

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
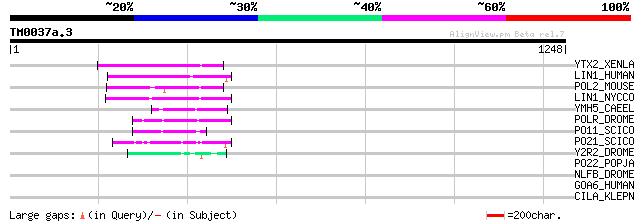
Query= TM0037a.3
(1248 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 124 2e-27
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 107 3e-22
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 105 1e-21
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 104 2e-21
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 62 1e-08
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 55 9e-07
PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type... 54 3e-06
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 52 8e-06
Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I retro... 49 7e-05
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 40 0.052
NLFB_DROME (Q9Y113) Negative elongation factor B homolog 37 0.34
GOA6_HUMAN (Q9NYA3) Golgi autoantigen, golgin subfamily A member... 35 1.3
CILA_KLEPN (P45413) Citrate lyase alpha chain (EC 4.1.3.6) (Citr... 35 1.3
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 124 bits (310), Expect = 2e-27
Identities = 78/286 (27%), Positives = 136/286 (47%), Gaps = 4/286 (1%)
Query: 197 KSRVAWLNHGDKNTRFFHTSTMVRRKRNKIEALTDEHGVSVTDPITLRSMAIDFFQHLYT 256
+SR+ L D+ +RFF+ + R +I L E G + DP +R A F+Q+L++
Sbjct: 359 RSRMQLLCDMDRGSRFFYALEKKKGNRKQITCLFAEDGTPLEDPEAIRDRARSFYQNLFS 418
Query: 257 SP--GTDVLYHVRNAFPTLPRDTLLEISNPLEADEIKAAVFSMRALKAPGPDGLNTLFFQ 314
D + + P + + P+ DE+ A+ M K+PG DGL FFQ
Sbjct: 419 PDPISPDACEELWDGLPVVSERRKERLETPITLDELSQALRLMPHNKSPGLDGLTIEFFQ 478
Query: 315 SQWDTIGSSVVQFIQDCIQRPEKIREVNDTLIVLIPKIDRPNLLSQYRLIGICNVIYKTL 374
WDT+G + + + ++ E ++ L+PK L+ +R + + + YK +
Sbjct: 479 FFWDTLGPDFHRVLTEAFKKGELPLSCRRAVLSLLPKKGDLRLIKNWRPVSLLSTDYKIV 538
Query: 375 TKALANRLKGVLGDLISPNQCSFIPGRQSSDNIIIAQEVFHSMRYLKRKKGWIAIKIDLE 434
KA++ RLK VL ++I P+Q +PGR DN+ + +++ H R +++ +D E
Sbjct: 539 AKAISLRLKSVLAEVIHPDQSYTVPGRTIFDNVFLIRDLLHFARRTGLSLAFLS--LDQE 596
Query: 435 KAYDRLEWSFLKDTLLEVGLDRNFCDLILDCVSLSSFQVLFNGSKT 480
KA+DR++ +L TL F + + + V N S T
Sbjct: 597 KAFDRVDHQYLIGTLQAYSFGPQFVGYLKTMYASAECLVKINWSLT 642
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 107 bits (266), Expect = 3e-22
Identities = 74/292 (25%), Positives = 146/292 (49%), Gaps = 16/292 (5%)
