPKKVNNVFMFGDGATQTIITFDRSVGLSPGTTTSLSGTVQVESEG 348
Query: 357 FIARDMTFENYAGPEKHQAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGT 416
F+A+ + F+N AGP +QAVA RV D AV++ C GYQDT+YV++ RQFYR + GT
Sbjct: 349 FMAKWIGFQNTAGPLGNQAVAFRVNGDRAVIFNCRFDGYQDTLYVNNGRQFYRNIVVSGT 408
Query: 417 VDFIFGNAAVVFQNCSLYARKPMAQQKNTITAQNR-KDPNQNTGISIHACRVLAAADLKA 475
VDFI G +A V QN + RK Q N +TA + K GI +H CR++A +L+A
Sbjct: 409 VDFINGKSATVIQNSLILCRKGSPGQTNHVTADGKQKGKAVKIGIVLHNCRIMADKELEA 468
Query: 476 SNESFETYLGRPWKLYSRTVYMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGPGAA 535
+ ++YLGRPWK ++ T + + IGD + GW EW T Y E+ N GPGA
Sbjct: 469 DRLTVKSYLGRPWKPFATTAVIGTEIGDLIQPTGWNEWQGEKFHLTATYVEFNNRGPGAN 528
Query: 536 VGQRVNWPGYRVITSALEASRFTVAQFILGATWLPSTGVAFLSGL 580
RV W ++ SA E RFTVA ++ A W+ V GL
Sbjct: 529 PAARVPWA--KMAKSAAEVERFTVANWLTPANWIQEANVTVQLGL 571
>PME_ASPAC (Q12535) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 331
Score = 107 bits (268), Expect = 7e-23
Identities = 86/292 (29%), Positives = 126/292 (42%), Gaps = 35/292 (11%)
Query: 262 SAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEENNLKIGRKRTNVMI 321
+A +VV+K G G TI +AI + + I++ G Y+E +
Sbjct: 22 TAPSGAIVVAKSG-GDYTTIGDAIDALSTSTTDTQTIFIEEGTYDEQ-----------VY 69
Query: 322 IGDGKGKTVITGGK----NVMDGLLTTFH-------------TASFAASGAGFIARDMTF 364
+ GK +I G + D L+T H TA+F G ++
Sbjct: 70 LPAMTGKVIIYGQTENTDSYADNLVTITHAISYEDAGESDDLTATFRNKAVGSQVYNLNI 129
Query: 365 ENYAGPEKHQAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFG-N 423
N G HQA+AL AD Y CN GYQDT+ + Q Y I G VDFIFG +
Sbjct: 130 ANTCGQACHQALALSAWADQQGYYGCNFTGYQDTLLAQTGNQLYINSYIEGAVDFIFGQH 189
Query: 424 AAVVFQNCSLYARKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADLKASNESFETY 483
A FQN + R +ITA R + I+ V A + ++ Y
Sbjct: 190 ARAWFQNVDI--RVVEGPTSASITANGRSSETDTSYYVINKSTVAAKEGDDVAEGTY--Y 245
Query: 484 LGRPWKLYSRTVYMMSYIGDHVHQRGWLEWNT-TFALDTLYYGEYMNYGPGA 534
LGRPW Y+R V+ + + + ++ GW EW+T T + + +GEY N G G+
Sbjct: 246 LGRPWSEYARVVFQQTSMTNVINSLGWTEWSTSTPNTEYVTFGEYANTGAGS 297
>PME_ASPTU (P17872) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 331
Score = 105 bits (263), Expect = 2e-22
Identities = 87/316 (27%), Positives = 137/316 (42%), Gaps = 23/316 (7%)
Query: 261 VSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEENNLKIGRKRTNVM 320
++A +VV+K G G TI+ A+ + + I++ G Y+E + I ++
Sbjct: 21 MTAPSGAIVVAKSG-GDYDTISAAVDALSTTSTETQTIFIEEGSYDEQ-VYIPALSGKLI 78
Query: 321 IIGDGKGKTVITGGK-NVMDGLLTTF-----HTASFAASGAGFIARDMTFENYAGPEKHQ 374
+ G + T T N+ + TA+ G ++ N G HQ
Sbjct: 79 VYGQTEDTTTYTSNLVNITHAIALADVDNDDETATLRNYAEGSAIYNLNIANTCGQACHQ 138
Query: 375 AVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFG-NAAVVFQNCSL 433
A+A+ A Y C GYQDT+ + Q Y I G VDFIFG +A F C +
Sbjct: 139 ALAVSAYASEQGYYACQFTGYQDTLLAETGYQVYAGTYIEGAVDFIFGQHARAWFHECDI 198
Query: 434 YARKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADLKASNESFETYLGRPWKLYSR 493
R +ITA R + ++ IH V AA S+ ++ YLGRPW Y+R
Sbjct: 199 --RVLEGPSSASITANGRSSESDDSYYVIHKSTVAAADGNDVSSGTY--YLGRPWSQYAR 254
Query: 494 TVYMMSYIGDHVHQRGWLEWNT-TFALDTLYYGEYMNYGPGAAVGQRVNWPGYRVITSAL 552
+ + + D ++ GW EW+T T + + + EY N G GA G R N+ +
Sbjct: 255 VCFQKTSMTDVINHLGWTEWSTSTPNTENVTFVEYGNTGTGAE-GPRANF--------SS 305
Query: 553 EASRFTVAQFILGATW 568
E + ++LG+ W
Sbjct: 306 ELTEPITISWLLGSDW 321
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.133 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 65,482,027
Number of Sequences: 164201
Number of extensions: 2690929
Number of successful extensions: 6632
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 25
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 6498
Number of HSP's gapped (non-prelim): 48
length of query: 582
length of database: 59,974,054
effective HSP length: 116
effective length of query: 466
effective length of database: 40,926,738
effective search space: 19071859908
effective search space used: 19071859908
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0039.5


