

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0037a.5
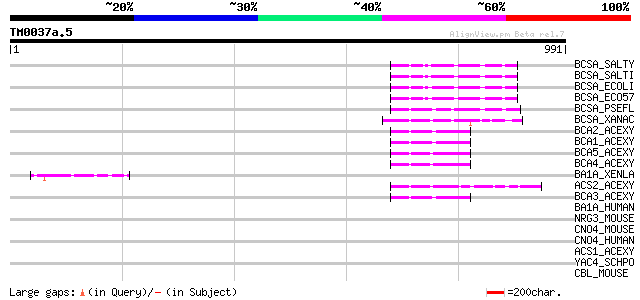
(991 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BCSA_SALTY (Q93IN2) Cellulose synthase catalytic subunit [UDP-fo... 68 1e-10
BCSA_SALTI (Q8Z291) Cellulose synthase catalytic subunit [UDP-fo... 67 2e-10
BCSA_ECOLI (P37653) Cellulose synthase catalytic subunit [UDP-fo... 66 5e-10
BCSA_ECO57 (Q8X5L7) Cellulose synthase catalytic subunit [UDP-fo... 66 5e-10
BCSA_PSEFL (P58931) Cellulose synthase catalytic subunit [UDP-fo... 55 9e-07
BCSA_XANAC (P58932) Cellulose synthase catalytic subunit [UDP-fo... 54 3e-06
BCA2_ACEXY (O82859) Cellulose synthase catalytic subunit [UDP-fo... 49 5e-05
BCA1_ACEXY (P19449) Cellulose synthase catalytic subunit [UDP-fo... 49 7e-05
BCA5_ACEXY (Q9WX75) Putative cellulose synthase 3 [Includes: Cel... 48 1e-04
BCA4_ACEXY (Q9RBJ2) Putative cellulose synthase 2 [Includes: Cel... 48 1e-04
BA1A_XENLA (Q8UVR5) ATP-utilizing chromatin assembly and remodel... 47 2e-04
ACS2_ACEXY (Q59167) Cellulose synthase 2 [Includes: Cellulose sy... 47 3e-04
BCA3_ACEXY (Q9WX61) Cellulose synthase 1 catalytic subunit [UDP-... 46 4e-04
BA1A_HUMAN (Q9NRL2) Bromodomain adjacent to zinc finger domain p... 39 0.053
NRG3_MOUSE (O35181) Pro-neuregulin-3 precursor (Pro-NRG3) [Conta... 39 0.069
CNO4_MOUSE (Q8BT14) CCR4-NOT transcription complex subunit 4 (EC... 38 0.12
CNO4_HUMAN (O95628) CCR4-NOT transcription complex subunit 4 (EC... 38 0.12
ACS1_ACEXY (P21877) Cellulose synthase 1 [Includes: Cellulose sy... 38 0.15
YAC4_SCHPO (Q09818) Putative general negative regulator of trans... 37 0.26
CBL_MOUSE (P22682) CBL E3 ubiquitin protein ligase (EC 6.3.2.-) ... 36 0.58
>BCSA_SALTY (Q93IN2) Cellulose synthase catalytic subunit
[UDP-forming] (EC 2.4.1.12)
Length = 874
Score = 68.2 bits (165), Expect = 1e-10
Identities = 63/229 (27%), Positives = 109/229 (47%), Gaps = 25/229 (10%)
Query: 681 GLSYGSIAADVLTSLKLHSRGWRSVYCMPKRAAFRGTAPINLTERLNQVLRWAVGSLEIL 740
G++ ++ D TSL+LH RG+ S Y +AA G A +L+ + Q +RWA G ++I
Sbjct: 448 GIAVETVTEDAHTSLRLHRRGYTSAYMRIPQAA--GLATESLSAHIGQRIRWARGMVQI- 504
Query: 741 FSRHCPIWYGFKEGRLKGLQRIAYINSTVYPFSSIPLLIYCLIPAICLLTDKFITPSVDT 800
F P+ F +G LK QR+ Y+N+ + S IP LI+ P LL +I
Sbjct: 505 FRLDNPL---FGKG-LKLAQRLCYLNAMFHFLSGIPRLIFLTAPLAFLLLHAYI-----I 555
Query: 801 FASMIIISLFI--SIFGSAILELRWSGVSLEEWWRSQQFWVIGSVSAHLFAVAQALMGGL 858
+A ++I+LF+ + +++ + G +W V+ + +A + L
Sbjct: 556 YAPALMIALFVIPHMVHASLTNSKIQGKYRHSFWSEIYETVLA------WYIAPPTLVAL 609
Query: 859 AKKVNKNFSVVSKAPDDEEFHELYTIRWTALLVPPTTIIIINLIGVVAG 907
F+V +K EE + + W + P ++++NL+GV AG
Sbjct: 610 INPHKGKFNVTAKGGLVEEKY----VDW-VISRPYIFLVLLNLLGVAAG 653
Score = 35.4 bits (80), Expect = 0.76
Identities = 37/137 (27%), Positives = 60/137 (43%), Gaps = 26/137 (18%)
Query: 211 GRLSPYRMMVVTRLLLLLLFIQYRIFH------PVPDAIGLWLISVVCEIWLTLSWIVDQ 264
GR S M++V L + +I +R PV GL L+ W+ L ++
