

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
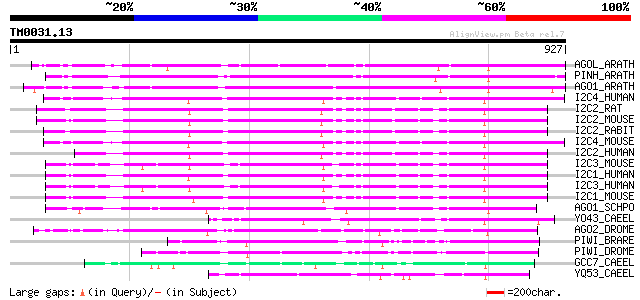
Query= TM0031.13
(927 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AGOL_ARATH (Q9SJK3) Argonaute-like protein At2g27880 525 e-148
PINH_ARATH (Q9XGW1) PINHEAD protein (ZWILLE protein) 493 e-139
AGO1_ARATH (O04379) Argonaute protein 480 e-135
I2C4_HUMAN (Q9HCK5) Eukaryotic translation initiation factor 2C ... 387 e-106
I2C2_RAT (Q9QZ81) Eukaryotic translation initiation factor 2C 2 ... 382 e-105
I2C2_MOUSE (Q8CJG0) Eukaryotic translation initiation factor 2C ... 380 e-105
I2C2_RABIT (O77503) Eukaryotic translation initiation factor 2C ... 379 e-104
I2C4_MOUSE (Q8CJF8) Eukaryotic translation initiation factor 2C ... 375 e-103
I2C2_HUMAN (Q9UKV8) Eukaryotic translation initiation factor 2C ... 375 e-103
I2C3_MOUSE (Q8CJF9) Eukaryotic translation initiation factor 2C ... 372 e-102
I2C1_HUMAN (Q9UL18) Eukaryotic translation initiation factor 2C ... 371 e-102
I2C3_HUMAN (Q9H9G7) Eukaryotic translation initiation factor 2C ... 370 e-102
I2C1_MOUSE (Q8CJG1) Eukaryotic translation initiation factor 2C ... 364 e-100
AGO1_SCHPO (O74957) Cell cycle control protein ago1 (RNA interfe... 313 1e-84
YO43_CAEEL (P34681) Hypothetical protein ZK757.3 in chromosome III 266 1e-70
AGO2_DROME (Q9VUQ5) Argonaute 2 protein 241 6e-63
PIWI_BRARE (Q8UVX0) Piwi protein 154 1e-36
PIWI_DROME (Q9VKM1) Piwi protein 135 5e-31
GCC7_CAEEL (Q21770) Germ cell expressed protein R06C7.1 97 2e-19
YQ53_CAEEL (Q09249) Hypothetical protein C16C10.3 in chromosome III 94 2e-18
>AGOL_ARATH (Q9SJK3) Argonaute-like protein At2g27880
Length = 997
Score = 525 bits (1351), Expect = e-148
Identities = 350/921 (38%), Positives = 499/921 (54%), Gaps = 84/921 (9%)
Query: 37 KVDLLDLPPEPVKKKLPTRLPIARKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALS 96
K ++ LPP K P+ R G G+ G K+ + NHF V VA+ D + Y V+++
Sbjct: 131 KPEMTSLPPASSKA---VTFPV-RPGRGTLGKKVMVRANHFLVQVADRD--LYHYDVSIN 184
Query: 97 YEDGRPVEGKGVGRKVIDKVQETY-GSELNGKDFAYDGEKTLFTIGSLARNKLEFTVVLE 155
E V K V R V+ + + Y S L GK AYDG K+L+T G L + EF V L
Sbjct: 185 PE----VISKTVNRNVMKLLVKNYKDSHLGGKSPAYDGRKSLYTAGPLPFDSKEFVVNLA 240
Query: 156 DVISNRNNGNCSPDGASTNDSDKKRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESE 215
+ + DG+S D + FKV + L + L ++ E
Sbjct: 241 EKRA---------DGSSGKD------------RPFKVAVKNVTSTDLYQLQQFLDRKQRE 279
Query: 216 NYQEAIRVLDIILRQHAAKQGCLLVRQSFFHNDPKNYADVGGGVLG-----CRGFHSSFR 270
+ I+VLD++LR + + V +SFFH A G G LG RG+ S R
Sbjct: 280 APYDTIQVLDVVLRDKPSND-YVSVGRSFFHTSLGKDARDGRGELGDGIEYWRGYFQSLR 338
Query: 271 TTQSGLSLNIDVSTTMIIQPGPVVDFLIANQNVRD---PF-SLDWAKAKRTLKNLRIKAS 326
TQ GLSLNIDVS +P V DF+ N+RD P D K K+ L+ L++K
Sbjct: 339 LTQMGLSLNIDVSARSFYEPIVVTDFISKFLNIRDLNRPLRDSDRLKVKKVLRTLKVKLL 398
Query: 327 PSN--QEYKITGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSA 384
N + KI+G+S LP +E FT++ K E TV +YF ++Y A
Sbjct: 399 HWNCTKSAKISGISSLPIRELRFTLEDKS-----------EKTVVQYFAEKYNYRVKYQA 447
Query: 385 DLPCINVGKPKRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQAL 444
LP I G RP Y+P+ELC + QRYTK L Q ++L++ + Q+P +R N + +
Sbjct: 448 -LPAIQTGSDTRPVYLPMELCQIDEGQRYTKRLNEKQVTALLKATCQRPPDRENSIKNLV 506
Query: 445 KTSNYGNEPMLKNCGITIASGFTQVEGRVLQAPRLKF---GNGEDFNPRNGRWNLNNKKV 501
+NY N+ + K G+++ + +E RVL P LK+ G + NPR G+WN+ +KK+
Sbjct: 507 VKNNY-NDDLSKEFGMSVTTQLASIEARVLPPPMLKYHDSGKEKMVNPRLGQWNMIDKKM 565
Query: 502 VRPAKIEHWAVVNFSARCDVRGLVRDLIKCARLKGIPIDEPYEEIFEENGQFRRAPPLVR 561
V AK+ W V+FS R D RGL ++ C +L G+ + + E + F PP
Sbjct: 566 VNGAKVTSWTCVSFSTRID-RGLPQEF--CKQLIGMCVSKGMEFKPQPAIPFISCPP-EH 621
Query: 562 VEKMFERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCISPTRVND--- 618
+E+ I K PG L+ +LP+ S YG K+ E GIV+QC P +VN
Sbjct: 622 IEEALLDIHKRAPGL-QLLIVILPDVTGS--YGKIKRICETELGIVSQCCQPRQVNKLNK 678
Query: 619 QYLTNVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAV 678
QY+ NV +KIN K GG N+VL + +IP+++ PTII+G DV+H PG+ PSIAAV
Sbjct: 679 QYMENVALKINVKTGGRNTVLNDAIRRNIPLITDRPTIIMGADVTHPQPGEDSSPSIAAV 738
Query: 679 VSSREWPLISKYRACVRTQSPKVEMIDNLFKQVSEKE----DEGIIRELLIDFYSSSGKR 734
V+S +WP I+KYR V Q+ + E+I +L+K V + + G+IRE I F ++G+
Sbjct: 739 VASMDWPEINKYRGLVSAQAHREEIIQDLYKLVQDPQRGLVHSGLIREHFIAFRRATGQI 798
Query: 735 KPDNIIIFRDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFF--Q 792
P II +RDGVSE QF+QVL E+ I +AC L E + P+ ++ QK HHT+ F Q
Sbjct: 799 -PQRIIFYRDGVSEGQFSQVLLHEMTAIRKACNSLQENYVPRVTFVIVQKRHHTRLFPEQ 857
Query: 793 PGSPD------NVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPD 846
G+ D N+ PGTV+D KICHP DFY+ +HAG+ GTSRP HYHVLLD+ GF+ D
Sbjct: 858 HGNRDMTDKSGNIQPGTVVDTKICHPNEFDFYLNSHAGIQGTSRPAHYHVLLDENGFTAD 917
Query: 847 ELQELVHSLSYVYQRSTTAISVVAPICYAHLAATQIGQFMKFEDKSDTSSSHGGLTAAGV 906
+LQ L ++L Y Y R T ++S+V P YAHLAA + +M+ E SD SS + GV
Sbjct: 918 QLQMLTNNLCYTYARCTKSVSIVPPAYYAHLAAFRARYYMESE-MSDGGSSRSRSSTTGV 976
Query: 907 APVVPQLPKLQDSVSSSMFFC 927
V+ QLP ++D+V MF+C
Sbjct: 977 GQVISQLPAIKDNVKEVMFYC 997
>PINH_ARATH (Q9XGW1) PINHEAD protein (ZWILLE protein)
Length = 988
Score = 493 bits (1270), Expect = e-139
Identities = 322/902 (35%), Positives = 487/902 (53%), Gaps = 78/902 (8%)
Query: 60 RKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQET 119
R G G+ GTK + NHF + D + QY V ++ E V K V R +I ++
Sbjct: 131 RPGFGTLGTKCIVKANHFLADLPTKDLN--QYDVTITPE----VSSKSVNRAIIAELVRL 184
Query: 120 YG-SELNGKDFAYDGEKTLFTIGSLARNKLEFTVVLEDVISNRNNGNCSPDGASTNDSDK 178
Y S+L + AYDG K+L+T G L EF+V + D NG
Sbjct: 185 YKESDLGRRLPAYDGRKSLYTAGELPFTWKEFSVKIVDEDDGIING-------------- 230
Query: 179 KRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHAAKQGCL 238
P +++KV I F A+ + + L G+ ++ QEA+++LDI+LR+ + K+ C
Sbjct: 231 -----PKRERSYKVAIKFVARANMHHLGEFLAGKRADCPQEAVQILDIVLRELSVKRFCP 285
Query: 239 LVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDF-- 296
+ R SFF D K +G G+ GF+ S R TQ GLSLNID+++ I+P PV++F
Sbjct: 286 VGR-SFFSPDIKTPQRLGEGLESWCGFYQSIRPTQMGLSLNIDMASAAFIEPLPVIEFVA 344
Query: 297 -LIANQNVRDPFS-LDWAKAKRTLKNLRIKASPS---NQEYKITGLSELPCKEQTFTMKK 351
L+ + P S D K K+ L+ ++++ + ++Y++ GL+ P +E F +
Sbjct: 345 QLLGKDVLSKPLSDSDRVKIKKGLRGVKVEVTHRANVRRKYRVAGLTTQPTRELMFPV-- 402
Query: 352 KGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTYVPVELCSLVSLQ 411
+E+ T + +V EYF ++++ LPC+ VG K+ +Y+P+E C +V Q
Sbjct: 403 ------DENCTMK--SVIEYFQEMYGFTIQHT-HLPCLQVGNQKKASYLPMEACKIVEGQ 453
Query: 412 RYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNYGNEPMLKNCGITIASGFTQVEG 471
RYTK L Q ++L++ + Q+P +R N + + ++ + Y +P K G+ I+ VE
Sbjct: 454 RYTKRLNEKQITALLKVTCQRPRDRENDILRTVQHNAYDQDPYAKEFGMNISEKLASVEA 513
Query: 472 RVLQAPRLKF---GNGEDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSARCD---VRGLV 525
R+L AP LK+ G +D P+ G+WN+ NKK++ + WA VNFS RG
Sbjct: 514 RILPAPWLKYHENGKEKDCLPQVGQWNMMNKKMINGMTVSRWACVNFSRSVQENVARGFC 573