Query: 220 RRKRNKIEALTDEHGVSVTDPITLRSMAIDFFQHLYTSPGTDV----LYHVRNAFPTLPR 275
+R++N+I+ + ++ G TDP +++ ++++HLY + ++ + P L +
Sbjct: 383 KREKNQIDTIKNDRGDITTDPTEIQTTIREYYKHLYANKLENLEEMDKFLDTYTLPRLNQ 442
Query: 276 DTLLEISNPLEADEIKAAVFSMRALKAPGPDGLNTLFFQSQWDTIGSSVVQFIQDCIQRP 335
+ + ++ P+ + EI+A + S+ K+PGP+G F+Q + + +++ Q +
Sbjct: 443 EEVESLNRPITSSEIEAIINSLPNKKSPGPEGFTAEFYQRYKEELVPFLLKLFQSIEKEG 502
Query: 336 EKIREVNDTLIVLIPKIDRPNLLSQ-YRLIGICNVIYKTLTKALANRLKGVLGDLISPNQ 394
+ I+LIPK R + +R I + N+ K L K LAN+++ + LI +Q
Sbjct: 503 ILPNSFYEASIILIPKPGRDTTKKENFRPISLMNIDAKILNKILANQIQQHIKKLIHHDQ 562
Query: 395 CSFIPGRQSSDNIIIAQEVFHSMRYLKRKK--GWIAIKIDLEKAYDRLEWSFLKDTLLEV 452
FIP Q NI ++ + ++++ R K + I ID EKA+D+++ F+ L ++
Sbjct: 563 VGFIPAMQGWFNI---RKSINIIQHINRTKDTNHMIISIDAEKAFDKIQQPFMLKPLNKL 619
Query: 453 GLDRNFCDLILDCVSLSSFQVLFNGSKTEFPSF-----QGHPPR*PIIPIFV 499
G+D + +I + ++ NG K E P QG P P++P V
Sbjct: 620 GIDGTYLKIIRAIYDKPTANIILNGQKLEAPPLKTGTRQGCPLS-PLLPNIV 670
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 105 bits (261), Expect = 1e-21
Identities = 83/281 (29%), Positives = 134/281 (47%), Gaps = 28/281 (9%)
Query: 217 TMVRRKRNKIEALTDEHGVSVTDPITLRSMAIDFFQHLYTSPGTDV----LYHVRNAFPT 272
T R + I + +E G TDP +++ F++ LY++ ++ + R P
Sbjct: 407 TKGHRDKILINKIRNEKGDITTDPEEIQNTIRSFYKRLYSTKLENLDEMDKFLDRYQVPK 466
Query: 273 LPRDTLLEISNPLEADEIKAAVFSMRALKAPGPDGLNTLFFQSQWDTIGSSVVQFIQDCI 332
L +D + +++P+ EI+A + S+ K+PGPDG + F+Q+ F +D I
Sbjct: 467 LNQDQVDHLNSPISPKEIEAVINSLPTKKSPGPDGFSAEFYQT-----------FKEDLI 515
Query: 333 QRPEKIR---EVNDTL--------IVLIPKIDR-PNLLSQYRLIGICNVIYKTLTKALAN 380
K+ EV TL I LIPK + P + +R I + N+ K L K LAN
Sbjct: 516 PILHKLFHKIEVEGTLPNSFYEATITLIPKPQKDPTKIENFRPISLMNIDAKILNKILAN 575
Query: 381 RLKGVLGDLISPNQCSFIPGRQSSDNIIIAQEVFHSMRYLKRKKGWIAIKIDLEKAYDRL 440
R++ + +I P+Q FIPG Q NI + V H + LK K I I +D EKA+D++
Sbjct: 576 RIQEHIKAIIHPDQVGFIPGMQGWFNIRKSINVIHYINKLKDKNHMI-ISLDAEKAFDKI 634
Query: 441 EWSFLKDTLLEVGLDRNFCDLILDCVSLSSFQVLFNGSKTE 481
+ F+ L G+ + ++I S + NG K E
Sbjct: 635 QHPFMIKVLERSGIQGPYLNMIKAIYSKPVANIKVNGEKLE 675
Score = 34.7 bits (78), Expect = 1.7
Identities = 32/122 (26%), Positives = 49/122 (39%), Gaps = 7/122 (5%)
Query: 9 SPNLSIRNFLWQELRRIAANTGEPWVVMGDFNTYLNAPD-----KWGGDPPNLLAMGKFR 63
+PN F+ L ++ A +++GDFNT L++ D K D L + K