Sbjct: 195 GRFSAL-MLIVLSLTVSCRYIWWRYTSTLNWDDPVSLVCGLILLFAETYAWIVL--VLGY 251
Query: 265 LPKWFPIDRETYLDRLSIRFEPENKPNMLSP---VDIFVTTVDPIKEPPLVTANTVLSIL 321
+P++R+ P P +S VDIFV T + E V NT+ + L
Sbjct: 252 FQVVWPLNRQ-----------PVPLPKEMSQWPTVDIFVPTYN---EDLNVVKNTIYASL 297
Query: 322 ALDYPAHKISCYVSDDG 338
+D+P K++ ++ DDG
Sbjct: 298 GIDWPKDKLNIWILDDG 314
>BCSA_SALTI (Q8Z291) Cellulose synthase catalytic subunit
[UDP-forming] (EC 2.4.1.12)
Length = 874
Score = 67.0 bits (162), Expect = 2e-10
Identities = 62/229 (27%), Positives = 109/229 (47%), Gaps = 25/229 (10%)
Query: 681 GLSYGSIAADVLTSLKLHSRGWRSVYCMPKRAAFRGTAPINLTERLNQVLRWAVGSLEIL 740
G++ ++ D TSL+LH RG+ S Y ++A G A +L+ + Q +RWA G ++I
Sbjct: 448 GIAVETVTEDAHTSLRLHRRGYTSAYMRIPQSA--GLATESLSAHIGQRIRWARGMVQI- 504
Query: 741 FSRHCPIWYGFKEGRLKGLQRIAYINSTVYPFSSIPLLIYCLIPAICLLTDKFITPSVDT 800
F P+ F +G LK QR+ Y+N+ + S IP LI+ P LL +I
Sbjct: 505 FRLDNPL---FGKG-LKLAQRLCYLNAMFHFLSGIPRLIFLTAPLAFLLLHAYI-----I 555
Query: 801 FASMIIISLFI--SIFGSAILELRWSGVSLEEWWRSQQFWVIGSVSAHLFAVAQALMGGL 858
+A ++I+LF+ + +++ + G +W V+ + +A + L
Sbjct: 556 YAPALMIALFVIPHMVHASLTNSKIQGKYRHSFWSEIYETVLA------WYIAPPTLVAL 609
Query: 859 AKKVNKNFSVVSKAPDDEEFHELYTIRWTALLVPPTTIIIINLIGVVAG 907
F+V +K EE + + W + P ++++NL+GV AG
Sbjct: 610 INPHKGKFNVTAKGGLVEEKY----VDW-VISRPYIFLVLLNLLGVAAG 653
Score = 35.4 bits (80), Expect = 0.76
Identities = 37/137 (27%), Positives = 60/137 (43%), Gaps = 26/137 (18%)
Query: 211 GRLSPYRMMVVTRLLLLLLFIQYRIFH------PVPDAIGLWLISVVCEIWLTLSWIVDQ 264
GR S M++V L + +I +R PV GL L+ W+ L ++
Sbjct: 195 GRFSAL-MLIVLSLTVSCRYIWWRYTSTLNWDDPVSLVCGLILLFAETYAWIVL--VLGY 251
Query: 265 LPKWFPIDRETYLDRLSIRFEPENKPNMLSP---VDIFVTTVDPIKEPPLVTANTVLSIL 321
+P++R+ P P +S VDIFV T + E V NT+ + L
Sbjct: 252 FQVVWPLNRQ-----------PVPLPKEMSQWPTVDIFVPTYN---EDLNVVKNTIYASL 297
Query: 322 ALDYPAHKISCYVSDDG 338
+D+P K++ ++ DDG
Sbjct: 298 GIDWPKDKLNIWILDDG 314
>BCSA_ECOLI (P37653) Cellulose synthase catalytic subunit
[UDP-forming] (EC 2.4.1.12)
Length = 872
Score = 65.9 bits (159), Expect = 5e-10
Identities = 61/230 (26%), Positives = 107/230 (46%), Gaps = 27/230 (11%)
Query: 681 GLSYGSIAADVLTSLKLHSRGWRSVYCMPKRAAFRGTAPINLTERLNQVLRWAVGSLEIL 740
G++ ++ D TSL+LH RG+ S Y +AA G A +L+ + Q +RWA G ++I
Sbjct: 448 GIAVETVTEDAHTSLRLHRRGYTSAYMRIPQAA--GLATESLSAHIGQRIRWARGMVQI- 504
Query: 741 FSRHCPIWYGFKEGR-LKGLQRIAYINSTVYPFSSIPLLIYCLIPAICLLTDKFITPSVD 799
F P+ G+ LK QR+ Y+N+ + S IP LI+ P LL +I
Sbjct: 505 FRLDNPL-----TGKGLKFAQRLCYVNAMFHFLSGIPRLIFLTAPLAFLLLHAYI----- 554
Query: 800 TFASMIIISLFI--SIFGSAILELRWSGVSLEEWWRSQQFWVIGSVSAHLFAVAQALMGG 857
+A ++I+LF+ + +++ + G +W V+ + +A +
Sbjct: 555 IYAPALMIALFVLPHMIHASLTNSKIQGKYRHSFWSEIYETVLA------WYIAPPTLVA 608
Query: 858 LAKKVNKNFSVVSKAPDDEEFHELYTIRWTALLVPPTTIIIINLIGVVAG 907
L F+V +K EE + + W + P ++++NL+GV G
Sbjct: 609 LINPHKGKFNVTAKGGLVEEEY----VDW-VISRPYIFLVLLNLVGVAVG 653
Score = 36.2 bits (82), Expect = 0.45
Identities = 38/135 (28%), Positives = 60/135 (44%), Gaps = 22/135 (16%)
