

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
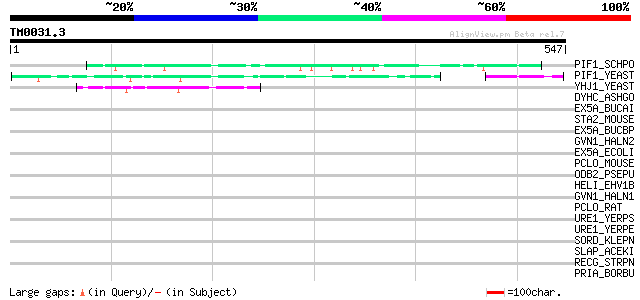
Query= TM0031.3
(547 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
PIF1_SCHPO (Q9UUA2) DNA repair and recombination protein pif1, m... 59 2e-08
PIF1_YEAST (P07271) DNA repair and recombination protein PIF1, m... 51 7e-06
YHJ1_YEAST (P38766) Hypothetical helicase in SLT2-PUT2 intergeni... 47 2e-04
DYHC_ASHGO (Q9C1M7) Dynein heavy chain, cytosolic (DYHC) 37 0.17
EX5A_BUCAI (P57530) Exodeoxyribonuclease V alpha chain (EC 3.1.1... 36 0.23
STA2_MOUSE (Q9WVL2) Signal transducer and activator of transcrip... 35 0.50
EX5A_BUCBP (Q89AB2) Exodeoxyribonuclease V alpha chain (EC 3.1.1... 34 0.86
GVN1_HALN2 (P57734) Gas vesicle protein gvpN 1 33 1.5
EX5A_ECOLI (P04993) Exodeoxyribonuclease V alpha chain (EC 3.1.1... 33 1.5
PCLO_MOUSE (Q9QYX7) Piccolo protein (Presynaptic cytomatrix prot... 33 2.5
ODB2_PSEPU (P09062) Lipoamide acyltransferase component of branc... 33 2.5
HELI_EHV1B (P28934) Probable helicase 33 2.5
GVN1_HALN1 (Q9HI16) Gas vesicle protein gvpN 1 33 2.5
PCLO_RAT (Q9JKS6) Piccolo protein (Multidomain presynaptic cytom... 32 4.3
URE1_YERPS (P52313) Urease alpha subunit (EC 3.5.1.5) (Urea amid... 32 5.6
URE1_YERPE (Q9ZFR9) Urease alpha subunit (EC 3.5.1.5) (Urea amid... 32 5.6
SORD_KLEPN (P37079) Sorbitol-6-phosphate 2-dehydrogenase (EC 1.1... 31 7.3
SLAP_ACEKI (P22258) Cell surface protein precursor (S-layer prot... 31 7.3
RECG_STRPN (Q54900) ATP-dependent DNA helicase recG (EC 3.6.1.-) 31 7.3
PRIA_BORBU (Q45032) Primosomal protein N' (Replication factor Y) 31 9.5
>PIF1_SCHPO (Q9UUA2) DNA repair and recombination protein pif1,
mitochondrial precursor
Length = 805
Score = 59.3 bits (142), Expect = 2e-08
Identities = 111/491 (22%), Positives = 195/491 (39%), Gaps = 86/491 (17%)
Query: 76 KNVLDDVLSDNS*FFFLYGFGGTGKTF----VWNILYAALRSRGLIVLNVASSGIASLLL 131
K +LD V+ FF G GTGK+ + +L + R + V AS+G+A+ +
Sbjct: 315 KRILDMVVEQQHSIFFT-GSAGTGKSVLLRKIIEVLKSKYRKQSDRVAVTASTGLAACNI 373
Query: 132 PGDRTAHSRFSIPISINEIS--TCNLRQGSPKAELLKKASLIIWDEAPMLNKHCIEALDR 189
G T HS + ++ + +++ + ++I DE M++ ++ L+