Query: 526 RDLIKCARLKGIPID-EPYEEIFEEN-GQFRRAPPLVRVEKMFERIQKELPGAPSFLLCL 583
+L + + G+ + EP I+ Q +A V M + KEL LL +
Sbjct: 574 NELGQMCEVSGMEFNPEPVIPIYSARPDQVEKALKHVYHTSMNKTKGKEL----ELLLAI 629
Query: 584 LPERKNSDLYGPWKKKNLAEYGIVTQCISPT---RVNDQYLTNVLMKINAKLGGLNSVLG 640
LP+ N LYG K+ E G+++QC +++ QYL NV +KIN K+GG N+VL
Sbjct: 630 LPDN-NGSLYGDLKRICETELGLISQCCLTKHVFKISKQYLANVSLKINVKMGGRNTVLV 688
Query: 641 VEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPK 700
++ IP+VS +PTII G DV+H G+ PSIAAVV+S++WP ++KY V Q+ +
Sbjct: 689 DAISCRIPLVSDIPTIIFGADVTHPENGEESSPSIAAVVASQDWPEVTKYAGLVCAQAHR 748
Query: 701 VEMIDNLFKQ----VSEKEDEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLN 756
E+I +L+K V G+IR+LLI F ++G+ KP II +RDGVSE QF QVL
Sbjct: 749 QELIQDLYKTWQDPVRGTVSGGMIRDLLISFRKATGQ-KPLRIIFYRDGVSEGQFYQVLL 807
Query: 757 IELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFFQPGSPD--------NVPPGTVIDNK 808
EL+ I +AC L+ + P IV QK HHT+ F D N+ PGTV+D K
Sbjct: 808 YELDAIRKACASLEPNYQPPVTFIVVQKRHHTRLFANNHRDKNSTDRSGNILPGTVVDTK 867
Query: 809 ICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAISV 868
ICHP DFY+C+HAG+ GTSRP HYHVL D+ F+ D +Q L ++L Y Y R T ++S+
Sbjct: 868 ICHPTEFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADGIQSLTNNLCYTYARCTRSVSI 927
Query: 869 VAPICYAHLAATQIGQFMK---FEDKSDTSSSHGGLTAAGVAPVVPQLPKLQDSVSSSMF 925
V P YAHLAA + +++ +D + T G V P LP L+++V MF
Sbjct: 928 VPPAYYAHLAAFRARFYLEPEIMQDNGSPGKKNTKTTTVGDVGVKP-LPALKENVKRVMF 986
Query: 926 FC 927
+C
Sbjct: 987 YC 988
>AGO1_ARATH (O04379) Argonaute protein
Length = 1048
Score = 480 bits (1235), Expect = e-135
Identities = 322/948 (33%), Positives = 492/948 (50%), Gaps = 87/948 (9%)
Query: 24 PPPPIVPADVEPVKVDL----LDLPPEPVKKKLPTRLPIARKGLGSKGTKLPLLTNHFKV 79
P P ++ E + V+ + P P K + P+ R G G G + + NHF
Sbjct: 144 PEPTVLAQQFEQLSVEQGAPSQAIQPIPSSSKA-FKFPM-RPGKGQSGKRCIVKANHFFA 201
Query: 80 TVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQETY-GSELNGKDFAYDGEKTLF 138
+ + D H Y V ++ E V +GV R V+ ++ + Y S L + AYDG K+L+
Sbjct: 202 ELPDKDLH--HYDVTITPE----VTSRGVNRAVMKQLVDNYRDSHLGSRLPAYDGRKSLY 255
Query: 139 TIGSLARNKLEFTVVLEDVISNRNNGNCSPDGASTNDSDKKRMRRPYHSKTFKVEISFAA 198
T G L N EF + L D ++R R FKV I A
Sbjct: 256 TAGPLPFNSKEFRINLLD--------------EEVGAGGQRRERE------FKVVIKLVA 295
Query: 199 KIPLQAIVNALRGQESENYQEAIRVLDIILRQHAAKQGCLLVRQSFFHNDPKNYADVGGG 258
+ L + L G++S+ QEA++VLDI+LR+ + + V +SF+ D +G G
Sbjct: 296 RADLHHLGMFLEGKQSDAPQEALQVLDIVLRELPTSR-YIPVGRSFYSPDIGKKQSLGDG 354
Query: 259 VLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDFL--IANQNVRD-PFS-LDWAKA 314
+ RGF+ S R TQ GLSLNID+S+T I+ PV+ F+ + N+++ P S D K
Sbjct: 355 LESWRGFYQSIRPTQMGLSLNIDMSSTAFIEANPVIQFVCDLLNRDISSRPLSDADRVKI 414
Query: 315 KRTLKNLRIKASPSN---QEYKITGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEY 371
K+ L+ ++++ + ++Y+I+GL+ + +E TF + D + +V EY
Sbjct: 415 KKALRGVKVEVTHRGNMRRKYRISGLTAVATRELTFPV----------DERNTQKSVVEY 464
Query: 372 FVNYRKIDLRYSADLPCINVGKPKRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQ 431
F ++++ LPC+ VG RP Y+P+E+C +V QRY+K L Q ++L++ + Q
Sbjct: 465 FHETYGFRIQHT-QLPCLQVGNSNRPNYLPMEVCKIVEGQRYSKRLNERQITALLKVTCQ 523
Query: 432 KPLERMNVLNQALKTSNYGNEPMLKNCGITIASGFTQVEGRVLQAPRLKF---GNGEDFN 488
+P++R + Q ++ ++Y + + GI I++ VE R+L P LK+ G
Sbjct: 524 RPIDREKDILQTVQLNDYAKDNYAQEFGIKISTSLASVEARILPPPWLKYHESGREGTCL 583
Query: 489 PRNGRWNLNNKKVVRPAKIEHWAVVNFSARCD---VRGLVRDLIKCARLKGIPIDEPYEE 545
P+ G+WN+ NKK++ + +W +NFS + R ++L + + G+ + E
Sbjct: 584 PQVGQWNMMNKKMINGGTVNNWICINFSRQVQDNLARTFCQELAQMCYVSGMAFNP--EP 641
Query: 546 IFEENGQFRRAPPLVRVEKMFERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYG 605
+ V + + K G LL ++ N LYG K+ E G
Sbjct: 642 VLPPVSARPEQVEKVLKTRYHDATSKLSQGKEIDLLIVILPDNNGSLYGDLKRICETELG 701
Query: 606 IVTQCISPTRV---NDQYLTNVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDV 662
IV+QC V + QY+ NV +KIN K+GG N+VL ++ IP+VS PTII G DV
Sbjct: 702 IVSQCCLTKHVFKMSKQYMANVALKINVKVGGRNTVLVDALSRRIPLVSDRPTIIFGADV 761
Query: 663 SHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDE----G 718
+H PG+ PSIAAVV+S++WP I+KY V Q+ + E+I +LFK+ + + G
Sbjct: 762 THPHPGEDSSPSIAAVVASQDWPEITKYAGLVCAQAHRQELIQDLFKEWKDPQKGVVTGG 821
Query: 719 IIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFL 778
+I+ELLI F S+G KP II +RDGVSE QF QVL EL+ I +AC L+ + P
Sbjct: 822 MIKELLIAFRRSTG-HKPLRIIFYRDGVSEGQFYQVLLYELDAIRKACASLEAGYQPPVT 880
Query: 779 VIVAQKNHHTKFFQPGSPD--------NVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSR 830
+V QK HHT+ F D N+ PGTV+D+KICHP DFY+C+HAG+ GTSR
Sbjct: 881 FVVVQKRHHTRLFAQNHNDRHSVDRSGNILPGTVVDSKICHPTEFDFYLCSHAGIQGTSR 940
Query: 831 PTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAISVVAPICYAHLAATQIGQFMKFED 890
P HYHVL D+ F+ D LQ L ++L Y Y R T ++S+V P YAHLAA + +M+ E
Sbjct: 941 PAHYHVLWDENNFTADGLQSLTNNLCYTYARCTRSVSIVPPAYYAHLAAFRARFYMEPET 1000
Query: 891 KSDTSSSHGGLTAAG-----------VAPVVPQLPKLQDSVSSSMFFC 927
S + G + G V V LP L+++V MF+C
Sbjct: 1001 SDSGSMASGSMARGGGMAGRSTRGPNVNAAVRPLPALKENVKRVMFYC 1048
>I2C4_HUMAN (Q9HCK5) Eukaryotic translation initiation factor 2C 4
(eIF2C 4) (eIF-2C 4) (Argonaute 4)
Length = 861
Score = 387 bits (993), Expect = e-106
Identities = 287/903 (31%), Positives = 454/903 (49%), Gaps = 90/903 (9%)
Query: 57 PIARKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKV 116
P R GLG+ G + LL NHF+V + D + Y V + E RP + V R+V+D +
Sbjct: 15 PPRRPGLGTVGKPIRLLANHFQVQIPKID--VYHYDVDIKPEK-RP---RRVNREVVDTM 68
Query: 117 QETYGSELNG-KDFAYDGEKTLFTIGSL--ARNKLEFTVVLEDVISNRNNGNCSPDGAST 173
+ ++ G + YDG++ ++T L R++++ V L +G
Sbjct: 69 VRHFKMQIFGDRQPGYDGKRNMYTAHPLPIGRDRVDMEVTLPG------------EGKD- 115
Query: 174 NDSDKKRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHAA 233
+TFKV + + + + LQ ++ AL G +E ++++ LD+I R H
Sbjct: 116 --------------QTFKVSVQWVSVVSLQLLLEALAGHLNEVPDDSVQALDVITR-HLP 160
Query: 234 KQGCLLVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPV 293
V +SFF Y +GGG GFH S R + LNIDVS T + P+
Sbjct: 161 SMRYTPVGRSFFSPPEGYYHPLGGGREVWFGFHQSVRPAMWNMMLNIDVSATAFYRAQPI 220
Query: 294 VDFL-----IANQNVRDPFSLDWAKAKRT--LKNLRIKASPSNQ---EYKITGLSELPCK 343
++F+ I N N + D + K T ++ L+++ + Q +Y++ ++ P
Sbjct: 221 IEFMCEVLDIQNINEQTKPLTDSQRVKFTKEIRGLKVEVTHCGQMKRKYRVCNVTRRPAS 280
Query: 344 EQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTYVPVE 403
QTF ++ + G E TV +YF + L+Y LPC+ VG+ ++ TY+P+E
Sbjct: 281 HQTFPLQLENGQ-------AMECTVAQYFKQKYSLQLKYP-HLPCLQVGQEQKHTYLPLE 332
Query: 404 LCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNY--GNEPMLKNCGIT 461
+C++V+ QR K LT Q S++++ + + +R +++ +K+++ G +P LK GI
Sbjct: 333 VCNIVAGQRCIKKLTDNQTSTMIKATARSAPDRQEEISRLVKSNSMVGGPDPYLKEFGIV 392
Query: 462 IASGFTQVEGRVLQAPRLKFG--NGEDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSARC 519
+ + T++ GRVL AP L++G N P G W++ K+ +I+ WAV F+ +