Sbjct: 142 APNARAATFIRDTLVKLKAYIAPHTIIVGDFNTPLSSKDRSWKQKLNRDTVKLTEVMKQM 201
Query: 64 DCLDDCYLSDLGFKGPPFTWEGRGVKERLDWALGNDQWLRSFPEASIFHLPQLKSDHKPL 123
D D KG F G ++D +G+ L + I +P + SDH L
Sbjct: 202 DLTDIYRTFYPKTKGYTFFSAPHGTFSKIDHIIGHKTGLNRYKNIEI--VPCILSDHHGL 259
Query: 124 LL 125
L
Sbjct: 260 RL 261
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 104 bits (259), Expect = 2e-21
Identities = 82/294 (27%), Positives = 142/294 (47%), Gaps = 17/294 (5%)
Query: 216 STMVRRKRNK--IEALTDEHGVSVTDPITLRSMAIDFFQHLYTSPGTDVL----YHVRNA 269
+ + R+KR K I ++ + + TDP ++ + ++++ LY+ ++ Y
Sbjct: 377 ANLTRKKRVKSLISSIRNGNDEITTDPSEIQKILNEYYKKLYSHKYENLKEIDQYLEACH 436
Query: 270 FPTLPRDTLLEISNPLEADEIKAAVFSMRALKAPGPDGLNTLFFQSQWDTIGSSVVQFIQ 329
P L + + ++ P+ + EI + + ++ K+PGPDG + F+Q T +V +
Sbjct: 437 LPRLSQKEVEMLNRPISSSEIASTIQNLPKKKSPGPDGFTSEFYQ----TFKEELVPILL 492
Query: 330 DCIQRPEKIREVNDTL----IVLIPKIDR-PNLLSQYRLIGICNVIYKTLTKALANRLKG 384
+ Q EK + +T I LIPK + P YR I + N+ K L K L NR++
Sbjct: 493 NLFQNIEKEGILPNTFYEANITLIPKPGKDPTRKENYRPISLMNIDAKILNKILTNRIQQ 552
Query: 385 VLGDLISPNQCSFIPGRQSSDNIIIAQEVFHSMRYLKRKKGWIAIKIDLEKAYDRLEWSF 444
+ +I +Q FIPG Q NI + V + LK K I + ID EKA+D ++ F
Sbjct: 553 HIKKIIHHDQVGFIPGSQGWFNIRKSINVIQHINKLKNKDHMI-LSIDAEKAFDNIQHPF 611
Query: 445 LKDTLLEVGLDRNFCDLILDCVSLSSFQVLFNGSKTE-FPSFQGHPPR*PIIPI 497
+ TL ++G++ F LI S + ++ NG K + FP G P+ P+
Sbjct: 612 MIRTLKKIGIEGTFLKLIEAIYSKPTANIILNGVKLKSFPLRSGTRQGCPLSPL 665
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 62.0 bits (149), Expect = 1e-08
Identities = 44/173 (25%), Positives = 78/173 (44%), Gaps = 9/173 (5%)
Query: 319 TIGSSVVQFIQDCIQRPEKIREVNDTLIVLIPKIDRPNLLSQYRLIGICNVIYKTLTKAL 378
T+G + F I K +I+ IPK P+ S YR I + + + + + +
Sbjct: 648 TLGKGMQSFSDSAIPNRWK-----HAVIIPIPKKGNPSSPSNYRPISLTDPFARIMERII 702
Query: 379 ANRLKGVLGDLISPNQCSFIPGRQSSDNIIIAQEVFHSMRYLKRKKGWIAIKIDLEKAYD 438
+R++ L+SP+Q F+ R +++ + ++HS+ LK +K + D KA+D
Sbjct: 703 CSRIRSEYSHLLSPHQHGFLNFRSCPSSLVRSISLYHSI--LKNEKSLDILFFDFAKAFD 760
Query: 439 RLEWSFLKDTLLEVGLDRNFCDLILDCVSLSSFQVLFNG--SKTEFPSFQGHP 489
++ L L GLD+ C + + L +F V N S +P G P
Sbjct: 761 KVSHPILLKKLALFGLDKLTCSWFKEFLHLRTFSVKINKFVSSNAYPISSGVP 813
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 55.5 bits (132), Expect = 9e-07