Query: 211 GRLSPYRMMVVTRLLLLLLFIQYRIFH------PVPDAIGLWLISVVCEIWLTLSWIVDQ 264
GR S M++V L + +I +R PV GL L+ W+ L ++
Sbjct: 195 GRFSAL-MLIVLSLTVSCRYIWWRYTSTLNWDDPVSLVCGLILLFAETYAWIVL--VLGY 251
Query: 265 LPKWFPIDRETYLDRLSIRFEPENKPNMLSP-VDIFVTTVDPIKEPPLVTANTVLSILAL 323
+P++R+ P K L P VDIFV T + E V NT+ + L +
Sbjct: 252 FQVVWPLNRQPV---------PLPKDMSLWPSVDIFVPTYN---EDLNVVKNTIYASLGI 299
Query: 324 DYPAHKISCYVSDDG 338
D+P K++ ++ DDG
Sbjct: 300 DWPKDKLNIWILDDG 314
>BCSA_ECO57 (Q8X5L7) Cellulose synthase catalytic subunit
[UDP-forming] (EC 2.4.1.12)
Length = 872
Score = 65.9 bits (159), Expect = 5e-10
Identities = 61/230 (26%), Positives = 107/230 (46%), Gaps = 27/230 (11%)
Query: 681 GLSYGSIAADVLTSLKLHSRGWRSVYCMPKRAAFRGTAPINLTERLNQVLRWAVGSLEIL 740
G++ ++ D TSL+LH RG+ S Y +AA G A +L+ + Q +RWA G ++I
Sbjct: 448 GIAVETVTEDAHTSLRLHRRGYTSAYMRIPQAA--GLATESLSAHIGQRIRWARGMVQI- 504
Query: 741 FSRHCPIWYGFKEGR-LKGLQRIAYINSTVYPFSSIPLLIYCLIPAICLLTDKFITPSVD 799
F P+ G+ LK QR+ Y+N+ + S IP LI+ P LL +I
Sbjct: 505 FRLDNPL-----TGKGLKFAQRLCYVNAMFHFLSGIPRLIFLTAPLAFLLLHAYI----- 554
Query: 800 TFASMIIISLFI--SIFGSAILELRWSGVSLEEWWRSQQFWVIGSVSAHLFAVAQALMGG 857
+A ++I+LF+ + +++ + G +W V+ + +A +
Sbjct: 555 IYAPALMIALFVLPHMIHASLTNSKIQGKYRHSFWSEIYETVLA------WYIAPPTLVA 608
Query: 858 LAKKVNKNFSVVSKAPDDEEFHELYTIRWTALLVPPTTIIIINLIGVVAG 907
L F+V +K EE + + W + P ++++NL+GV G
Sbjct: 609 LINPHKGKFNVTAKGGLVEEEY----VDW-VISRPYIFLVLLNLVGVAVG 653
Score = 36.2 bits (82), Expect = 0.45
Identities = 38/135 (28%), Positives = 60/135 (44%), Gaps = 22/135 (16%)
Query: 211 GRLSPYRMMVVTRLLLLLLFIQYRIFH------PVPDAIGLWLISVVCEIWLTLSWIVDQ 264
GR S M++V L + +I +R PV GL L+ W+ L ++
Sbjct: 195 GRFSAL-MLIVLSLTVSCRYIWWRYTSTLNWDDPVSLVCGLILLFAETYAWIVL--VLGY 251
Query: 265 LPKWFPIDRETYLDRLSIRFEPENKPNMLSP-VDIFVTTVDPIKEPPLVTANTVLSILAL 323
+P++R+ P K L P VDIFV T + E V NT+ + L +
Sbjct: 252 FQVVWPLNRQPV---------PLPKDMSLWPSVDIFVPTYN---EDLNVVKNTIYASLGI 299
Query: 324 DYPAHKISCYVSDDG 338
D+P K++ ++ DDG
Sbjct: 300 DWPKDKLNIWILDDG 314
>BCSA_PSEFL (P58931) Cellulose synthase catalytic subunit
[UDP-forming] (EC 2.4.1.12)
Length = 739
Score = 55.1 bits (131), Expect = 9e-07
Identities = 57/233 (24%), Positives = 99/233 (42%), Gaps = 23/233 (9%)
Query: 681 GLSYGSIAADVLTSLKLHSRGWRSVYCMPKRAAFRGTAPINLTERLNQVLRWAVGSLEIL 740
G++ ++ D T+LKL+ G+ + Y +AA G A +L+ +NQ +RWA G +I
Sbjct: 334 GVAVETVTEDAHTALKLNRLGYNTAYLAIPQAA--GLATESLSRHINQRIRWARGMAQIF 391
Query: 741 FSRHCPIWYGFKEGRLKGLQRIAYINSTVYPFSSIPLLIYCLIPAICLLTDKFITPSVDT 800
+ + + G K G QRI Y N+ + F +P L++ P L+ F
Sbjct: 392 RTDNPLLGKGLKWG-----QRICYANAMQHFFYGLPRLVFLTAPLAYLI---FGAEIFHA 443
Query: 801 FASMIIISLFISIFGSAILELRWSGVSLEEWWRSQQFWVIGSVSAHLFAVAQALMGGLAK 860
A MI+ + + S++ R G +W V+ + + ++ L
Sbjct: 444 SALMIVAYVLPHLVHSSLTNSRIQGRFRHSFWNEVYETVLA------WYILPPVLVALVN 497
Query: 861 KVNKNFSVVSKAP-DDEEFHELYTIRWTALLVPPTTIIIINLIGVVAGFTDAI 912
F+V K D++F + W L P ++ +NLIG+ G I