Sbjct: 393 VHNEMTELTGRVLPAPMLQYGGRNKTVATPNQGVWDMRGKQFYAGIEIKVWAVACFAPQK 452
Query: 520 DVR-----GLVRDLIKCARLKGIPIDEPYEEIFEENGQFRRAPPLVRVEKMFERIQKELP 574
R L K ++ G+PI + F + Q + VE MF+ ++
Sbjct: 453 QCREDLLKSFTDQLRKISKDAGMPIQG--QPCFCKYAQGADS-----VEPMFKHLKMTYV 505
Query: 575 GAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCISPTRV---NDQYLTNVLMKINAK 631
G ++ +LP + + +Y K+ G+ TQC+ V + Q L+N+ +KINAK
Sbjct: 506 GL-QLIVVILPGK--TPVYAEVKRVGDTLLGMATQCVQVKNVVKTSPQTLSNLCLKINAK 562
Query: 632 LGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPLISKYR 691
LGG+N+VL PS V + P I LG DV+H G PSIAAVV S + S+Y
Sbjct: 563 LGGINNVLVPHQRPS---VFQQPVIFLGADVTHPPAGDGKKPSIAAVVGSMDGHP-SRYC 618
Query: 692 ACVRTQSPKVEMIDNL-FKQVSEKEDEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQ 750
A VR Q+ + E+ L + Q ++ ++RELLI FY S+ + KP II +R GVSE Q
Sbjct: 619 ATVRVQTSRQEISQELLYSQEVIQDLTNMVRELLIQFYKST-RFKPTRIIYYRGGVSEGQ 677
Query: 751 FNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFF------QPGSPDNVPPGTV 804
QV EL I +AC L+E + P IV QK HHT+ F + G NVP GT
Sbjct: 678 MKQVAWPELIAIRKACISLEEDYRPGITYIVVQKRHHTRLFCADKTERVGKSGNVPAGTT 737
Query: 805 IDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTT 864
+D+ I HP DFY+C+HAG+ GTSRP+HY VL DD F+ DELQ L + L + Y R T
Sbjct: 738 VDSTITHPSEFDFYLCSHAGIQGTSRPSHYQVLWDDNCFTADELQLLTYQLCHTYVRCTR 797
Query: 865 AISVVAPICYAHLAATQIGQFMKFEDKSDTSSSHGGLTAAGVAP-VVPQLPKLQDSVSSS 923
++S+ AP YA L A + + +D SH + G P + + ++ +
Sbjct: 798 SVSIPAPAYYARLVAFRARYHLVDKDHDSAEGSHVSGQSNGRDPQALAKAVQIHHDTQHT 857
Query: 924 MFF 926
M+F
Sbjct: 858 MYF 860
>I2C2_RAT (Q9QZ81) Eukaryotic translation initiation factor 2C 2
(eIF2C 2) (eIF-2C 2) (Golgi ER protein 95 kDa) (GERp95)
Length = 860
Score = 382 bits (981), Expect = e-105
Identities = 274/886 (30%), Positives = 440/886 (48%), Gaps = 99/886 (11%)
Query: 45 PEPVKKKLPTRL--PIARKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRP 102
P P +P P R G+ G + L N F++ + D + ++ + +P
Sbjct: 12 PAPTTSPIPGYAFKPPPRPDFGTTGRTIKLQANFFEMDIPKIDIYHYELDI-------KP 64
Query: 103 VE-GKGVGRKVIDKVQETYGSELNG-KDFAYDGEKTLFTIGSL--ARNKLEFTVVLEDVI 158
+ + V R++++ + + + +++ G + +DG K L+T L R+K+E V L
Sbjct: 65 EKCPRRVNREIVEHMVQHFKTQIFGDRKPVFDGRKNLYTAMPLPIGRDKVELEVTLPGEG 124
Query: 159 SNRNNGNCSPDGASTNDSDKKRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQ 218
+R FKV I + + + LQA+ +AL G+
Sbjct: 125 KDR---------------------------IFKVSIKWVSCVSLQALHDALSGRLPSVPF 157
Query: 219 EAIRVLDIILRQHAAKQGCLLVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSL 278
E I+ LD+++R H V +SFF +GGG GFH S R + + L
Sbjct: 158 ETIQALDVVMR-HLPSMRYTPVGRSFFTASEGCSNPLGGGREVWFGFHQSVRPSLWKMML 216
Query: 279 NIDVSTTMIIQPGPVVDFLIA-------NQNVRDPFSLDWAKAKRTLKNLRIKASPSNQ- 330
NIDVS T + PV++F+ + + K + +K L+++ + Q
Sbjct: 217 NIDVSATAFYKAQPVIEFVCEVLDFKSIEEQQKPLTDSQRVKFTKEIKGLKVEITHCGQM 276
Query: 331 --EYKITGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPC 388
+Y++ ++ P QTF ++++ G E TV +YF + K+ LRY LPC
Sbjct: 277 KRKYRVCNVTRRPASHQTFPLQQESGQT-------VECTVAQYFKDRHKLVLRYP-HLPC 328
Query: 389 INVGKPKRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSN 448
+ VG+ ++ TY+P+E+C++V+ QR K LT Q S+++ + + +R +++ +++++
Sbjct: 329 LQVGQEQKHTYLPLEVCNIVAGQRCIKKLTDNQTSTMIRATARSAPDRQEEISKLMRSAS 388
Query: 449 YGNEPMLKNCGITIASGFTQVEGRVLQAPRLKFG--NGEDFNPRNGRWNLNNKKVVRPAK 506
+ +P ++ GI + T V GRVLQ P + +G N P G W++ NK+ +
Sbjct: 389 FNTDPYVREFGIMVKDEMTDVTGRVLQPPSILYGGRNKAIATPVQGVWDMRNKQFHTGIE 448
Query: 507 IEHWAVVNFSAR-----CDVRGLVRDLIKCARLKGIPIDEPYEEIFEENGQFRRAPPLVR 561
I+ WA+ F+ + ++ L K +R G+PI + F + Q +
Sbjct: 449 IKVWAIACFAPQRQCTEVHLKSFTEQLRKISRDAGMPIQG--QPCFCKYAQGADS----- 501
Query: 562 VEKMFERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCI---SPTRVND 618
VE MF ++ G ++ +LP + + +Y K+ G+ TQC+ + R
Sbjct: 502 VEPMFRHLKNTYAGL-QLVVVILPGK--TPVYAEVKRVGDTVLGMATQCVQMKNVQRTTP 558
Query: 619 QYLTNVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAV 678
Q L+N+ +KIN KLGG+N++L + P V + P I LG DV+H G PSIAAV
Sbjct: 559 QTLSNLCLKINVKLGGVNNIL---LPQGRPPVFQQPVIFLGADVTHPPAGDGKKPSIAAV 615
Query: 679 VSSREWPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRELLIDFYSSSGKRKPDN 738
V S + ++Y A VR Q + E+I +L ++RELLI FY S+ + KP
Sbjct: 616 VGSMD-AHPNRYCATVRVQQHRQEIIQDL---------AAMVRELLIQFYKST-RFKPTR 664
Query: 739 IIIFRDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFF------Q 792
II +RDGVSE QF QVL+ EL I EAC L++ + P IV QK HHT+ F +
Sbjct: 665 IIFYRDGVSEGQFQQVLHHELLAIREACIKLEKEYQPGITFIVVQKRHHTRLFCTDKNER 724
Query: 793 PGSPDNVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELV 852
G N+P GT +D KI HP DFY+C+HAG+ GTSRP+HYHVL DD FS DELQ L
Sbjct: 725 VGKSGNIPAGTTVDTKITHPTEFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILT 784
Query: 853 HSLSYVYQRSTTAISVVAPICYAHLAATQIGQFMKFEDKSDTSSSH 898
+ L + Y R T ++S+ AP YAHL A + + ++ SH
Sbjct: 785 YQLCHTYVRCTRSVSIPAPAYYAHLVAFRARYHLVDKEHDSAEGSH 830
>I2C2_MOUSE (Q8CJG0) Eukaryotic translation initiation factor 2C 2
(eIF2C 2) (eIF-2C 2) (Piwi/argonaute family protain
meIF2C2)
Length = 860
Score = 380 bits (976), Expect = e-105
Identities = 273/885 (30%), Positives = 438/885 (48%), Gaps = 97/885 (10%)
Query: 45 PEPVKKKLPTRL--PIARKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRP 102
P P +P P R G+ G + L N F++ + D + ++ + + RP
Sbjct: 12 PAPTTSPIPGYAFKPPPRPDFGTTGRTIKLQANFFEMDIPKIDIYHYELDIK---PEKRP 68
Query: 103 VEGKGVGRKVIDKVQETYGSELNG-KDFAYDGEKTLFTIGSL--ARNKLEFTVVLEDVIS 159
+ V R++++ + + + +++ G + +DG K L+T L R+K+E V L
Sbjct: 69 ---RRVNREIVEHMVQHFKTQIFGDRKPVFDGRKNLYTAMPLPIGRDKVELEVTLPGEGK 125
Query: 160 NRNNGNCSPDGASTNDSDKKRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQE 219
+R KV I + + + LQA+ +AL G+ E
Sbjct: 126 DR---------------------------ILKVSIKWVSCVSLQALHDALSGRLPSVPFE 158
Query: 220 AIRVLDIILRQHAAKQGCLLVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLN 279
I+ LD+++R H V +SFF +GGG GFH S R + + LN
Sbjct: 159 TIQALDVVMR-HLPSMRYTPVGRSFFTASEGCSNPLGGGREVWFGFHQSVRPSLWKMMLN 217
Query: 280 IDVSTTMIIQPGPVVDFLIA-------NQNVRDPFSLDWAKAKRTLKNLRIKASPSNQ-- 330
IDVS T + PV++F+ + + K + +K L+++ + Q
Sbjct: 218 IDVSATAFYKAQPVIEFVCEVLDFKSIEEQQKPLTDSQRVKFTKEIKGLKVEITHCGQMK 277
Query: 331 -EYKITGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCI 389
+Y++ ++ P QTF ++++ G E TV +YF + K+ LRY LPC+
Sbjct: 278 RKYRVCNVTRRPASHQTFPLQQESGQT-------VECTVAQYFKDRHKLVLRYP-HLPCL 329
Query: 390 NVGKPKRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNY 449
VG+ ++ TY+P+E+C++V+ QR K LT Q S+++ + + +R +++ ++++++
Sbjct: 330 QVGQEQKHTYLPLEVCNIVAGQRCIKKLTDDQTSTMIRATARSAPDRQEEISKLMRSASF 389
Query: 450 GNEPMLKNCGITIASGFTQVEGRVLQAPRLKFG--NGEDFNPRNGRWNLNNKKVVRPAKI 507
+P ++ GI + T V GRVLQ P + +G N P G W++ NK+ +I
Sbjct: 390 NTDPYVREFGIMVKDEMTDVTGRVLQPPSILYGGRNKAIATPVQGVWDMRNKQFHTGIEI 449