Sbjct: 374 -GGVTLHSFAGVGLARESVDLLVSKIKKNKKCVNRWLRTRVLIIDEVSMVDAELMDKLEE 432
Query: 190 SLNDIMKTQSTHGYDIPFGGKVVVLGDDFGQILPVISKGSRSEIVGSAINSSYLWKHCKV 249
I K PFGG +VL DF Q+ PV G S+ S WK
Sbjct: 433 VARVIRKDSK------PFGGIQLVLTGDFFQLPPVPENGKESKF----CFESQTWKSA-- 480
Query: 250 MKLTVNMSLQNATSTSSATEIKEFVDWLL-QVGDGTV-------KTIDEEETLIE---IP 298
L + L + +K + L ++ D +V +TI+ E+ L+ P
Sbjct: 481 --LDFTIGLTHVFRQKDEEFVKMLNELRLGKLSDESVRKFKVLNRTIEYEDGLLPTELFP 538
Query: 299 PDLLIEQCKEPLLELVN---FAYPKLAHNLQKSSFFQERAI---LAPTLESL-------- 344
+E+ + ++ +N + + + F++R + +AP L
Sbjct: 539 TRYEVERSNDMRMQQINQNPVTFTAIDSGTVRDKEFRDRLLQGCMAPATLVLKVNAQVML 598
Query: 345 -----EEINNFMLAMIPG---DETEYLSCDTPCKSDEDSGDNAE*FTSEVLNDFKCSEIP 396
+++ N L + G DET + K E G NA + S ++ F ++
Sbjct: 599 IKNIDDQLVNGSLGKVIGFIDDETYQME----KKDAEMQGRNAFEYDSLDISPFDLPDVK 654
Query: 397 NHAIKLKAGVPIMLIRNIDQAAGLCNGTRMIVNALTKYIIVVTVLNENKMGETTFIPRMS 456
KL A R + K+ +V L GE T + +
Sbjct: 655 QKKYKLIA-------------------MRKASSTAIKWPLVRFKLPNG--GERTIVVQRE 693
Query: 457 LTPSNSDIP---FKFQRRQFSVTLCFVMTINKSQGQSLSHVGLYLPRPVFTHGQPYVALY 513
N ++P + R Q + L + ++I+K+QGQ+L V + L R VF GQ YVAL
Sbjct: 694 TW--NIELPNGEVQASRSQIPLILAYAISIHKAQGQTLDRVKVDLGR-VFEKGQAYVALS 750
Query: 514 RVKSRKMLKML 524
R +++ L++L
Sbjct: 751 RATTQEGLQVL 761
>PIF1_YEAST (P07271) DNA repair and recombination protein PIF1,
mitochondrial precursor
Length = 857
Score = 51.2 bits (121), Expect = 7e-06
Identities = 98/437 (22%), Positives = 168/437 (38%), Gaps = 87/437 (19%)
Query: 2 EELQNLCMIEIEKILQGNERSLKEFP-------CLPYPKFSEIHNFEVIFVADELNYNRA 54
+ELQN + + + NE K+ P F+E+ N + D+ N N
Sbjct: 172 DELQNNSISQERSLEMINENEKKKMQFGEKIAVLTQRPSFTELQNDQ-----DDSNLNPH 226
Query: 55 EMVKIHDEFVSSLTSEQEIFYKNVLDDVLSDNS*FFFLYGFGGTGKTFVWNILYAALRSR 114
VK+ + L+ EQE K L++N F G GTGK+ + + L+
Sbjct: 227 NGVKV--KIPICLSKEQESIIK------LAENGHNIFYTGSAGTGKSILLREMIKVLK-- 276
Query: 115 GLI----VLNVASSGIASLLLPGDRTAHSRFSIPISINEISTCNLRQGSPKAELLKK--- 167
G+ V AS+G+A+ + G T HS F+ + + + G + L++
Sbjct: 277 GIYGRENVAVTASTGLAACNI-GGITIHS-FAGILGKGDADKLYKKVGRRSRKHLRRWEN 334
Query: 168 ASLIIWDEAPMLNKHCIEALDRSLNDIMKTQSTHGYDIPFGGKVVVLGDDFGQILPVISK 227
++ DE ML+ ++ LD I K PFGG ++ DF Q+ PV