Sbjct: 498 PKAGGFNVTDKGGIIDKQFFD-----W-KLARPYLVLLAVNLIGLGFGIHQLI 544
Score = 32.0 bits (71), Expect = 8.4
Identities = 21/84 (25%), Positives = 38/84 (45%), Gaps = 20/84 (23%)
Query: 255 WLTLSWIVDQLPKWFPIDRETYLDRLSIRFEPENKPNMLSPVDIFVTTVDPIKEPPLVTA 314
++ +W + + P W ++ EPE P VD+F+ T + E +
Sbjct: 137 YVQTAWPLRRTPVW-------------LKTEPEEWPT----VDVFIPTYN---EALSIVK 176
Query: 315 NTVLSILALDYPAHKISCYVSDDG 338
T+ + A+D+P K+ +V DDG
Sbjct: 177 LTIFAAQAMDWPKDKLRVHVLDDG 200
>BCSA_XANAC (P58932) Cellulose synthase catalytic subunit
[UDP-forming] (EC 2.4.1.12)
Length = 729
Score = 53.5 bits (127), Expect = 3e-06
Identities = 62/253 (24%), Positives = 107/253 (41%), Gaps = 29/253 (11%)
Query: 667 SCRYEDRTLWGYEVGLSYGSIAADVLTSLKLHSRGWRSVYCMPKRAAFRGTAPINLTERL 726
SC RT G++ ++ D T+LKL RG+R+ Y +AA G A +L+ +
Sbjct: 314 SCAVIKRTALEEVGGVAVETVTEDAHTALKLQRRGYRTAYLAVPQAA--GLATESLSGHV 371
Query: 727 NQVLRWAVGSLEILFSRHCPIWYGFKEGRLKGLQRIAYINSTVYPFSSIPLLIYCLIPAI 786
Q +RWA G +I + + G LK QR+ Y+N+ ++ F +P +IY P
Sbjct: 372 AQRIRWARGMAQIARIDNPLLGRG-----LKLSQRLCYLNAMLHFFYGVPRIIYLTAPLA 426
Query: 787 CLLTDKFITPSVDTFASMIIISLFISIFGSAILEL----RWSGVSLEEWWRSQQFWVIGS 842
L F + A MI+ I + + L R+ + E + + W I
Sbjct: 427 YLF---FGAHVIQASALMILAYALPHILQANLTNLRVQSRFRHLLWNEVYETTLAWYI-- 481
Query: 843 VSAHLFAVAQALMGGLAKKVNKNFSVVSKAPDDEEFHELYTIRWTALLVPPTTIIIINLI 902
L A+ +G V +V+++ D + + Y ++++N++
Sbjct: 482 FRPTLVALLNPKLGKF--NVTPKGGLVARSYFDAQIAKPYLF-----------LLLLNVV 528
Query: 903 GVVAGFTDAINSG 915
G+VAG I G
Sbjct: 529 GMVAGVLRLIYVG 541
Score = 35.4 bits (80), Expect = 0.76
Identities = 18/43 (41%), Positives = 27/43 (61%), Gaps = 3/43 (6%)
Query: 296 VDIFVTTVDPIKEPPLVTANTVLSILALDYPAHKISCYVSDDG 338
VD+F+ T + EP V TVL+ +D+PA KI+ ++ DDG
Sbjct: 155 VDVFIPTYN---EPLSVVRTTVLAASVIDWPAGKITIHLLDDG 194
>BCA2_ACEXY (O82859) Cellulose synthase catalytic subunit
[UDP-forming] (EC 2.4.1.12)
Length = 756
Score = 49.3 bits (116), Expect = 5e-05
Identities = 38/144 (26%), Positives = 66/144 (45%), Gaps = 8/144 (5%)
Query: 681 GLSYGSIAADVLTSLKLHSRGWRSVYCMPKRAAFRGTAPINLTERLNQVLRWAVGSLEIL 740
G + ++ D T+L++ RGW + Y A+ G A LT + Q +RWA G ++I
Sbjct: 326 GFAVETVTEDAHTALRMQRRGWSTAYLRIPVAS--GLATERLTTHIGQRMRWARGMIQIF 383
Query: 741 FSRHCPIWYGFKEGRLKGLQRIAYINSTVYPFSSIPLLIYCLIPAICLLTDKFITPSVD- 799
+ + G K G QR+ Y+++ F +IP +I+ P L + I +
Sbjct: 384 RVDNPMLGRGLKLG-----QRLCYLSAMTSFFFAIPRVIFLASPLAFLFAGQNIIAAAPL 438
Query: 800 TFASMIIISLFISIFGSAILELRW 823
A+ + +F SI +A + W
Sbjct: 439 AVAAYALPHMFHSIATAAKVNKGW 462
Score = 35.8 bits (81), Expect = 0.58
Identities = 18/43 (41%), Positives = 25/43 (57%), Gaps = 3/43 (6%)
Query: 296 VDIFVTTVDPIKEPPLVTANTVLSILALDYPAHKISCYVSDDG 338
VDIF+ T D E + TVL L +D+P K++ Y+ DDG
Sbjct: 151 VDIFIPTYD---EQLSIVRLTVLGALGIDWPPDKVNVYILDDG 190
>BCA1_ACEXY (P19449) Cellulose synthase catalytic subunit
[UDP-forming] (EC 2.4.1.12)
Length = 754
Score = 48.9 bits (115), Expect = 7e-05