Query: 508 EHWAVVNFSAR-----CDVRGLVRDLIKCARLKGIPIDEPYEEIFEENGQFRRAPPLVRV 562
+ WA+ F+ + ++ L K +R G+PI + F + Q + V
Sbjct: 450 KVWAIACFAPQRQCTEVHLKSFTEQLRKISRDAGMPIQG--QPCFCKYAQGADS-----V 502
Query: 563 EKMFERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCI---SPTRVNDQ 619
E MF ++ G ++ +LP + + +Y K+ G+ TQC+ + R Q
Sbjct: 503 EPMFRHLKNTYAGL-QLVVVILPGK--TPVYAEVKRVGDTVLGMATQCVQMKNVQRTTPQ 559
Query: 620 YLTNVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVV 679
L+N+ +KIN KLGG+N++L + P V + P I LG DV+H G PSIAAVV
Sbjct: 560 TLSNLCLKINVKLGGVNNIL---LPQGRPPVFQQPVIFLGADVTHPPAGDGKKPSIAAVV 616
Query: 680 SSREWPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRELLIDFYSSSGKRKPDNI 739
S + ++Y A VR Q + E+I +L ++RELLI FY S+ + KP I
Sbjct: 617 GSMD-AHPNRYCATVRVQQHRQEIIQDL---------AAMVRELLIQFYKST-RFKPTRI 665
Query: 740 IIFRDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFF------QP 793
I +RDGVSE QF QVL+ EL I EAC L++ + P IV QK HHT+ F +
Sbjct: 666 IFYRDGVSEGQFQQVLHHELLAIREACIKLEKDYQPGITFIVVQKRHHTRLFCTDKNERV 725
Query: 794 GSPDNVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVH 853
G N+P GT +D KI HP DFY+C+HAG+ GT RP+HYHVL DD FS DELQ L +
Sbjct: 726 GKSGNIPAGTTVDTKITHPTEFDFYLCSHAGIQGTGRPSHYHVLWDDNRFSSDELQILTY 785
Query: 854 SLSYVYQRSTTAISVVAPICYAHLAATQIGQFMKFEDKSDTSSSH 898
L + Y R T ++S+ AP YAHL A + + ++ SH
Sbjct: 786 QLCHTYVRCTRSVSIPAPAYYAHLVAFRARYHLVDKEHDSAEGSH 830
>I2C2_RABIT (O77503) Eukaryotic translation initiation factor 2C 2
(eIF2C 2) (eIF-2C 2) (Fragment)
Length = 840
Score = 379 bits (972), Expect = e-104
Identities = 270/872 (30%), Positives = 434/872 (48%), Gaps = 97/872 (11%)
Query: 57 PIARKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVE-GKGVGRKVIDK 115
P R G+ G + L N F++ + D + ++ + +P + + V R++++
Sbjct: 6 PPPRPDFGTSGRTIKLQANFFEMDIPKIDIYHYELDI-------KPEKCPRRVNREIVEH 58
Query: 116 VQETYGSELNG-KDFAYDGEKTLFTIGSL--ARNKLEFTVVLEDVISNRNNGNCSPDGAS 172
+ + + +++ G + +DG K L+T L R K+E V L +R
Sbjct: 59 MVQHFKAQIFGDRKPVFDGRKNLYTAMPLPIGREKVELEVTLPGEGKDR----------- 107
Query: 173 TNDSDKKRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHA 232
FKV I + + + LQA+ +AL G+ E I+ LD+++R H
Sbjct: 108 ----------------IFKVSIKWVSCVSLQALHDALSGRLPSVPFETIQALDVVMR-HL 150
Query: 233 AKQGCLLVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGP 292
V +SFF +GGG GFH S R + + LNIDVS T + P
Sbjct: 151 PSMRYTPVGRSFFTASEGCSNPLGGGREVWFGFHQSVRPSLWKMMLNIDVSATAFYKAQP 210
Query: 293 VVDFLIA-------NQNVRDPFSLDWAKAKRTLKNLRIKASPSNQ---EYKITGLSELPC 342
V++F+ + + K + +K L+++ + Q +Y++ ++ P
Sbjct: 211 VIEFVCEVLDFKSIEEQQKPLTDSQRVKFTKEIKGLKVEITHCGQMKRKYRVCNVTRRPA 270
Query: 343 KEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTYVPV 402
QTF ++++ G E TV +YF + K+ LRY LPC+ VG+ ++ TY+P+
Sbjct: 271 SHQTFPLQQESGQT-------VECTVAQYFKDRHKLVLRYP-HLPCLQVGQEQKHTYLPL 322
Query: 403 ELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNYGNEPMLKNCGITI 462
E+C++V+ QR K LT Q S+++ + + +R +++ ++++++ +P ++ GI +
Sbjct: 323 EVCNIVAGQRCIKKLTDNQTSTMIRATARSAPDRQEEISKLMRSASFNTDPYVREFGIMV 382
Query: 463 ASGFTQVEGRVLQAPRLKFG--NGEDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSAR-- 518
T V GRVLQ P + +G N P G W++ NK+ +I+ WA+ F+ +
Sbjct: 383 KDEMTDVTGRVLQPPSILYGGRNKAIATPVQGVWDMRNKQFHTGIEIKVWAIACFAPQRQ 442
Query: 519 ---CDVRGLVRDLIKCARLKGIPIDEPYEEIFEENGQFRRAPPLVRVEKMFERIQKELPG 575
++ L K +R G+PI + F + Q + V MF ++ G
Sbjct: 443 CTEVHLKSFTEQLRKISRDAGMPIQG--QPCFCKYAQGADS-----VGPMFRHLKNTYAG 495
Query: 576 APSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCI---SPTRVNDQYLTNVLMKINAKL 632
++ +LP + + +Y K+ G+ TQC+ + R Q L+N+ +KIN KL
Sbjct: 496 L-QLVVVILPGK--TPVYAEVKRVGDTVLGMATQCVQMKNVQRTTPQTLSNLCLKINVKL 552
Query: 633 GGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPLISKYRA 692
GG+N++L + P V + P I LG DV+H G PSIAAVV S + ++Y A
Sbjct: 553 GGVNNIL---LPQGRPPVFQQPVIFLGADVTHPPAGDGKKPSIAAVVGSMD-AHPNRYCA 608
Query: 693 CVRTQSPKVEMIDNLFKQVSEKEDEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFN 752
VR Q + E+I +L ++RELLI FY S+ + KP II +RDGVSE QF
Sbjct: 609 TVRVQQHRQEIIQDL---------AAMVRELLIQFYKST-RFKPTRIIFYRDGVSEGQFQ 658
Query: 753 QVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFF------QPGSPDNVPPGTVID 806
QVL+ EL I EAC L++ + P IV QK HHT+ F + G N+P GT +D
Sbjct: 659 QVLHHELLAIREACIKLEKDYQPGITFIVVQKRHHTRLFCTDKNERVGKSGNIPAGTTVD 718
Query: 807 NKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAI 866
KI HP DFY+C+HAG+ GTSRP+HYHVL DD FS DELQ L + L + Y R T ++
Sbjct: 719 TKITHPTEFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRCTRSV 778
Query: 867 SVVAPICYAHLAATQIGQFMKFEDKSDTSSSH 898
S+ AP YAHL A + + ++ SH
Sbjct: 779 SIPAPAYYAHLVAFRARYHLVDKEHDSAEGSH 810
>I2C4_MOUSE (Q8CJF8) Eukaryotic translation initiation factor 2C 4
(eIF2C 4) (eIF-2C 4) (Piwi/argonaute family protain
meIF2C4)
Length = 861
Score = 375 bits (963), Expect = e-103
Identities = 281/903 (31%), Positives = 451/903 (49%), Gaps = 90/903 (9%)
Query: 57 PIARKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKV 116
P R GLG+ G + LL NHF+V + D + Y V + E RP + V R+V+D +
Sbjct: 15 PPRRPGLGTVGKPIRLLANHFQVQIPKID--VYHYDVDIKPEK-RP---RRVNREVVDTM 68
Query: 117 QETYGSELNG-KDFAYDGEKTLFTIGSL--ARNKLEFTVVLEDVISNRNNGNCSPDGAST 173
+ ++ G + YDG++ ++T L R++++ V L +G
Sbjct: 69 VRHFKMQIFGDRQPGYDGKRNMYTAHPLPIGRDRIDMEVTLPG------------EGKD- 115
Query: 174 NDSDKKRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHAA 233
+TFKV + + + LQ ++ AL G +E ++++ LD+I R H
Sbjct: 116 --------------QTFKVSVQWVSVASLQLLLEALAGHLNEVPDDSVQALDVITR-HLP 160
Query: 234 KQGCLLVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPV 293
V +SFF Y +GGG GFH S R + LNIDVS T + P+
Sbjct: 161 SMRYTPVGRSFFSPPEGYYHPLGGGREVWFGFHQSVRPAMWNMMLNIDVSATAFYRAQPI 220
Query: 294 VDFL-----IANQNVRDPFSLDWAKAKRT--LKNLRIKASPSNQ---EYKITGLSELPCK 343
++F+ I N N + D + K T ++ L+++ + Q +Y++ ++ P
Sbjct: 221 IEFMCEVLDIQNINEQTKPLTDSQRVKFTKEIRGLKVEVTHCGQMKRKYRVCNVTRRPAS 280
Query: 344 EQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTYVPVE 403
QTF ++ + G E TV +YF + L++ LPC+ VG+ ++ TY+P+E
Sbjct: 281 HQTFPLQLENGQ-------AMECTVAQYFKQKYSLQLKHP-HLPCLQVGQEQKHTYLPLE 332
Query: 404 LCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNY--GNEPMLKNCGIT 461
+C++V+ QR K LT Q S++++ + + +R +++ +K+++ G +P LK GI
Sbjct: 333 VCNIVAGQRCIKKLTDNQTSTMIKATARSAPDRQEEISRLVKSNSMVGGPDPYLKEFGIV 392
Query: 462 IASGFTQVEGRVLQAPRLKFG--NGEDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSARC 519
+ + T++ GRVL AP L++G N P G W++ K+ +I+ WAV F+ +
Sbjct: 393 VHNEMTELTGRVLPAPMLQYGGRNKTVATPSQGVWDMRGKQFYAGIEIKVWAVACFAPQK 452
Query: 520 DVR-----GLVRDLIKCARLKGIPIDEPYEEIFEENGQFRRAPPLVRVEKMFERIQKELP 574
R L K ++ G+PI + F + Q + VE MF+ ++
Sbjct: 453 QCREDLLKSFTDQLRKISKDAGMPIQG--QPCFCKYAQGADS-----VEPMFKHLKMTYV 505
Query: 575 GAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCI---SPTRVNDQYLTNVLMKINAK 631