Identities = 57/223 (25%), Positives = 96/223 (42%), Gaps = 12/223 (5%)
Query: 277 TLLEISNPLEADEIKAAVFSMRALKAPGPDGLNTLFFQSQWDTIGSSVVQFIQDCIQRPE 336
+L + + + +++A+ S+ + +PGPDG+ + I ++ I C P
Sbjct: 336 SLERVWSAITEQDLRASRVSLSS--SPGPDGITPKSAREVPSGIMLRIMNLILWCGNLPH 393
Query: 337 KIREVNDTLIVLIPKIDRPNLLSQYRLIGICNVIYKTLTKALANRLKGVLGDLISPNQCS 396
IR V IPK +R I + +V+ + L LA RL + P Q
Sbjct: 394 SIRLART---VFIPKTVTAKRPQDFRPISVPSVLVRQLNAILATRLNSSIN--WDPRQRG 448
Query: 397 FIPGRQSSDNIIIAQEVF-HSMRYLKRKKGWIAIKIDLEKAYDRLEWSFLKDTLLEVGLD 455
F+P +DN I V HS ++ + +IA +D+ KA+D L + + DTL G
Sbjct: 449 FLPTDGCADNATIVDLVLRHSHKHFR--SCYIA-NLDVSKAFDSLSHASIYDTLRAYGAP 505
Query: 456 RNFCDLILDCVSLSSFQVLFNG-SKTEFPSFQGHPPR*PIIPI 497
+ F D + + + +G S EF +G P+ PI
Sbjct: 506 KGFVDYVQNTYEGGGTSLNGDGWSSEEFVPARGVKQGDPLSPI 548
>PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type I
retrotransposable element R1 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1004
Score = 53.5 bits (127), Expect = 3e-06
Identities = 49/178 (27%), Positives = 82/178 (45%), Gaps = 19/178 (10%)
Query: 276 DTLLEISNPL----EADEIKAAVFSMRALKAPGPDGLNTLFFQSQWDTIGSSVVQFIQDC 331
D L I+ PL E DE+ +V + K+PGPDG+ ++ W I + + C
Sbjct: 392 DRLKAIARPLPPDLEMDEVSDSVRRCKVRKSPGPDGIVGEMVRAVWGAIPEYMFCLYKQC 451
Query: 332 IQR---PEKIREVNDTLIVLIPKIDR-PNLLSQYRLIGICNVIYKTLTKALANRLKGVLG 387
+ P+K + +L++L+ +DR + YR I + + + K L + RL L
Sbjct: 452 LLESYFPQKWKIA--SLVILLKLLDRIRSDPGSYRPICLLDNLGKVLEGIMVKRLDQKLM 509
Query: 388 DL-ISPNQCSFIPGRQSSDNIIIAQEVFH--SMRYLKRKKGWIAIKIDLEKAYDRLEW 442
D+ +SP Q +F G+ + D Q M+Y+ I + ID + A+D L W
Sbjct: 510 DVEVSPYQFAFTYGKSTEDAWRCVQRHVECSEMKYV------IGLNIDFQGAFDNLGW 561
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 52.4 bits (124), Expect = 8e-06
Identities = 66/273 (24%), Positives = 114/273 (41%), Gaps = 21/273 (7%)
Query: 231 DEHGVSVTDPITLRSMAIDFFQHLYTSPGTDVLYHVRNAFPTLPRDTLLEISNPLEADEI 290
DE+ T +R M D+++ ++ + G ++ A P +TL + P+ EI
Sbjct: 109 DENDKIATKIPPVRHM-FDYWKDVFATGGGSAATNINRAPPAPHMETLWD---PVSLIEI 164
Query: 291 KAAVFSMRALKAPGPDGLNTLFFQSQWDTIGSSVVQFIQDCIQRPEKIRE-VNDTLIVLI 349
K+A S K GPDG+ W+ + + + + ++ + + V
Sbjct: 165 KSARASNE--KGAGPDGVTP----RSWNALDDRYKRLLYNIFVFYGRVPSPIKGSRTVFT 218
Query: 350 PKIDRPNLLSQYRLIGICNVIYKTLTKALANRLKGVLGDLISPNQCSFIPGRQSSDNIII 409
PKI+ +R + IC+VI + K LA R Q +++P N+ +