Sbjct: 335 IGALVVDEISMLDAELLDKLDFIARKIRKNHQ------PFGGIQLIFCGDFFQLPPVSKD 388
Query: 228 GSRSEIVGSAINSSYLWKHCKVMKLTVNMSLQNATSTSSATEIKEFVDWLLQVGDGTVKT 287
+R + + ++ K K V M++ ++K F++ L ++ G +
Sbjct: 389 PNRP--------TKFAFE-SKAWKEGVKMTIMLQKVFRQRGDVK-FIEMLNRMRLGNIDD 438
Query: 288 IDEEETLIEIPPDLLIEQCKEPLLELVNFAYPKLAHNLQKSSFFQERAILAPTLESLEEI 347
E E + KL+ L A L T +E
Sbjct: 439 ETERE-------------------------FKKLSRPLPDDEIIP--AELYSTRMEVERA 471
Query: 348 NNFMLAMIPGDETEYLSCDTPCKSDEDSGDNAE*FTSEVLNDFKCSEIPNHAIKLKAGVP 407
NN L+ +PG + + D DE+ +L +F + + LK G
Sbjct: 472 NNSRLSKLPGQVHIFNAIDGGALEDEE-------LKERLLQNF----LAPKELHLKVGAQ 520
Query: 408 IMLIRNIDQAAGLCNGT 424
+M+++N+D A L NG+
Sbjct: 521 VMMVKNLD--ATLVNGS 535
Score = 44.7 bits (104), Expect = 6e-04
Identities = 32/77 (41%), Positives = 45/77 (57%), Gaps = 8/77 (10%)
Query: 470 RRQFSVTLCFVMTINKSQGQSLSHVGLYLPRPVFTHGQPYVALYRVKSRKMLKMLIIDDE 529
R Q + L + ++I+KSQGQ+L V + L R VF GQ YVAL R SR+ L++L D
Sbjct: 691 RVQLPLMLAWSLSIHKSQGQTLPKVKVDLRR-VFEKGQAYVALSRAVSREGLQVLNFD-- 747
Query: 530 *VVSNTTRNVVYQEVLD 546
TR +Q+V+D
Sbjct: 748 -----RTRIKAHQKVID 759
>YHJ1_YEAST (P38766) Hypothetical helicase in SLT2-PUT2 intergenic
region
Length = 723
Score = 46.6 bits (109), Expect = 2e-04
Identities = 53/186 (28%), Positives = 84/186 (44%), Gaps = 20/186 (10%)
Query: 67 LTSEQEIFYKNVLDDVLSDNS*FFFLYGFGGTGKTFVWNILYAALRS---RGLIVLNVAS 123
LT EQE V++ ++ + F+ G GTGK+ + + L S + I + AS
Sbjct: 232 LTMEQE----RVVNLIVKKRTNVFYT-GSAGTGKSVILQTIIRQLSSLYGKESIAIT-AS 285
Query: 124 SGIASLLLPGDRTAHSRFSIPISINEISTCNLRQGSPKAELL--KKASLIIWDEAPMLNK 181
+G+A++ + G T H I I I + S K L + ++I DE M++
Sbjct: 286 TGLAAVTIGGS-TLHKWSGIGIGNKTIDQLVKKIQSQKDLLAAWRYTKVLIIDEISMVDG 344
Query: 182 HCIEALDRSLNDIMKTQSTHGYDIPFGGKVVVLGDDFGQILPVISKGSRSEIVGSAINSS 241
+ ++ L++ I K D PFGG +VL DF Q+ PV K + V S
Sbjct: 345 NLLDKLEQIARRIRKN------DDPFGGIQLVLTGDFFQLPPVAKKDEHN--VVKFCFES 396
Query: 242 YLWKHC 247
+WK C
Sbjct: 397 EMWKRC 402
Score = 41.6 bits (96), Expect = 0.005
Identities = 25/68 (36%), Positives = 41/68 (59%), Gaps = 4/68 (5%)
Query: 463 DIPFK---FQRRQFSVTLCFVMTINKSQGQSLSHVGLYLPRPVFTHGQPYVALYRVKSRK 519