G ++ +LP + + +Y K+ G+ TQC+ + + + Q L+N+ +K+NAK
Sbjct: 506 GL-QLIVVILPGK--TPVYAEVKRVGDTLLGMATQCVQIKNVVKTSPQTLSNLCLKMNAK 562
Query: 632 LGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPLISKYR 691
LGG+N+V PS V + P I LG DV+H G PSIAAVV S + S+Y
Sbjct: 563 LGGINNVPVPHQRPS---VFQQPVIFLGADVTHPPAGDGKKPSIAAVVGSMDGHP-SRYC 618
Query: 692 ACVRTQSPKVEMIDNL-FKQVSEKEDEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQ 750
A V Q+ + E+ L + Q ++ + RELLI FY S+ + KP II +R GVSE Q
Sbjct: 619 ATVWVQTSRQEIAQELLYSQEVVQDLTSMARELLIQFYKST-RFKPTRIIYYRGGVSEGQ 677
Query: 751 FNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFF------QPGSPDNVPPGTV 804
QV EL I +AC L+E + P IV QK HHT+ F + G NVP GT
Sbjct: 678 MKQVAWPELIAIRKACISLEEDYRPGITYIVVQKRHHTRLFCADKMERVGKSGNVPAGTT 737
Query: 805 IDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTT 864
+D+ + HP DFY+C+HAG+ GTSRP+HY VL DD F+ DELQ L + L + Y R T
Sbjct: 738 VDSTVTHPSEFDFYLCSHAGIQGTSRPSHYQVLWDDNCFTADELQLLTYQLCHTYVRCTR 797
Query: 865 AISVVAPICYAHLAATQIGQFMKFEDKSDTSSSHGGLTAAGVAP-VVPQLPKLQDSVSSS 923
++S+ AP YA L A + + +D SH + G P + + ++ +
Sbjct: 798 SVSIPAPAYYARLVAFRARYHLVDKDHDSAEGSHVSGQSNGRDPQALAKAVQIHHDTQHT 857
Query: 924 MFF 926
M+F
Sbjct: 858 MYF 860
>I2C2_HUMAN (Q9UKV8) Eukaryotic translation initiation factor 2C 2
(eIF2C 2) (eIF-2C 2)
Length = 851
Score = 375 bits (962), Expect = e-103
Identities = 262/820 (31%), Positives = 415/820 (49%), Gaps = 89/820 (10%)
Query: 108 VGRKVIDKVQETYGSELNG-KDFAYDGEKTLFTIGSL--ARNKLEFTVVLEDVISNRNNG 164
V R++++ + + + +++ G + +DG K L+T L R+K+E V L +R
Sbjct: 62 VNREIVEHMVQHFKTQIFGDRKPVFDGRKNLYTAMPLPIGRDKVELEVTLPGEGKDR--- 118
Query: 165 NCSPDGASTNDSDKKRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQEAIRVL 224
FKV I + + + LQA+ +AL G+ E I+ L
Sbjct: 119 ------------------------IFKVSIKWVSCVSLQALHDALSGRLPSVPFETIQAL 154
Query: 225 DIILRQHAAKQGCLLVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDVST 284
D+++R H V +SFF +GGG GFH S R + + LNIDVS
Sbjct: 155 DVVMR-HLPSMRYTPVGRSFFTASEGCSNPLGGGREVWFGFHQSVRPSLWKMMLNIDVSA 213
Query: 285 TMIIQPGPVVDFLIA-------NQNVRDPFSLDWAKAKRTLKNLRIKASPSNQ---EYKI 334
T + PV++F+ + + K + +K L+++ + Q +Y++
Sbjct: 214 TAFYKAQPVIEFVCEVLDFKSIEEQQKPLTDSQRVKFTKEIKGLKVEITHCGQMKRKYRV 273
Query: 335 TGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGKP 394
++ P QTF ++++ G E TV +YF + K+ LRY LPC+ VG+
Sbjct: 274 CNVTRRPASHQTFPLQQESGQT-------VECTVAQYFKDRHKLVLRYP-HLPCLQVGQE 325
Query: 395 KRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNYGNEPM 454
++ TY+P+E+C++V+ QR K LT Q S+++ + + +R +++ ++++++ +P
Sbjct: 326 QKHTYLPLEVCNIVAGQRCIKKLTDNQTSTMIRATARSAPDRQEEISKLMRSASFNTDPY 385
Query: 455 LKNCGITIASGFTQVEGRVLQAPRLKFG--NGEDFNPRNGRWNLNNKKVVRPAKIEHWAV 512
++ GI + T V GRVLQ P + +G N P G W++ NK+ +I+ WA+
Sbjct: 386 VREFGIMVKDEMTDVTGRVLQPPSILYGGRNKAIATPVQGVWDMRNKQFHTGIEIKVWAI 445
Query: 513 VNFSAR-----CDVRGLVRDLIKCARLKGIPIDEPYEEIFEENGQFRRAPPLVRVEKMFE 567
F+ + ++ L K +R G+PI + F + Q + VE MF
Sbjct: 446 ACFAPQRQCTEVHLKSFTEQLRKISRDAGMPIQG--QPCFCKYAQGADS-----VEPMFR 498
Query: 568 RIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCI---SPTRVNDQYLTNV 624
++ G ++ +LP + + +Y K+ G+ TQC+ + R Q L+N+
Sbjct: 499 HLKNTYAGL-QLVVVILPGK--TPVYAEVKRVGDTVLGMATQCVQMKNVQRTTPQTLSNL 555
Query: 625 LMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREW 684
+KIN KLGG+N++L + P V + P I LG DV+H G PSIAAVV S +
Sbjct: 556 CLKINVKLGGVNNIL---LPQGRPPVFQQPVIFLGADVTHPPAGDGKKPSIAAVVGSMD- 611
Query: 685 PLISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRELLIDFYSSSGKRKPDNIIIFRD 744
++Y A VR Q + E+I +L ++RELLI FY S+ + KP II +RD
Sbjct: 612 AHPNRYCATVRVQQHRQEIIQDL---------AAMVRELLIQFYKST-RFKPTRIIFYRD 661
Query: 745 GVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFF------QPGSPDN 798
GVSE QF QVL+ EL I EAC L++ + P IV QK HHT+ F + G N
Sbjct: 662 GVSEGQFQQVLHHELLAIREACIKLEKDYQPGITFIVVQKRHHTRLFCTDKNERVGKSGN 721
Query: 799 VPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYV 858
+P GT +D KI HP DFY+C+HAG+ GTSRP+HYHVL DD FS DELQ L + L +
Sbjct: 722 IPAGTTVDTKITHPTEFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQLCHT 781
Query: 859 YQRSTTAISVVAPICYAHLAATQIGQFMKFEDKSDTSSSH 898
Y R T ++S+ AP YAHL A + + ++ SH
Sbjct: 782 YVRCTRSVSIPAPAYYAHLVAFRARYHLVDKEHDSAEGSH 821
>I2C3_MOUSE (Q8CJF9) Eukaryotic translation initiation factor 2C 3
(eIF2C 3) (eIF-2C 3) (Piwi/argonaute family protain
meIF2C3)
Length = 860
Score = 372 bits (955), Expect = e-102
Identities = 276/878 (31%), Positives = 426/878 (48%), Gaps = 106/878 (12%)
Query: 60 RKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVID-KVQE 118
R G G+ G + LL N F+V + D + ++ + D P + V R+V+D KVQ
Sbjct: 20 RPGYGTMGKPIKLLANCFQVEIPKIDVYLYEVDIK---PDKCP---RRVNREVVDSKVQH 73
Query: 119 TYGSELNGKDFAYDGEKTLFTIGSLARNKLEFTVVLEDVISNRNNGNCSPDGASTNDSDK 178
+ + YDG+++L+T L + T V DV G P
Sbjct: 74 FKVTIFGDRRPVYDGKRSLYTANPL---PVATTGVDLDVTLPGEGGKDRP---------- 120
Query: 179 KRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQE--------AIRVLDIILRQ 230
FKV + F +++ + AL G E + +D++LR
Sbjct: 121 -----------FKVSVKFVSRVSWHLLHEALAGGTLPEPLELDKPVSTNPVHAVDVVLR- 168
Query: 231 HAAKQGCLLVRQSFFHNDPKNY-ADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQ 289
H V +SFF + P+ Y +GGG GFH S R + LNIDVS T +
Sbjct: 169 HLPSMKYTPVGRSFF-SAPEGYDHPLGGGREVWFGFHQSVRPAMWKMMLNIDVSATAFYK 227
Query: 290 PGPVVDFLI-------ANQNVRDPFSLDWAKAKRTLKNLRIKAS---PSNQEYKITGLSE 339
PV+ F+ ++ R K + +K L+++ + ++Y++ ++
Sbjct: 228 AQPVIQFMCEVLDIHNIDEQPRPLTDSHRVKFTKEIKGLKVEVTHCGTMRRKYRVCNVTR 287
Query: 340 LPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTY 399
P QTF ++ + G E TV +YF + L+Y LPC+ VG+ ++ TY
Sbjct: 288 RPASHQTFPLQLENGQTVER-------TVAQYFREKYTLQLKY-PHLPCLQVGQEQKHTY 339
Query: 400 VPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNYGNEPMLKNCG 459
+P+E+C++V+ QR K LT Q S++++ + + +R +++ ++++NY +P ++
Sbjct: 340 LPLEVCNIVAGQRCIKKLTDNQTSTMIKATARSAPDRQEEISRLVRSANYETDPFVQEFQ 399
Query: 460 ITIASGFTQVEGRVLQAPRLKFG--NGEDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSA 517
+ + V GRVL AP L++G N P +G W++ K+ +I+ WA+ F+
Sbjct: 400 LKVRDEMAHVTGRVLPAPMLQYGGRNRTVATPSHGVWDMRGKQFHTGVEIKMWAIACFAT 459
Query: 518 RCDVR-----GLVRDLIKCARLKGIPIDEPYEEIFEENGQ---FRRAPPLVRVEKMFERI 569
+ R G L K ++ G+PI GQ + A VE MF +
Sbjct: 460 QRQCREEILKGFTDQLRKISKDAGMPI----------QGQPCFCKYAQGADSVEPMFRHL 509
Query: 570 QKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCISPTRV---NDQYLTNVLM 626
+ G ++ +LP + + +Y K+ G+ TQC+ V + Q L+N+ +
Sbjct: 510 KNTYSGL-QLIIVILPGK--TPVYAEVKRVGDTLLGMATQCVQVKNVIKTSPQTLSNLCL 566
Query: 627 KINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPL 686
KIN KLGG+N++L PS V + P I LG DV+H G PSIAAVV S +
Sbjct: 567 KINVKLGGINNILVPHQRPS---VFQQPVIFLGADVTHPPAGDGKKPSIAAVVGSMD-AH 622