Identities = 39/144 (27%), Positives = 64/144 (44%), Gaps = 8/144 (5%)
Query: 681 GLSYGSIAADVLTSLKLHSRGWRSVYCMPKRAAFRGTAPINLTERLNQVLRWAVGSLEIL 740
G + ++ D T+L++ RGW + Y A+ G A LT + Q +RWA G ++I
Sbjct: 324 GFAVETVTEDAHTALRMQRRGWSTAYLRIPVAS--GLATERLTTHIGQRMRWARGMIQIF 381
Query: 741 FSRHCPIWYGFKEGRLKGLQRIAYINSTVYPFSSIPLLIYCLIP-AICLLTDKFITPSVD 799
+ + G K G QR+ Y+++ F +IP +I+ P A I S
Sbjct: 382 RVDNPMLGGGLKLG-----QRLCYLSAMTSFFFAIPRVIFLASPLAFLFFGQNIIAASPL 436
Query: 800 TFASMIIISLFISIFGSAILELRW 823
+ I +F SI +A + W
Sbjct: 437 AVLAYAIPHMFHSIATAAKVNKGW 460
Score = 35.8 bits (81), Expect = 0.58
Identities = 18/43 (41%), Positives = 25/43 (57%), Gaps = 3/43 (6%)
Query: 296 VDIFVTTVDPIKEPPLVTANTVLSILALDYPAHKISCYVSDDG 338
VDIF+ T D E + TVL L +D+P K++ Y+ DDG
Sbjct: 151 VDIFIPTYD---EQLSIVRLTVLGALGIDWPPDKVNVYILDDG 190
>BCA5_ACEXY (Q9WX75) Putative cellulose synthase 3 [Includes:
Cellulose synthase catalytic subunit [UDP-forming] (EC
2.4.1.12); Cyclic di-GMP binding domain (Cellulose
synthase 3 regulatory subunit)]
Length = 1518
Score = 48.1 bits (113), Expect = 1e-04
Identities = 38/144 (26%), Positives = 66/144 (45%), Gaps = 8/144 (5%)
Query: 681 GLSYGSIAADVLTSLKLHSRGWRSVYCMPKRAAFRGTAPINLTERLNQVLRWAVGSLEIL 740
G + ++ D T+LK+ RGW + Y AA G A L + Q +RWA G ++I+
Sbjct: 321 GFATETVTEDAHTALKMQRRGWGTAYLREPLAA--GLATERLILHIGQRVRWARGMIQIM 378
Query: 741 FSRHCPIWYGFKEGRLKGLQRIAYINSTVYPFSSIPLLIYCLIP-AICLLTDKFITPSVD 799
+ + G L+ QR+ Y+++ + +IP L + + P A L I S
Sbjct: 379 RLDNPMLGAG-----LRWEQRLCYLSAMSHFLFAIPRLTFLVSPLAFLFLGQNIIAASPL 433
Query: 800 TFASMIIISLFISIFGSAILELRW 823
+ + +F S+ + +E RW
Sbjct: 434 AISVYALPHIFHSVITLSRIEGRW 457
Score = 33.5 bits (75), Expect = 2.9
Identities = 17/43 (39%), Positives = 27/43 (62%), Gaps = 3/43 (6%)
Query: 296 VDIFVTTVDPIKEPPLVTANTVLSILALDYPAHKISCYVSDDG 338
VD+FV + + E + +TVL L LD+PA +++ Y+ DDG
Sbjct: 148 VDVFVPSYN---EELSLVRSTVLGALDLDWPADRLNVYILDDG 187
>BCA4_ACEXY (Q9RBJ2) Putative cellulose synthase 2 [Includes:
Cellulose synthase catalytic subunit [UDP-forming] (EC
2.4.1.12); Cyclic di-GMP binding domain (Cellulose
synthase 2 regulatory subunit)]
Length = 1518
Score = 48.1 bits (113), Expect = 1e-04
Identities = 38/144 (26%), Positives = 66/144 (45%), Gaps = 8/144 (5%)
Query: 681 GLSYGSIAADVLTSLKLHSRGWRSVYCMPKRAAFRGTAPINLTERLNQVLRWAVGSLEIL 740
G + ++ D T+LK+ RGW + Y AA G A L + Q +RWA G ++I+
Sbjct: 321 GFATETVTEDAHTALKMQRRGWGTAYLREPLAA--GLATERLILHIGQRVRWARGMIQIM 378
Query: 741 FSRHCPIWYGFKEGRLKGLQRIAYINSTVYPFSSIPLLIYCLIP-AICLLTDKFITPSVD 799
+ + G L+ QR+ Y+++ + +IP L + + P A L I S
Sbjct: 379 RLDNPMLGAG-----LRWEQRLCYLSAMSHFLFAIPRLTFLVSPLAFLFLGQNIIAASPL 433
Query: 800 TFASMIIISLFISIFGSAILELRW 823
+ + +F S+ + +E RW
Sbjct: 434 AISVYALPHIFHSVITLSRIEGRW 457
Score = 33.5 bits (75), Expect = 2.9
Identities = 17/43 (39%), Positives = 27/43 (62%), Gaps = 3/43 (6%)
Query: 296 VDIFVTTVDPIKEPPLVTANTVLSILALDYPAHKISCYVSDDG 338
VD+FV + + E + +TVL L LD+PA +++ Y+ DDG