DIP + +R Q + LC+ ++I+K+QGQ++ + + L R +F GQ YVAL R +
Sbjct: 636 DIPRENVGLERTQIPLMLCWALSIHKAQGQTIQRLKVDL-RRIFEAGQVYVALSRAVTMD 694
Query: 520 MLKMLIID 527
L++L D
Sbjct: 695 TLQVLNFD 702
>DYHC_ASHGO (Q9C1M7) Dynein heavy chain, cytosolic (DYHC)
Length = 4083
Score = 36.6 bits (83), Expect = 0.17
Identities = 20/74 (27%), Positives = 41/74 (55%), Gaps = 7/74 (9%)
Query: 425 RMIVNALTKYIIVVTVLNENKMGETTFIPRMSLTPSNSDIPFKFQRRQFSVTLCFVMTIN 484
+M++ T+ + +++E+ + ET+F+ RM+ +NSD+P F+ ++ L +
Sbjct: 2795 QMLLRCGTESEKICLIIDESNILETSFLERMNTLLANSDVPGLFEADEYEALLSKI---- 2850
Query: 485 KSQGQSLSHVGLYL 498
GQ +S +GL L
Sbjct: 2851 ---GQRISQLGLLL 2861
>EX5A_BUCAI (P57530) Exodeoxyribonuclease V alpha chain (EC
3.1.11.5)
Length = 602
Score = 36.2 bits (82), Expect = 0.23
Identities = 38/134 (28%), Positives = 58/134 (42%), Gaps = 25/134 (18%)
Query: 405 GVPIMLIRNIDQAAGLCNGTRMIVNALTKYIIVVTVLNENKMGETTFIPRMSLTPSNSDI 464
G PIM+I N ++A + NG I N I+ V+ L EN + ++I
Sbjct: 481 GKPIMIINN-NRALNVSNGNIGITNINKNGILQVSFLKENN--------------TINNI 525
Query: 465 PFKFQRRQFSVTLCFVMTINKSQGQSLSHVGLYLPR---PVFTHGQPYVALYRVKSRKML 521
P K R + + +T++KSQG + L LP + Y + R SRK+L
Sbjct: 526 PVKILR---NYKTAWAITVHKSQGSEFMNTALILPNFNSHILNKDTLYTGITR--SRKIL 580
Query: 522 KMLIIDDE*VVSNT 535
I D+ + NT
Sbjct: 581 S--IFSDKKIFLNT 592
>STA2_MOUSE (Q9WVL2) Signal transducer and activator of
transcription 2
Length = 923
Score = 35.0 bits (79), Expect = 0.50
Identities = 26/87 (29%), Positives = 43/87 (48%), Gaps = 8/87 (9%)
Query: 292 ETLIEIPPDLLIEQCKEPLLELVNFAYPKLAHNLQKSSFFQERAILAPTLESLEEINNFM 351
+ L+E+ P +L+E + LLEL + AH LQ+ S + E L++I+
Sbjct: 821 QVLLELAPQVLLEPAPQVLLELAPQVQLEPAHLLQQPS-------ESDLPEDLQQISVED 873
Query: 352 LAMIPGDETEYLSC-DTPCKSDEDSGD 377
L + TEY++ + P + E SGD
Sbjct: 874 LKKLSNPSTEYITTNENPMLAGESSGD 900
>EX5A_BUCBP (Q89AB2) Exodeoxyribonuclease V alpha chain (EC
3.1.11.5)
Length = 618
Score = 34.3 bits (77), Expect = 0.86
Identities = 36/145 (24%), Positives = 63/145 (42%), Gaps = 35/145 (24%)
Query: 405 GVPIMLIRNIDQAAGLCNGTRMIVNALTKYIIVVTVLNENKMGETTFIPRMSLTPSNSDI 464
G PI++++N ++ L NG +T+L+ NK + F+P
Sbjct: 487 GKPILILKN-NEEMNLFNGE-----------CGLTLLDANKKLKVFFLP----------- 523