Sbjct: 219 PKIEGGPDPGVFRPLSICSVILREFNKILARRFVSCY--TYDERQTAYLPIDGVCINVSM 276
Query: 410 AQEVFHSMRYLKRKKGWIAIKIDLEKAYDRLEWSFLKDTLLEVGLDRNFCDLILDCVSLS 469
+ + L RK+ IAI +DL KA++ + S L D + E G D I D +
Sbjct: 277 LTAIIAEAKRL-RKELHIAI-LDLVKAFNSVYHSALIDAITEAGCPPGVVDYIADMYNNV 334
Query: 470 SFQVLFNGSKTEFPS-----FQGHPPR*PIIPI 497
++ F G K E S +QG P P+ +
Sbjct: 335 ITEMQFEG-KCELASILAGVYQGDPLSGPLFTL 366
>Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I
retrotransposable element R1DM (ORF 2)
Length = 1021
Score = 49.3 bits (116), Expect = 7e-05
Identities = 57/239 (23%), Positives = 93/239 (38%), Gaps = 44/239 (18%)
Query: 266 VRNAFPTL----PRDTLLEISNPLEADEIKAAVFSMRALKAPGPDGLNTLFFQSQWDTIG 321
+RN FP P E+ LE E+ V +++ ++PG DG+N ++ W I
Sbjct: 411 LRNFFPVAESEAPTAIAEEVPPALEVFEVDTCVARLKSRRSPGLDGINGTICKAVWRAIP 470
Query: 322 SSVVQFIQDCIQR---PEKIREVNDTLIVLIPKIDRPNLLSQYRLIGICNVIYKTLTKAL 378
+ CI+ P + + ++ P D+ S YR I + V K L +
Sbjct: 471 EHLASLFSRCIRLGYFPAEWKCPRVVSLLKGPDKDKCE-PSSYRGICLLPVFGKVLEAIM 529
Query: 379 ANRLKGVLGDLISPNQC----SFIPGRQSSDNIIIAQEVFHSMRYLKRKKGWIAIK---- 430
NR++ VL P C F GR D + R++K G A +
Sbjct: 530 VNRVREVL-----PEGCRWQFGFRQGRCVED----------AWRHVKSSVGASAAQYVLG 574
Query: 431 --IDLEKAYDRLEWSFLKDTLLEVGLDRNFCDLILDCVSLSSFQVLFNGSKTEFPSFQG 487
+D + A+D +EWS L ++G C + +Q F+G + S G
Sbjct: 575 TFVDFKGAFDNVEWSAALSRLADLG-----------CREMGLWQSFFSGRRAVIRSSSG 622
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 39.7 bits (91), Expect = 0.052
Identities = 48/198 (24%), Positives = 84/198 (42%), Gaps = 11/198 (5%)
Query: 284 PLEADEIKAAVFSMRALKAPGPDGLNTLFFQSQWDTIGSSVVQFIQDCIQRPEKIREVND 343
P+ +EI+ A+ + APG DGL Q T F+Q + R
Sbjct: 5 PIAREEIQCAIKGWKP-SAPGSDGLTV-----QAITRTRLPRNFVQLHLLRGHVPTPWTA 58
Query: 344 TLIVLIPKIDRPNLLSQYRLIGICNVIYKTLTKALANRLKGVLGDLISPNQCSFIPGRQS 403
LIPK S +R I I + + + L + LA RL+ + + P Q + +
Sbjct: 59 MRTTLIPKDGDLENPSNWRPITIASALQRLLHRILAKRLEAAVE--LHPAQKGYARIDGT 116
Query: 404 SDNIIIAQEVFHSMRYLKRKKGWIAIKIDLEKAYDRLEWSFLKDTLLEVGLDRNFCDLIL 463
N ++ S R +++K + + +D+ KA+D + S + L +G+D + I
Sbjct: 117 LVNSLLLDTYISSRR--EQRKTYNVVSLDVRKAFDTVSHSSICRALQRLGIDEGTSNYIT 174
Query: 464 DCVSLSSFQVLFN-GSKT 480
+S S+ + GS+T
Sbjct: 175 GSLSDSTTTIRVGPGSQT 192
>NLFB_DROME (Q9Y113) Negative elongation factor B homolog
Length = 594