Sbjct: 148 VDVFVPSYN---EELSLVRSTVLGALDLDWPADRLNVYILDDG 187
>BA1A_XENLA (Q8UVR5) ATP-utilizing chromatin assembly and remodely
factor 1 (xACF1) (Fragment)
Length = 627
Score = 47.4 bits (111), Expect = 2e-04
Identities = 48/189 (25%), Positives = 85/189 (44%), Gaps = 32/189 (16%)
Query: 37 CEICGDSVGLTVDGDLFVACEECG----FPVCRPCYEYERREGTQGCPQCHTRYK--RIK 90
C++C DG+ V C+ C RP +Y EG CP+CH + + R+
Sbjct: 225 CKVCRKKG----DGESMVLCDGCDRGHHIYCVRPKLKYVP-EGDWFCPECHPKQRSHRLP 279
Query: 91 GSPRVSGDEDEDDVDDIEQEFKMEEEKYKLKQEEMLQGKMKHGDD--DENAKPLLVNGEL 148
R S D DE++ ++++Q+ EEE+ + +QEE+ + + + D+ +E + P ++
Sbjct: 280 SRHRYSMDSDEEEEEELDQK---EEEEEEEEQEELSESENEQEDEMSEEESPPKRGRAKV 336
Query: 149 PISSYSIVEPAGGE---KLDDKEKTDDWKLNQGNLWPETAAPVDPEKNMNDETRQPLSRK 205
+ ++ GG+ KL K KT P+ P + +TR S +
Sbjct: 337 QLP----LKMRGGKATGKLGPKPKTGKQST------PKNTQPAPEGRGQGKKTRSAPSLE 386
Query: 206 VAIPSGRLS 214
P+ RLS
Sbjct: 387 ---PTSRLS 392
>ACS2_ACEXY (Q59167) Cellulose synthase 2 [Includes: Cellulose
synthase catalytic subunit [UDP-forming] (EC 2.4.1.12);
Cyclic di-GMP binding domain (Cellulose synthase 2
regulatory domain)]
Length = 1596
Score = 46.6 bits (109), Expect = 3e-04
Identities = 66/271 (24%), Positives = 112/271 (40%), Gaps = 25/271 (9%)
Query: 681 GLSYGSIAADVLTSLKLHSRGWRSVYCMPKRAAFRGTAPINLTERLNQVLRWAVGSLEIL 740
G + ++ D T+LK+ GW + Y AA G + L + Q +RWA G L+I+
Sbjct: 322 GFATETVTEDAHTALKMQREGWHTAYLRQPLAA--GLSTERLMLHIGQRVRWARGMLQIM 379
Query: 741 FSRHCPIWYGFKEGRLKGLQRIAYINSTVYPFSSIPLLIYCLIPAICLLTDKFITPSVDT 800
+ + G L+ QR+ Y+++ + +IP L++ P L + I +
Sbjct: 380 RLDNPLLGSG-----LRWQQRLCYLSAMSHFLFAIPRLVFLASPLAFLFLGQNII-AASP 433
Query: 801 FASMIIISLFISIFGSAILELRWSGVSLEEWWRSQQFWVIGSVSAHLFAVAQALMGGLAK 860
FA I++ F +F S R +E WR + I + LF V +M L
Sbjct: 434 FA--ILVYAFPHVFHSIGTLSR-----VEGRWRYSFWSEIYETTLALFLVRVTIMTLLNP 486
Query: 861 KVNKNFSVVSKAP-DDEEFHELYTIRWTALLVPPTTIIIINLIGVVAGFTDAINSGAHSY 919
+ + F+V K E+ +L + P + +I + +V G + H
Sbjct: 487 RKGE-FNVTDKGGLLQSEYFDLNAV------YPNVILAVILALALVRGI-GGMMWEYHDR 538
Query: 920 GALLGKLFFSLWVIAHLYPFLKGL-MGRQNR 949
AL +LWV L L + +GR+ R
Sbjct: 539 LALQSFALNTLWVAVSLIIVLASIAVGRETR 569
Score = 41.6 bits (96), Expect = 0.011
Identities = 31/128 (24%), Positives = 57/128 (44%), Gaps = 27/128 (21%)
Query: 218 MMVVTRLLLLLLFIQYRIFHPVPDAIGLWLISV-------VCEIWLTLSWIVDQLPKWFP 270
++V R ++ L + P+ A+ L L++ +C + +SW +D+ P P
Sbjct: 81 LLVSLRYMVWRLTTTLELHSPLQAALSLLLVAAELYALLTLCLSYFQMSWPLDRKPLPLP 140
Query: 271 IDRETYLDRLSIRFEPENKPNMLSPVDIFVTTVDPIKEPPLVTANTVLSILALDYPAHKI 330
D + VD++V + + E + +TVL LA+D+PA K+
Sbjct: 141 ADTTDW-----------------PVVDVYVPSYN---EELSLVRSTVLGALAIDWPADKL 180
Query: 331 SCYVSDDG 338
+ Y+ DDG
Sbjct: 181 NVYILDDG 188
>BCA3_ACEXY (Q9WX61) Cellulose synthase 1 catalytic subunit
[UDP-forming] (EC 2.4.1.12)
Length = 745
Score = 46.2 bits (108), Expect = 4e-04
Identities = 37/144 (25%), Positives = 63/144 (43%), Gaps = 8/144 (5%)