Query: 465 PFKFQRRQFSVTLCFV--------MTINKSQGQSLSHVGLYLPRPVFTHGQPYVALYRVK 516
K Q + +S+ + V MT++KSQG S V L LP + T +Y
Sbjct: 524 --KNQEQIYSIPIHLVPEHQTNWTMTVHKSQGSEFSEVVLILP-TIMTSILTKELIYTAV 580
Query: 517 SRKMLKMLIIDDE*V-VSNTTRNVV 540
+R K+ I DE + + + +N++
Sbjct: 581 TRSKKKLTIYSDENIFIKSLKKNII 605
>GVN1_HALN2 (P57734) Gas vesicle protein gvpN 1
Length = 347
Score = 33.5 bits (75), Expect = 1.5
Identities = 39/204 (19%), Positives = 80/204 (39%), Gaps = 27/204 (13%)
Query: 51 YNRAEMVKIHDEFVSSLTSEQEIFYKNVLDDVLSDNS*FFFLYGFGGTGKTFVWNILYAA 110
Y E + HD+F+ ++ ++I +D+ L+ G T V+N
Sbjct: 130 YAEKERISEHDQFIHNVVKSKDIIRDRWVDNPLTLAV---------QEGATLVYNEFSRT 180
Query: 111 LRSRGLIVLNVASSGIASLLLPGDR------TAHSRFSIPISINEISTCNLRQGSPKAEL 164
++L+V G+ L LPG R H F ++ N + + + P+ L
Sbjct: 181 KPVANNVLLSVFEEGV--LELPGKRGKSRYVDVHPEFRTILTSNSVEYAGVHE--PQDAL 236
Query: 165 LKKASLIIWD------EAPMLNKHCIEALDRSLNDIMKTQSTHG--YDIPFGGKVVVLGD 216
L + I D E ++ H ++ D ++ DI++ G DI G + ++ +
Sbjct: 237 LDRLIGIYMDFYDLDTEIEIVRAHVDKSADTNVEDIVRVLRELGERLDITVGTRAAIMAN 296
Query: 217 DFGQILPVISKGSRSEIVGSAINS 240
+ + + + ++I + S
Sbjct: 297 EGATTVDTVDQAVLTDICTDVLAS 320
>EX5A_ECOLI (P04993) Exodeoxyribonuclease V alpha chain (EC
3.1.11.5) (Exodeoxyribonuclease V 67 kDa polypeptide)
Length = 608
Score = 33.5 bits (75), Expect = 1.5
Identities = 37/120 (30%), Positives = 49/120 (40%), Gaps = 22/120 (18%)
Query: 405 GVPIMLIRNIDQAAGLCNGTRMIVNALTKYIIVVTVLNENKMGETTFIPRMSLTPSNSDI 464
G P+M+ RN D A GL NG I AL + G+ T R+ + +I
Sbjct: 479 GRPVMIARN-DSALGLFNGDIGI--ALDR-------------GQGT---RVWFAMPDGNI 519
Query: 465 PFKFQRRQFSVTLCFVMTINKSQGQSLSHVGLYLP---RPVFTHGQPYVALYRVKSRKML 521
R + MT++KSQG H L LP PV T Y A+ R + R L
Sbjct: 520 KSVQPSRLPEHETTWAMTVHKSQGSEFDHAALILPSQRTPVVTRELVYTAVTRARRRLSL 579
>PCLO_MOUSE (Q9QYX7) Piccolo protein (Presynaptic cytomatrix protein)
(Aczonin) (Brain-derived HLMN protein)
Length = 5038
Score = 32.7 bits (73), Expect = 2.5
Identities = 29/113 (25%), Positives = 49/113 (42%), Gaps = 9/113 (7%)
Query: 249 VMKLTVNMSLQNATSTSSATEIKEFVDWLLQVGDGTVKTIDEEETLIEIPPDLLIEQ--- 305
V + +N SLQ T + E + V L + + + EE TL+ +P + +Q
Sbjct: 3055 VTSIDINASLQTITMETLPAETMDSVPTLTTASEVFSEVVGEESTLLIVPDEDKQQQQLD 3114