Score = 37.0 bits (84), Expect = 0.34
Identities = 44/175 (25%), Positives = 74/175 (42%), Gaps = 21/175 (12%)
Query: 121 KPLLLQLSPIDIDHSQRPNRQVFGEIGRRKRHLMRRLEGINSRLRMQHIPYLEKLQKRLW 180
K LL++ P+ S RP V I R +H+ + I R R + ++++++W
Sbjct: 103 KELLVKSFPVVRVKSLRP---VVMAILRNTQHIDDKYLKILVRDRELYADTDTEVKRQIW 159
Query: 181 KEYQTTLIQE-----ELLWRQKSRVAWLNHGDKNTRFFHTSTMVRRKRNKIEALTDEHGV 235
++ Q+ E R+K + + +H + N FFH + VRR+ ++ L + G
Sbjct: 160 RDNQSLFGDEVSPLLSQYIREKEHILF-DHTNLNNLFFHPTPKVRRQGEVVQKLANMIGT 218
Query: 236 SVTDPITLRSMAIDFFQHLYTSPGTDVLYHVRNAFPTLPRDTLLEISNPLEADEI 290
SV L M + F + L+ RN R LL + LE EI
Sbjct: 219 SV----KLYDMVLQFLRTLF--------LRTRNVHYCTLRAELLMALHDLEVQEI 261
>GOA6_HUMAN (Q9NYA3) Golgi autoantigen, golgin subfamily A member 6
(Golgin linked to PML) (Golgin-like protein)
Length = 693
Score = 35.0 bits (79), Expect = 1.3
Identities = 22/65 (33%), Positives = 34/65 (51%), Gaps = 1/65 (1%)
Query: 137 RPNRQVFGEIGRRKRHLMRRLEGINSRLRMQHIPYLEKLQKRLWKEYQTTLIQEELLWRQ 196
R ++ E G R R +RL RLR + L+K +KRLW + + +EE L +Q
Sbjct: 368 REQQKTLQEQGERLRKQEQRLRKQEERLRKEE-ERLQKQEKRLWDQEERLWKKEERLQKQ 426
Query: 197 KSRVA 201
+ R+A
Sbjct: 427 EERLA 431
>CILA_KLEPN (P45413) Citrate lyase alpha chain (EC 4.1.3.6) (Citrase
alpha chain) (Citrate (pro-3S)-lyase alpha chain)
(Citrate CoA-transferase subunit) (EC 2.8.3.10)
Length = 508
Score = 35.0 bits (79), Expect = 1.3
Identities = 19/47 (40%), Positives = 28/47 (59%), Gaps = 2/47 (4%)
Query: 1010 GSLSDLVDEKRVGLQPNPLQLHSHLGFVEVSRNGADSIALSF--APC 1054
G L +E GL NP+Q+HSH G V++ ++G SI ++F PC
Sbjct: 119 GLRGKLGEEISAGLMENPVQIHSHGGRVQLIQSGELSIDVAFLGVPC 165
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.343 0.152 0.539
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 143,650,250
Number of Sequences: 164201
Number of extensions: 6025869
Number of successful extensions: 15202
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 15171
Number of HSP's gapped (non-prelim): 19
length of query: 1248
length of database: 59,974,054
effective HSP length: 122
effective length of query: 1126
effective length of database: 39,941,532
effective search space: 44974165032
effective search space used: 44974165032
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 72 (32.3 bits)
Lotus: description of TM0037a.3