Query: 681 GLSYGSIAADVLTSLKLHSRGWRSVYCMPKRAAFRGTAPINLTERLNQVLRWAVGSLEIL 740
G + ++ D T+L++ +GW + Y A+ G A L + Q +RWA G ++I
Sbjct: 324 GFATETVTEDAHTALRMQRKGWSTAYLRIPLAS--GLATERLITHIGQRMRWARGMIQIF 381
Query: 741 FSRHCPIWYGFKEGRLKGLQRIAYINSTVYPFSSIPLLIYCLIP-AICLLTDKFITPSVD 799
+ + G K G QR+ Y+++ F +IP +I+ P A + I S
Sbjct: 382 RVDNPMLGSGLKLG-----QRLCYLSAMTSFFFAIPRVIFLASPLAFLFFSQNIIAASPL 436
Query: 800 TFASMIIISLFISIFGSAILELRW 823
I +F SI +A + W
Sbjct: 437 AVGVYAIPHMFHSIATAAKVNKGW 460
Score = 36.6 bits (83), Expect = 0.34
Identities = 19/49 (38%), Positives = 27/49 (54%), Gaps = 3/49 (6%)
Query: 290 PNMLSPVDIFVTTVDPIKEPPLVTANTVLSILALDYPAHKISCYVSDDG 338
P+ VDIF+ T D E + TVL L +D+P K++ Y+ DDG
Sbjct: 145 PDEWPTVDIFIPTYD---EALSIVRLTVLGALGIDWPPDKVNVYILDDG 190
>BA1A_HUMAN (Q9NRL2) Bromodomain adjacent to zinc finger domain
protein 1A (ATP-utilizing chromatin assembly and
remodeling factor 1) (hACF1) (ATP-dependent chromatin
remodelling protein) (Williams syndrome transcription
factor-related chromatin remodelin
Length = 1556
Score = 39.3 bits (90), Expect = 0.053
Identities = 30/110 (27%), Positives = 51/110 (46%), Gaps = 19/110 (17%)
Query: 30 KNLDGQLCEICGDSVGLTVDGDLFVACEECGFPVCRPCYEYERR---EGTQGCPQCHTRY 86
K++ C+IC D + V C+ C C + + EG CP+C +
Sbjct: 1144 KSILNARCKICRKKG----DAENMVLCDGCDRGHHTYCVRPKLKTVPEGDWFCPECRPKQ 1199
Query: 87 KRIKGS----PRVSGDED--------EDDVDDIEQEFKMEEEKYKLKQEE 124
+ + S P + DED +D+VD E+E + EEE+Y+++Q+E
Sbjct: 1200 RSRRLSSRQRPSLESDEDVEDSMGGEDDEVDGDEEEGQSEEEEYEVEQDE 1249
>NRG3_MOUSE (O35181) Pro-neuregulin-3 precursor (Pro-NRG3)
[Contains: Neuregulin-3 (NRG-3)]
Length = 713
Score = 38.9 bits (89), Expect = 0.069
Identities = 13/43 (30%), Positives = 33/43 (76%)
Query: 588 KASKRQREVQVHSKQDESGEDGSIKEATDEDKQLLKSHMNVEN 630
K+ K+ +++Q H K+ ++G++ S+K ++ + + L+KSH++++N
Sbjct: 386 KSKKQAKQIQEHLKESQNGKNYSLKASSTKSESLMKSHVHLQN 428
>CNO4_MOUSE (Q8BT14) CCR4-NOT transcription complex subunit 4 (EC
6.3.2.-) (E3 ubiquitin protein ligase CNOT4)
(CCR4-associated factor 4) (Potential transcriptional
repressor NOT4Hp)
Length = 575
Score = 38.1 bits (87), Expect = 0.12
Identities = 24/88 (27%), Positives = 39/88 (44%), Gaps = 6/88 (6%)
Query: 37 CEICGDSVGLTVDGDLFVACEECGFPVCRPCYEYERREGTQGCPQCHTRYKRIKGSPRVS 96
C +C + L +D F C CG+ +CR C+ R + CP C Y P V
Sbjct: 14 CPLCMEP--LEIDDINFFPCT-CGYQICRFCWHRIRTDENGLCPACRKPYPE---DPAVY 67
Query: 97 GDEDEDDVDDIEQEFKMEEEKYKLKQEE 124
++++ I+ E K ++ + K K E
Sbjct: 68 KPLSQEELQRIKNEKKQKQNERKQKISE 95
>CNO4_HUMAN (O95628) CCR4-NOT transcription complex subunit 4 (EC
6.3.2.-) (E3 ubiquitin protein ligase CNOT4)
(CCR4-associated factor 4) (Potential transcriptional
repressor NOT4Hp)
Length = 575
Score = 38.1 bits (87), Expect = 0.12
Identities = 24/88 (27%), Positives = 39/88 (44%), Gaps = 6/88 (6%)
Query: 37 CEICGDSVGLTVDGDLFVACEECGFPVCRPCYEYERREGTQGCPQCHTRYKRIKGSPRVS 96
C +C + L +D F C CG+ +CR C+ R + CP C Y P V
Sbjct: 14 CPLCMEP--LEIDDINFFPCT-CGYQICRFCWHRIRTDENGLCPACRKPYPE---DPAVY 67