Query: 306 CKEPLLELVNFAYPKLAHNL----QKSSFF--QERAILAPTLESLEEINNFML 352
+ LLEL + A L Q+ F QE+ ++ LE L+ + +L
Sbjct: 3115 LERELLELEKIKQQRFAEELEWERQEIQRFREQEKIMVQKKLEELQSMKQHLL 3167
>ODB2_PSEPU (P09062) Lipoamide acyltransferase component of
branched-chain alpha-keto acid dehydrogenase complex (EC
2.3.1.168) (Dihydrolipoyllysine-residue
(2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide
branched chain transacylase)
Length = 423
Score = 32.7 bits (73), Expect = 2.5
Identities = 19/53 (35%), Positives = 30/53 (55%)
Query: 184 IEALDRSLNDIMKTQSTHGYDIPFGGKVVVLGDDFGQILPVISKGSRSEIVGS 236
I A D+ + D+M ++T P GKV+ LG G+++ V S+ R E+ GS
Sbjct: 30 IIAEDQVVADVMTDKATVEIPSPVSGKVLALGGQPGEVMAVGSELIRIEVEGS 82
>HELI_EHV1B (P28934) Probable helicase
Length = 881
Score = 32.7 bits (73), Expect = 2.5
Identities = 18/43 (41%), Positives = 23/43 (52%)
Query: 481 MTINKSQGQSLSHVGLYLPRPVFTHGQPYVALYRVKSRKMLKM 523
MTI +SQG SL V + PR YVA+ R S + L+M
Sbjct: 807 MTIARSQGLSLDKVAICFPRNNLRINSVYVAMSRTVSSRFLRM 849
>GVN1_HALN1 (Q9HI16) Gas vesicle protein gvpN 1
Length = 347
Score = 32.7 bits (73), Expect = 2.5
Identities = 38/204 (18%), Positives = 80/204 (38%), Gaps = 27/204 (13%)
Query: 51 YNRAEMVKIHDEFVSSLTSEQEIFYKNVLDDVLSDNS*FFFLYGFGGTGKTFVWNILYAA 110
Y E + HD+F+ ++ ++I +D+ L+ G T V+N
Sbjct: 130 YAEKERISEHDQFIHNVVKSKDIIRDRWVDNPLTLAV---------QEGATLVYNEFSRT 180
Query: 111 LRSRGLIVLNVASSGIASLLLPGDR------TAHSRFSIPISINEISTCNLRQGSPKAEL 164
++L+V G+ L LPG R H F ++ N + + + P+ L
Sbjct: 181 KPVANNVLLSVFEEGV--LELPGKRGKSRYVDVHPEFRTILTSNSVEYAGVHE--PQDAL 236
Query: 165 LKKASLIIWD------EAPMLNKHCIEALDRSLNDIMKT--QSTHGYDIPFGGKVVVLGD 216
L + I D E ++ H ++ D ++ DI++ + DI G + ++ +
Sbjct: 237 LDRLIGIYMDFYDLDTEIEIVRAHVDKSADTNVEDIVRVLRELRERLDITVGTRAAIMAN 296
Query: 217 DFGQILPVISKGSRSEIVGSAINS 240
+ + + + ++I + S
Sbjct: 297 EGATTVDTVDQAVLTDICTDVLAS 320
>PCLO_RAT (Q9JKS6) Piccolo protein (Multidomain presynaptic cytomatrix
protein)
Length = 5085
Score = 32.0 bits (71), Expect = 4.3
Identities = 29/113 (25%), Positives = 49/113 (42%), Gaps = 9/113 (7%)
Query: 249 VMKLTVNMSLQNATSTSSATEIKEFVDWLLQVGDGTVKTIDEEETLIEIPPDLLIEQ--- 305