Query: 687 ISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRELLIDFYSSSGKRKPDNIIIFRDGV 746
S+Y A VR Q P+ E+I +L ++RELLI FY S+ + KP II +RDGV
Sbjct: 623 PSRYCATVRVQRPRQEIIQDL---------ASMVRELLIQFYKST-RFKPTRIIFYRDGV 672
Query: 747 SESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFF------QPGSPDNVP 800
SE QF QVL EL I EAC L++ + P IV QK HHT+ F + G N+P
Sbjct: 673 SEGQFRQVLYYELLAIREACISLEKDYQPGITYIVVQKRHHTRLFCADRTERVGRSGNIP 732
Query: 801 PGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQ 860
GT +D I HP DFY+C+HAG+ GTSRP+HYHVL DD F+ DELQ L + L + Y
Sbjct: 733 AGTTVDTDITHPYEFDFYLCSHAGIQGTSRPSHYHVLWDDNFFTADELQLLTYQLCHTYV 792
Query: 861 RSTTAISVVAPICYAHLAATQIGQFMKFEDKSDTSSSH 898
R T ++S+ AP YAHL A + + ++ SH
Sbjct: 793 RCTRSVSIPAPAYYAHLVAFRARYHLVDKEHDSAEGSH 830
>I2C1_HUMAN (Q9UL18) Eukaryotic translation initiation factor 2C 1
(eIF2C 1) (eIF-2C 1) (Putative RNA-binding protein Q99)
Length = 857
Score = 371 bits (953), Expect = e-102
Identities = 266/868 (30%), Positives = 433/868 (49%), Gaps = 95/868 (10%)
Query: 60 RKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQET 119
R G+G+ G + LL N+F+V + D + ++ + D P + V R+V++ + +
Sbjct: 26 RPGIGTVGKPIKLLANYFEVDIPKIDVYHYEVDIK---PDKCP---RRVNREVVEYMVQH 79
Query: 120 YGSELNG-KDFAYDGEKTLFTIGSL--ARNKLEFTVVLEDVISNRNNGNCSPDGASTNDS 176
+ ++ G + YDG+K ++T+ +L +++F V + +R
Sbjct: 80 FKPQIFGDRKPVYDGKKNIYTVTALPIGNERVDFEVTIPGEGKDR--------------- 124
Query: 177 DKKRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHAAKQG 236
FKV I + A + + + AL + E+++ LD+ +R H A
Sbjct: 125 ------------IFKVSIKWLAIVSWRMLHEALVSGQIPVPLESVQALDVAMR-HLASMR 171
Query: 237 CLLVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDF 296
V +SFF Y +GGG GFH S R + LNIDVS T + PV++F
Sbjct: 172 YTPVGRSFFSPPEGYYHPLGGGREVWFGFHQSVRPAMWKMMLNIDVSATAFYKAQPVIEF 231
Query: 297 L-----IANQNVRDPFSLDWAKAKRT--LKNLRIKASPSNQ---EYKITGLSELPCKEQT 346
+ I N N + D + + T +K L+++ + Q +Y++ ++ P QT
Sbjct: 232 MCEVLDIRNINEQPKPLTDSQRVRFTKEIKGLKVEVTHCGQMKRKYRVCNVTRRPASHQT 291
Query: 347 FTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTYVPVELCS 406
F ++ + G E TV +YF + L+Y LPC+ VG+ ++ TY+P+E+C+
Sbjct: 292 FPLQLESGQT-------VECTVAQYFKQKYNLQLKYP-HLPCLQVGQEQKHTYLPLEVCN 343
Query: 407 LVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNYGNEPMLKNCGITIASGF 466
+V+ QR K LT Q S++++ + + +R +++ +K ++Y +P ++ GI +
Sbjct: 344 IVAGQRCIKKLTDNQTSTMIKATARSAPDRQEEISRLMKNASYNLDPYIQEFGIKVKDDM 403
Query: 467 TQVEGRVLQAPRLKFG--NGEDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSARCDVR-- 522
T+V GRVL AP L++G N P G W++ K+ +I+ WA+ F+ + R
Sbjct: 404 TEVTGRVLPAPILQYGGRNRAIATPNQGVWDMRGKQFYNGIEIKVWAIACFAPQKQCREE 463
Query: 523 ---GLVRDLIKCARLKGIPIDEPYEEIFEENGQFRRAPPLVRVEKMFERIQKELPGAPSF 579
L K ++ G+PI + F + Q + VE MF ++ G
Sbjct: 464 VLKNFTDQLRKISKDAGMPIQG--QPCFCKYAQGADS-----VEPMFRHLKNTYSGL-QL 515
Query: 580 LLCLLPERKNSDLYGPWKKKNLAEYGIVTQCISPTRV---NDQYLTNVLMKINAKLGGLN 636
++ +LP + + +Y K+ G+ TQC+ V + Q L+N+ +KIN KLGG+N
Sbjct: 516 IIVILPGK--TPVYAEVKRVGDTLLGMATQCVQVKNVVKTSPQTLSNLCLKINVKLGGIN 573
Query: 637 SVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPLISKYRACVRT 696
++L + V + P I LG DV+H G PSI AVV S + S+Y A VR
Sbjct: 574 NILVPHQRSA---VFQQPVIFLGADVTHPPAGDGKKPSITAVVGSMD-AHPSRYCATVRV 629
Query: 697 QSPKVEMIDNLFKQVSEKEDEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLN 756
Q P+ E+I++L ++RELLI FY S+ + KP II +RDGV E Q Q+L+
Sbjct: 630 QRPRQEIIEDL---------SYMVRELLIQFYKST-RFKPTRIIFYRDGVPEGQLPQILH 679
Query: 757 IELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFF------QPGSPDNVPPGTVIDNKIC 810
EL I +AC L++ + P IV QK HHT+ F + G N+P GT +D I
Sbjct: 680 YELLAIRDACIKLEKDYQPGITYIVVQKRHHTRLFCADKNERIGKSGNIPAGTTVDTNIT 739
Query: 811 HPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAISVVA 870
HP DFY+C+HAG+ GTSRP+HY+VL DD F+ DELQ L + L + Y R T ++S+ A
Sbjct: 740 HPFEFDFYLCSHAGIQGTSRPSHYYVLWDDNRFTADELQILTYQLCHTYVRCTRSVSIPA 799
Query: 871 PICYAHLAATQIGQFMKFEDKSDTSSSH 898
P YA L A + + ++ SH
Sbjct: 800 PAYYARLVAFRARYHLVDKEHDSGEGSH 827
>I2C3_HUMAN (Q9H9G7) Eukaryotic translation initiation factor 2C 3
(eIF2C 3) (eIF-2C 3) (Argonaute 3)
Length = 860
Score = 370 bits (951), Expect = e-102
Identities = 274/878 (31%), Positives = 426/878 (48%), Gaps = 106/878 (12%)
Query: 60 RKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQET 119
R G G+ G + LL N F+V + D + ++ + D P + V R+V+D + +
Sbjct: 20 RPGYGAMGKPIKLLANCFQVEIPKIDVYLYEVDIK---PDKCP---RRVNREVVDSMVQH 73
Query: 120 YGSELNG-KDFAYDGEKTLFTIGSLARNKLEFTVVLEDVISNRNNGNCSPDGASTNDSDK 178
+ + G + YDG+++L+T L + T V DV G P
Sbjct: 74 FKVTIFGDRRPVYDGKRSLYTANPL---PVATTGVDLDVTLPGEGGKDRP---------- 120
Query: 179 KRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQE--------AIRVLDIILRQ 230
FKV I F +++ + L G+ E + +D++LR
Sbjct: 121 -----------FKVSIKFVSRVSWHLLHEVLTGRTLPEPLELDKPISTNPVHAVDVVLR- 168
Query: 231 HAAKQGCLLVRQSFFHNDPKNY-ADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQ 289
H V +SFF + P+ Y +GGG GFH S R + LNIDVS T +
Sbjct: 169 HLPSMKYTPVGRSFF-SAPEGYDHPLGGGREVWFGFHQSVRPAMWKMMLNIDVSATAFYK 227
Query: 290 PGPVVDFLI-------ANQNVRDPFSLDWAKAKRTLKNLRIKAS---PSNQEYKITGLSE 339
PV+ F+ ++ R K + +K L+++ + ++Y++ ++
Sbjct: 228 AQPVIQFMCEVLDIHNIDEQPRPLTDSHRVKFTKEIKGLKVEVTHCGTMRRKYRVCNVTR 287
Query: 340 LPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTY 399
P QTF ++ + G E TV +YF + L+Y LPC+ VG+ ++ TY
Sbjct: 288 RPASHQTFPLQLENGQTVER-------TVAQYFREKYTLQLKY-PHLPCLQVGQEQKHTY 339
Query: 400 VPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNYGNEPMLKNCG 459
+P+E+C++V+ QR K LT Q S++++ + + +R +++ ++++NY +P ++
Sbjct: 340 LPLEVCNIVAGQRCIKKLTDNQTSTMIKATARSAPDRQEEISRLVRSANYETDPFVQEFQ 399
Query: 460 ITIASGFTQVEGRVLQAPRLKFG--NGEDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSA 517
+ V GRVL AP L++G N P +G W++ K+ +I+ WA+ F+
Sbjct: 400 FKVRDEMAHVTGRVLPAPMLQYGGRNRTVATPSHGVWDMRGKQFHTGVEIKMWAIACFAT 459
Query: 518 RCDVR-----GLVRDLIKCARLKGIPIDEPYEEIFEENGQ---FRRAPPLVRVEKMFERI 569
+ R G L K ++ G+PI GQ + A VE MF +
Sbjct: 460 QRQCREEILKGFTDQLRKISKDAGMPI----------QGQPCFCKYAQGADSVEPMFRHL 509
Query: 570 QKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCISPTRV---NDQYLTNVLM 626
+ G ++ +LP + + +Y K+ G+ TQC+ V + Q L+N+ +
Sbjct: 510 KNTYSGL-QLIIVILPGK--TPVYAEVKRVGDTLLGMATQCVQVKNVIKTSPQTLSNLCL 566
Query: 627 KINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPL 686
KIN KLGG+N++L PS V + P I LG DV+H G PSIAAVV S +
Sbjct: 567 KINVKLGGINNILVPHQRPS---VFQQPVIFLGADVTHPPAGDGKKPSIAAVVGSMD-AH 622
Query: 687 ISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRELLIDFYSSSGKRKPDNIIIFRDGV 746
S+Y A VR Q P+ E+I +L ++RELLI FY S+ + KP II +RDGV
Sbjct: 623 PSRYCATVRVQRPRQEIIQDL---------ASMVRELLIQFYKST-RFKPTRIIFYRDGV 672
Query: 747 SESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFF------QPGSPDNVP 800
SE QF QVL EL I EAC L++ + P IV QK HHT+ F + G N+P