Query: 97 GDEDEDDVDDIEQEFKMEEEKYKLKQEE 124
++++ I+ E K ++ + K K E
Sbjct: 68 KPLSQEELQRIKNEKKQKQNERKQKISE 95
>ACS1_ACEXY (P21877) Cellulose synthase 1 [Includes: Cellulose
synthase catalytic domain [UDP-forming] (EC 2.4.1.12);
Cyclic di-GMP binding domain (Cellulose synthase 1
regulatory domain)]
Length = 1550
Score = 37.7 bits (86), Expect = 0.15
Identities = 34/118 (28%), Positives = 52/118 (43%), Gaps = 7/118 (5%)
Query: 667 SCRYEDRTLWGYEVGLSYGSIAADVLTSLKLHSRGWRSVYCMPKRAAFRGTAPINLTERL 726
SC RT G + ++ D T+LK+ GW + Y A G A L +
Sbjct: 310 SCAILRRTAIEQIGGFATQTVTEDAHTALKMQRLGWSTAYLRIPLAG--GLATERLILHI 367
Query: 727 NQVLRWAVGSLEILFSRHCPIWYGFKEGRLKGLQRIAYINSTVYPFSSIPLLIYCLIP 784
Q +RWA G L+I F P+ F G G QR+ Y+++ ++P +I+ P
Sbjct: 368 GQRVRWARGMLQI-FRIDNPL---FGRGLSWG-QRLCYLSAMTSFLFAVPRVIFLSSP 420
Score = 32.3 bits (72), Expect = 6.4
Identities = 18/49 (36%), Positives = 26/49 (52%), Gaps = 3/49 (6%)
Query: 290 PNMLSPVDIFVTTVDPIKEPPLVTANTVLSILALDYPAHKISCYVSDDG 338
P+ VDIFV T + E + TVL L +D+P K+ ++ DDG
Sbjct: 145 PDEWPTVDIFVPTYN---EELSIVRLTVLGSLGIDWPPEKVRVHILDDG 190
>YAC4_SCHPO (Q09818) Putative general negative regulator of
transcription C16C9.04c
Length = 489
Score = 37.0 bits (84), Expect = 0.26
Identities = 22/91 (24%), Positives = 46/91 (50%), Gaps = 6/91 (6%)
Query: 33 DGQLCEICGDSVGLTVDGDLFVACEECGFPVCRPCYEYERREGTQGCPQCHTRY--KRIK 90
D C +C + + ++ F C+ CG+ VCR C+ + + + CP C Y + ++
Sbjct: 14 DDMCCPLCMEEIDISDKN--FKPCQ-CGYRVCRFCWHHIKEDLNGRCPACRRLYTEENVQ 70
Query: 91 GSPRVSGDEDEDDVDDIEQEFKMEEEKYKLK 121
P V+ +E + D+ + K E+E+ +++
Sbjct: 71 WRP-VTAEEWKMDLHRKNERKKREKERKEVE 100
>CBL_MOUSE (P22682) CBL E3 ubiquitin protein ligase (EC 6.3.2.-)
(Signal transduction protein CBL) (Proto-oncogene c-CBL)
(Casitas B-lineage lymphoma proto-oncogene)
Length = 913
Score = 35.8 bits (81), Expect = 0.58
Identities = 27/102 (26%), Positives = 40/102 (38%), Gaps = 29/102 (28%)
Query: 35 QLCEICGDSVGLTVDGDLFVACEECGFPVCRPCYEYERREGTQGCPQCHTRYK------- 87
QLC+IC ++ D V E CG +C C + QGCP C K
Sbjct: 377 QLCKICAEN-------DKDVKIEPCGHLMCTSCLTSWQESEGQGCPFCRCEIKGTEPIVV 429
Query: 88 --------------RIKGSPRVSGDEDEDDVDDIEQEFKMEE 115
+G+P + D+D+D+ D + F M+E
Sbjct: 430 DPFDPRGSGSLLRQGAEGAPSPNYDDDDDERAD-DSLFMMKE 470
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.138 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 120,912,192
Number of Sequences: 164201
Number of extensions: 5401924
Number of successful extensions: 17127
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 62
Number of HSP's that attempted gapping in prelim test: 16784
Number of HSP's gapped (non-prelim): 183
length of query: 991
length of database: 59,974,054
effective HSP length: 120
effective length of query: 871
effective length of database: 40,269,934
effective search space: 35075112514
effective search space used: 35075112514
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0037a.5