V + +N SLQ T + E + V L + + + EE TL+ +P + +Q
Sbjct: 3101 VTGIDINASLQTITMETLPAETMDSVPTLTTASEVFSEVVGEESTLLIVPDEDKQQQQLD 3160
Query: 306 CKEPLLELVNFAYPKLAHNL----QKSSFF--QERAILAPTLESLEEINNFML 352
+ LLEL + A L Q+ F QE+ ++ LE L+ + +L
Sbjct: 3161 LERELLELEKIKQQRFAEELEWERQEIQRFREQEKIMVQKKLEELQSMKQHLL 3213
>URE1_YERPS (P52313) Urease alpha subunit (EC 3.5.1.5) (Urea
amidohydrolase)
Length = 571
Score = 31.6 bits (70), Expect = 5.6
Identities = 40/150 (26%), Positives = 58/150 (38%), Gaps = 15/150 (10%)
Query: 27 PCLPYPKFSEIHNFEVIFVADELNYNRAEMVKIHDEFVSSLTSEQEIFYKNVLDD--VLS 84
P LPY S+ F++I V LN N V F S + I +NVL D V+S
Sbjct: 304 PTLPYGVNSQAELFDMIMVCHNLNPN----VPADVSFAESRVRPETIAAENVLHDMGVIS 359
Query: 85 DNS*FFFLYGFGGTGKTFVWNILYAALRSRGLIVLNVASSGIASLLLPGDRTAHSRFSIP 144
S G G + A +RG + + A + +L R+
Sbjct: 360 MFSSDSQAMGRVGENWLRILQTADAMKAARGKLPEDAAGNDNFRVL---------RYVAK 410
Query: 145 ISINEISTCNLRQGSPKAELLKKASLIIWD 174
I+IN T + E+ K A L++WD
Sbjct: 411 ITINPAITQGVSHVIGSVEVGKMADLVLWD 440
>URE1_YERPE (Q9ZFR9) Urease alpha subunit (EC 3.5.1.5) (Urea
amidohydrolase)
Length = 571
Score = 31.6 bits (70), Expect = 5.6
Identities = 40/150 (26%), Positives = 58/150 (38%), Gaps = 15/150 (10%)
Query: 27 PCLPYPKFSEIHNFEVIFVADELNYNRAEMVKIHDEFVSSLTSEQEIFYKNVLDD--VLS 84
P LPY S+ F++I V LN N V F S + I +NVL D V+S
Sbjct: 304 PTLPYGVNSQAELFDMIMVCHNLNPN----VPADVSFAESRVRPETIAAENVLHDMGVIS 359
Query: 85 DNS*FFFLYGFGGTGKTFVWNILYAALRSRGLIVLNVASSGIASLLLPGDRTAHSRFSIP 144
S G G + A +RG + + A + +L R+
Sbjct: 360 MFSSDSQAMGRVGENWLRILQTADAMKAARGKLPEDAAGNDNFRVL---------RYVAK 410
Query: 145 ISINEISTCNLRQGSPKAELLKKASLIIWD 174
I+IN T + E+ K A L++WD
Sbjct: 411 ITINPAITQGVSHVIGSVEVGKMADLVLWD 440
>SORD_KLEPN (P37079) Sorbitol-6-phosphate 2-dehydrogenase (EC
1.1.1.140) (Glucitol-6-phosphate dehydrogenase)
(Ketosephosphate reductase)
Length = 267
Score = 31.2 bits (69), Expect = 7.3
Identities = 23/79 (29%), Positives = 38/79 (47%), Gaps = 8/79 (10%)
Query: 261 ATSTSSATEIKEFVDWLLQVG---DGTVKTIDEEETLIEIPPDLLIEQCKEPLLELVNFA 317
+T SSATE+++ +D ++Q DG V + P L+ E+ EL A
Sbjct: 57 STDISSATEVQQTIDAIIQRWSRIDGLVNNAG-----VNFPRLLVDEKAPAGRYELNEAA 111
Query: 318 YPKLAHNLQKSSFFQERAI 336