Sbjct: 673 SEGQFRQVLYYELLAIREACISLEKDYQPGITYIVVQKRHHTRLFCADRTERVGRSGNIP 732
Query: 801 PGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQ 860
GT +D I HP DFY+C+HAG+ GTSRP+HYHVL DD F+ DELQ L + L + Y
Sbjct: 733 AGTTVDTDITHPYEFDFYLCSHAGIQGTSRPSHYHVLWDDNCFTADELQLLTYQLCHTYV 792
Query: 861 RSTTAISVVAPICYAHLAATQIGQFMKFEDKSDTSSSH 898
R T ++S+ AP YAHL A + + ++ SH
Sbjct: 793 RCTRSVSIPAPAYYAHLVAFRARYHLVDKEHDSAEGSH 830
>I2C1_MOUSE (Q8CJG1) Eukaryotic translation initiation factor 2C 1
(eIF2C 1) (eIF-2C 1) (Piwi/argonaute family protain
meIF2C1)
Length = 857
Score = 364 bits (934), Expect = e-100
Identities = 261/868 (30%), Positives = 434/868 (49%), Gaps = 95/868 (10%)
Query: 60 RKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQET 119
R G+G+ G + LL N+F+V + D + ++ + D P + V R+V++ + +
Sbjct: 26 RPGIGTVGKPIKLLANYFEVDIPKIDVYHYEVDIK---PDKCP---RRVNREVVEYMVQH 79
Query: 120 YGSELNG-KDFAYDGEKTLFTIGSL--ARNKLEFTVVLEDVISNRNNGNCSPDGASTNDS 176
+ ++ G + YDG++ ++T+ +L +++F V + +R
Sbjct: 80 FKPQIFGDRKPVYDGKENIYTVTALPIGNERVDFEVTIPGEGKDR--------------- 124
Query: 177 DKKRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHAAKQG 236
FKV I + A + + + AL + E+++ LD+ +R H A
Sbjct: 125 ------------IFKVSIKWLAIVSWRMLHEALVSGQIPVPLESVQALDVAMR-HLASMR 171
Query: 237 CLLVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDF 296
V +SFF Y +GGG GFH S R + LNIDVS T + PV++F
Sbjct: 172 YTPVGRSFFSPPEGYYHPLGGGREVWFGFHQSVRPAMWKMMLNIDVSATAFYKAQPVIEF 231
Query: 297 LIANQNVRD----PFSLDWAKAKR---TLKNLRIKASPSNQ---EYKITGLSELPCKEQT 346
+ ++R+ P L ++ R +K L+++ + Q +Y++ ++ P QT
Sbjct: 232 MCEVLDIRNIDEQPKPLTDSQRVRFTKEIKGLKVEVTHCGQMKRKYRVCNVTRRPASHQT 291
Query: 347 FTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTYVPVELCS 406
F ++ + G E TV ++F + L+Y LPC+ VG+ ++ TY+P+E+C+
Sbjct: 292 FPLQLESGQT-------VECTVAQHFKQKYNLQLKYP-HLPCLQVGQEQKHTYLPLEVCN 343
Query: 407 LVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNYGNEPMLKNCGITIASGF 466
+V+ QR K LT Q S++++ + + +R +++ +K ++ +P ++ GI +
Sbjct: 344 IVAGQRCIKKLTDNQTSTMIKATARSAPDRQEEISRLMKNASCNLDPYIQEFGIKVKDDM 403
Query: 467 TQVEGRVLQAPRLKFG--NGEDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSARCDVR-- 522
T+V GRVL AP L++G N P G W++ K+ +I+ WA+ F+ + R
Sbjct: 404 TEVTGRVLPAPILQYGGRNRAIATPNQGVWDMRGKQFYNGIEIKVWAIACFAPQKQCREE 463
Query: 523 ---GLVRDLIKCARLKGIPIDEPYEEIFEENGQFRRAPPLVRVEKMFERIQKELPGAPSF 579
L K ++ G+PI + F + Q + VE MF ++ G
Sbjct: 464 VLKNFTDQLRKISKDAGMPIQG--QPCFCKYAQGADS-----VEPMFRHLKNTYSGL-QL 515
Query: 580 LLCLLPERKNSDLYGPWKKKNLAEYGIVTQCI---SPTRVNDQYLTNVLMKINAKLGGLN 636
++ +LP + + +Y K+ G+ TQC+ + + + Q L+N+ +KIN KLGG+N
Sbjct: 516 IIVILPGK--TPVYAEVKRVGDTLLGMATQCVQVKNAVKTSPQTLSNLCLKINVKLGGIN 573
Query: 637 SVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPLISKYRACVRT 696
++L + V + P I LG DV+H G PSI AVV S + S+Y A VR
Sbjct: 574 NILVPHQRSA---VFQQPVIFLGADVTHPPAGDGKKPSITAVVGSMD-AHPSRYCATVRV 629
Query: 697 QSPKVEMIDNLFKQVSEKEDEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLN 756
Q P+ E+I++L ++RELLI FY S+ + KP II +RDGV E Q Q+L+
Sbjct: 630 QRPRQEIIEDL---------SYMVRELLIQFYKST-RFKPTRIIFYRDGVPEGQLPQILH 679
Query: 757 IELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFF------QPGSPDNVPPGTVIDNKIC 810
EL I +AC L++ + P IV QK HHT+ F + G N+P GT +D I
Sbjct: 680 YELLAIRDACIKLEKDYQPGITYIVVQKRHHTRLFCADKNERIGKSGNIPAGTTVDTNIT 739
Query: 811 HPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAISVVA 870
HP DFY+C+HAG+ GTSRP+HY+VL DD F+ DELQ L + L + Y R T ++S+ A
Sbjct: 740 HPFEFDFYLCSHAGIQGTSRPSHYYVLWDDNRFTADELQILTYQLCHTYVRCTRSVSIPA 799
Query: 871 PICYAHLAATQIGQFMKFEDKSDTSSSH 898
P YA L A + + ++ SH
Sbjct: 800 PAYYARLVAFRARYHLVDKEHDSGEGSH 827
>AGO1_SCHPO (O74957) Cell cycle control protein ago1 (RNA
interference pathway protein ago1)
Length = 834
Score = 313 bits (803), Expect = 1e-84
Identities = 252/862 (29%), Positives = 409/862 (47%), Gaps = 124/862 (14%)
Query: 60 RKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVID----- 114
R G G G ++ L N F++ ++ + QY V + +G V RK
Sbjct: 12 RPGYGGLGKQITLKANFFQI-ISLPNETINQYHVIVG-------DGSRVPRKQSQLIWNS 63
Query: 115 -KVQETYGSELNGKDFAYDGEKTLFTIGSLARNKLEFTVVLEDVISNRNNGNCSPDGAST 173
+V++ +GS + + YDG ++ G +A ++ + E
Sbjct: 64 KEVKQYFGS--SWMNSVYDGRSMCWSKGDIADGTIKVNIGSES----------------- 104
Query: 174 NDSDKKRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQ--EAIRVLDIILRQH 231
H + + I ++KI L + + + S + Q +I LD++L++
Sbjct: 105 ------------HPREIEFSIQKSSKINLHTLSQFVNSKYSSDPQVLSSIMFLDLLLKKK 152
Query: 232 AAKQGCLLVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPG 291
++ SFF + N +GGGV +GF+ S R Q +S+N+D+S++ +
Sbjct: 153 PSET-LFGFMHSFFTGE--NGVSLGGGVEAWKGFYQSIRPNQGFMSVNVDISSSAFWRND 209
Query: 292 PVVDFLIAN---QNVRDPFSLDWAKAKRTLKNLRIKASP--------SNQEYKITGLSEL 340
++ L+ NVRD D + R + L++ +N+ Y I G S
Sbjct: 210 SLLQILMEYTDCSNVRDLTRFDLKRLSRKFRFLKVTCQHRNNVGTDLANRVYSIEGFSSK 269
Query: 341 PCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTYV 400
+ F + NGEE ++I+V EYF+ + L+Y +LPCI V K +
Sbjct: 270 SASDSFFVRRL----NGEE----QKISVAEYFLENHNVRLQYP-NLPCILV---KNGAML 317
Query: 401 PVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNYGNEPMLKNCGI 460
P+E C +V QRYT L + Q ++++ + Q+P ER+ ++ + ++ +P L G+
Sbjct: 318 PIEFCFVVKGQRYTAKLNSDQTANMIRFAVQRPFERVQQIDDFVHQMDWDTDPYLTQYGM 377
Query: 461 TIASGFTQVEGRVLQAPRLKFGNGEDFNPRNGRWNLNNKKVVRP--AKIEHWAVVNFSA- 517
I +V RVL+ P +++G P +GRWNL K+ + P A I WAV+ F++
Sbjct: 378 KIQKKMLEVPARVLETPSIRYGGDCIERPVSGRWNLRGKRFLDPPRAPIRSWAVMCFTST 437
Query: 518 -RCDVRGLVRDLIKCARLKGIPIDEPYEEIFEENGQ--FRRAPPLV------RVEKMFER 568
R +RG+ L + Y + G + PP++ VE++
Sbjct: 438 RRLPMRGIENFL------------QTYVQTLTSLGINFVMKKPPVLYADIRGSVEELCIT 485
Query: 569 IQK---ELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCISPTRV---NDQYLT 622
+ K ++ AP L + ++ + + YG K+ G+ +QC + QY
Sbjct: 486 LYKKAEQVGNAPPDYLFFILDKNSPEPYGSIKRVCNTMLGVPSQCAISKHILQSKPQYCA 545
Query: 623 NVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSR 682
N+ MKIN K+GG+N L + NP + VPT+ILG DV H G + + SIA++V+S
Sbjct: 546 NLGMKINVKVGGINCSLIPKSNP----LGNVPTLILGGDVYHPGVGATGV-SIASIVASV 600
Query: 683 EWPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRELLIDFYSSSGKRKPDNIIIF 742
+ KY A R+Q E+I+ + + I+ LL F + + K++P II F
Sbjct: 601 DLNGC-KYTAVSRSQPRHQEVIEGM---------KDIVVYLLQGFRAMT-KQQPQRIIYF 649
Query: 743 RDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFFQPGSPD----- 797
RDG SE QF V+N EL+QI EAC L +NPK LV QK HH +FF D
Sbjct: 650 RDGTSEGQFLSVINDELSQIKEACHSLSPKYNPKILVCTTQKRHHARFFIKNKSDGDRNG 709
Query: 798 NVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSY 857
N PGT+I+ + HP DFY+ +H + G S P HY VL D+I PD+ Q L ++L Y
Sbjct: 710 NPLPGTIIEKHVTHPYQYDFYLISHPSLQGVSVPVHYTVLHDEIQMPPDQFQTLCYNLCY 769
Query: 858 VYQRSTTAISVVAPICYAHLAA 879
VY R+T+A+S+V P+ YAHL +
Sbjct: 770 VYARATSAVSLVPPVYYAHLVS 791