+ K+ + QK FF +A+
Sbjct: 112 FEKMVNINQKGVFFMSQAV 130
>SLAP_ACEKI (P22258) Cell surface protein precursor (S-layer
protein)
Length = 762
Score = 31.2 bits (69), Expect = 7.3
Identities = 30/108 (27%), Positives = 46/108 (41%), Gaps = 3/108 (2%)
Query: 26 FPCLPYPKFSEIHNFEVIFVADELNYNRAEMVKIHDEFVSSLTSEQEIFYKNVLDDVLSD 85
+ L + N V VAD NYN A++V ++ S T+ Y DV +
Sbjct: 587 YKALKLTDLKDATNLNVKIVAD--NYNVAKVVVFNNASFVSTTTSTVYAYVTGTADVYVN 644
Query: 86 NS*FFFLYGF-GGTGKTFVWNILYAALRSRGLIVLNVASSGIASLLLP 132
S F L G KT+ N A + +VL + ++ IA++ LP
Sbjct: 645 GSTFTRLTVLENGQTKTYDANAQLATNYTHKAVVLTLTNAKIANIALP 692
>RECG_STRPN (Q54900) ATP-dependent DNA helicase recG (EC 3.6.1.-)
Length = 671
Score = 31.2 bits (69), Expect = 7.3
Identities = 25/94 (26%), Positives = 44/94 (46%), Gaps = 11/94 (11%)
Query: 49 LNYNRAEMVKIHDEFVSSLTSEQEIFYKNVLDDVLSDNS*FFFLYGFGGTGKTFVWNILY 108
LN+++ ++ + +LT QE + +L D+ SD+ L G G+GKT V
Sbjct: 235 LNWSQEKVTAVKVSLPFALTQAQEKSLQEILTDMKSDHHMNRLLQGDVGSGKTVV----- 289
Query: 109 AALRSRGLIVLNVASSGI-ASLLLPGDRTAHSRF 141
GL + ++G A+L++P + A F
Sbjct: 290 -----AGLAMFAAVTAGYQAALMVPTEILAEQHF 318
>PRIA_BORBU (Q45032) Primosomal protein N' (Replication factor Y)
Length = 660
Score = 30.8 bits (68), Expect = 9.5
Identities = 20/62 (32%), Positives = 34/62 (54%), Gaps = 6/62 (9%)
Query: 67 LTSEQEIFYKNVLDDVLSDNS*FFFLYGFGGTGKTFVWNIL---YAALRSRGLIVLNVAS 123
L +EQ+ YK ++ S+ + F+L+G G+GKT ++ L Y AL + L ++ S
Sbjct: 134 LNNEQQNIYKEIIG---SEKTNVFYLFGIPGSGKTEIFIKLCEYYLALEQQVLFLIPEIS 190
Query: 124 SG 125
G
Sbjct: 191 LG 192
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.139 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 61,038,228
Number of Sequences: 164201
Number of extensions: 2524223
Number of successful extensions: 5995
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 5978
Number of HSP's gapped (non-prelim): 31
length of query: 547
length of database: 59,974,054
effective HSP length: 115
effective length of query: 432
effective length of database: 41,090,939
effective search space: 17751285648
effective search space used: 17751285648
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0031.3