>YO43_CAEEL (P34681) Hypothetical protein ZK757.3 in chromosome III
Length = 1040
Score = 266 bits (681), Expect = 1e-70
Identities = 183/610 (30%), Positives = 312/610 (51%), Gaps = 68/610 (11%)
Query: 332 YKITGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINV 391
YK+ L +LP + F +E+ + +V +YF + + L+Y LPC++V
Sbjct: 419 YKVNSL-QLPADKLMF-------QGIDEEGRQVVCSVADYF-SEKYGPLKYPK-LPCLHV 468
Query: 392 GKPKRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNYGN 451
G P R ++P+E C + S Q+Y K ++ Q S++++ + +R + + Q +++G
Sbjct: 469 GPPTRNIFLPMEHCLIDSPQKYNKKMSEKQTSAIIKAAAVDATQREDRIKQLAAQASFGT 528
Query: 452 EPMLKNCGITIASGFTQVEGRVLQAPRLKFG-NGEDFNP----RNGRWNLNNKKVVRPAK 506
+P LK G+ ++S Q RV+Q P + FG N NP ++G W ++N+ + PA
Sbjct: 529 DPFLKEFGVAVSSQMIQTTARVIQPPPIMFGGNNRSVNPVVFPKDGSWTMDNQTLYMPAT 588
Query: 507 IEHW---AVVNFSARCDVRGLVRDLIKCARLKGIPIDEPYEEIFEENGQFRRAPPLVRVE 563
+ A+V+ + ++ + L A G+ F R P LV+
Sbjct: 589 CRSYSMIALVDPRDQTSLQTFCQSLTMKATAMGM--------------NFPRWPDLVKYG 634
Query: 564 K-------MFERIQKELPGAPSFLLCLLP--ERKNSDLYGPWKKKNLAEYGIVTQCI--- 611
+ +F I E + C++ + KNSD+Y K+++ +GI++QC+
Sbjct: 635 RSKEDVCTLFTEIADEYRVTNTVCDCIIVVLQSKNSDIYMTVKEQSDIVHGIMSQCVLMK 694
Query: 612 SPTRVNDQYLTNVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQS- 670
+ +R N+++K+N K+GG+NS + + + +V + PT+++G+DV+H + +
Sbjct: 695 NVSRPTPATCANIVLKLNMKMGGINSRIVADKITNKYLVDQ-PTMVVGIDVTHPTQAEMR 753
Query: 671 -DIPSIAAVVSSREWPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRELLIDFYS 729
++PS+AA+V++ + L Y A V+ Q E + L IRE +I FY
Sbjct: 754 MNMPSVAAIVANVDL-LPQSYGANVKVQKKCRESVVYLLDA---------IRERIITFYR 803
Query: 730 SSGKRKPDNIIIFRDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTK 789
+ K+KP II++RDGVSE QF++VL E+ I AC + E + P IV QK HH +
Sbjct: 804 HT-KQKPARIIVYRDGVSEGQFSEVLREEIQSIRTACLAIAEDFRPPITYIVVQKRHHAR 862
Query: 790 FF------QPGSPDNVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGF 843
F G NVPPGT +D I P DFY+C+H G+ GTSRP YHVLLD+ F
Sbjct: 863 IFCKYQNDMVGKAKNVPPGTTVDTGIVSPEGFDFYLCSHYGVQGTSRPARYHVLLDECKF 922
Query: 844 SPDELQELVHSLSYVYQRSTTAISVVAPICYAHLAATQ----IGQFMKFEDKSDTSSSHG 899
+ DE+Q + + + + Y R T ++S+ P+ YA L AT+ + + + D +D ++
Sbjct: 923 TADEIQSITYGMCHTYGRCTRSVSIPTPVYYADLVATRARCHVKRKLGLADNNDCDTNSR 982
Query: 900 GLTAAGVAPV 909
T A + V
Sbjct: 983 SSTLASLLNV 992
Score = 37.4 bits (85), Expect = 0.19
Identities = 27/93 (29%), Positives = 45/93 (48%), Gaps = 2/93 (2%)
Query: 52 LPTRLPIARKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRK 111
L R +AR GLG+ G ++P+ +N F + + N QY V + + R ++ K R
Sbjct: 81 LNMREVVARPGLGTIGRQIPVKSNFFAMDLKNPKMVVIQYHVEIHHPGCRKLD-KDEMRI 139
Query: 112 VIDKVQETYGSELNGK-DFAYDGEKTLFTIGSL 143
+ K + + + K AYDG L+T+ L
Sbjct: 140 IFWKAVSDHPNIFHNKFALAYDGAHQLYTVARL 172
>AGO2_DROME (Q9VUQ5) Argonaute 2 protein
Length = 1214
Score = 241 bits (615), Expect = 6e-63
Identities = 215/866 (24%), Positives = 391/866 (44%), Gaps = 107/866 (12%)
Query: 41 LDLPPEPVKKKLPTRLPIARKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDG 100
L LPP+P I R +G G + N+ + ++ + Y V + E
Sbjct: 395 LPLPPQPAGS-------IKRGTIGKPGQ---VGINYLDLDLSKMPSVAYHYDVKIMPE-- 442
Query: 101 RPVEGKGVGRKVIDKVQETYGSELNGKDFAYDGEKTLFTIGSLARNKLEFTVVLEDVISN 160
RP K R+ ++ + +L G AYDG+ + +++ L N V + D
Sbjct: 443 RP---KKFYRQAFEQFRV---DQLGGAVLAYDGKASCYSVDKLPLNSQNPEVTVTD---- 492
Query: 161 RNNGNCSPDGASTNDSDKKRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQEA 220
NG + +R T +++ + + I L+++ + + + A
Sbjct: 493 -RNG--------------RTLRY-----TIEIKETGDSTIDLKSLTTYMNDRIFDKPMRA 532
Query: 221 IRVLDIILRQHAAKQGCLLVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLNI 280
++ ++++L + + R F +DP N ++ G G + +F LN+
Sbjct: 533 MQCVEVVLASPCHNKAIRVGRSFFKMSDPNNRHELDDGYEALVGLYQAFMLGDRPF-LNV 591
Query: 281 DVSTTMIIQPGPVVDFL---IANQNVRDPFSLDWAKA--KRTLKNLRIKASPSN------ 329
D+S P++++L + + +LD+++ + L+ + + +P
Sbjct: 592 DISHKSFPISMPMIEYLERFSLKAKINNTTNLDYSRRFLEPFLRGINVVYTPPQSFQSAP 651
Query: 330 QEYKITGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCI 389
+ Y++ GLS P +TF E D +++T+ YF + R L++ L C+
Sbjct: 652 RVYRVNGLSRAPASSETF----------EHDG--KKVTIASYF-HSRNYPLKFP-QLHCL 697
Query: 390 NVGKPKRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNY 449
NVG + +P+ELCS+ Q + Q +++++ + R + L+ +
Sbjct: 698 NVGSSIKSILLPIELCSIEEGQALNRKDGATQVANMIKYAATSTNVRKRKIMNLLQYFQH 757
Query: 450 GNEPMLKNCGITIASGFTQVEGRVLQAPRLKFGNGEDFNPRNGRWNLNNKKVVRPAKIEH 509
+P + GI IA+ F V RVL P++++ + +NG W ++ K + P H
Sbjct: 758 NLDPTISRFGIRIANDFIVVSTRVLSPPQVEYHSKRFTMVKNGSWRMDGMKFLEPKPKAH 817
Query: 510 WAVVNFSARCDVR-GLVRDLIKCARLKGIPIDE--PYEEIFEENGQFRRAPPLVRVEKMF 566
V + CD R G + + + I + + + +R P E+
Sbjct: 818 KCAVLY---CDPRSGRKMNYTQLNDFGNLIISQGKAVNISLDSDVTYR---PFTDDERSL 871
Query: 567 ERIQKELPGAPSFL-LCLLPERKNSDLYGPWKKKNLAEYGIVTQCISPTRV----NDQYL 621
+ I +L + L + ++P+ + S Y K+K ++GI+TQCI V N+Q +
Sbjct: 872 DTIFADLKRSQHDLAIVIIPQFRIS--YDTIKQKAELQHGILTQCIKQFTVERKCNNQTI 929
Query: 622 TNVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSS 681
N+L+KIN+KL G+N ++ +P +P++ T+ +G DV+H SP Q +IPS+ V +S
Sbjct: 930 GNILLKINSKLNGINHK--IKDDPRLPMMKN--TMYIGADVTHPSPDQREIPSVVGVAAS 985
Query: 682 REWPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRELLIDFYSSSGKRKPDNIII 741
+ P + Y R Q +E I+++F E + Y PD+II
Sbjct: 986 HD-PYGASYNMQYRLQRGALEEIEDMFSITLEH----------LRVYKEYRNAYPDHIIY 1034
Query: 742 FRDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFFQPGSP----- 796
+RDGVS+ QF ++ N EL I +AC + PK ++ K HHT+FF G
Sbjct: 1035 YRDGVSDGQFPKIKNEELRCIKQACDKVG--CKPKICCVIVVKRHHTRFFPSGDVTTSNK 1092
Query: 797 -DNVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSL 855
+NV PGTV+D I HP F+M +H + GT++PT Y+V+ + D LQ+L ++L
Sbjct: 1093 FNNVDPGTVVDRTIVHPNEMQFFMVSHQAIQGTAKPTRYNVIENTGNLDIDLLQQLTYNL 1152
Query: 856 SYVYQRSTTAISVVAPICYAHLAATQ 881
+++ R ++S AP AHL A +
Sbjct: 1153 CHMFPRCNRSVSYPAPAYLAHLVAAR 1178
>PIWI_BRARE (Q8UVX0) Piwi protein
Length = 858
Score = 154 bits (389), Expect = 1e-36
Identities = 161/642 (25%), Positives = 283/642 (44%), Gaps = 70/642 (10%)
Query: 264 GFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDFLIANQNVRDPFSLDWAKAKRTLKNLRI 323
GF ++ +S + L DVS +++ V+DF+ + + A K L L I
Sbjct: 249 GFMTTILQYESSIMLCSDVSHK-VLRSETVLDFMYSLRQQCGDQRFPEACTKE-LVGLII 306
Query: 324 KASPSNQEYKITGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYS 383
+N+ Y+I ++ TF KKG + EI+ YF + +D+
Sbjct: 307 LTKYNNKTYRIDDIAWDHTPNNTF---KKG---------DTEISFKNYFKSQYGLDITDG 354
Query: 384 ADLPCINVGK--------PKRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLE 435
+ ++ K P P + E C L L +A + + L +R P +
Sbjct: 355 NQVLLVSHVKRLGPSGRPPPGPAMLVPEFCYLTGLTDKMRADFNIMKD-LASHTRLSPEQ 413
Query: 436 RMNVLNQALKTSNYGNEPM--LKNCGITIASGFTQVEGRVLQAPRL-KFGNGEDFNPRNG 492
R +N+ + N + L G++ + + GRVL + R+ + G ++NP
Sbjct: 414 REGRINRLISNINRNGDVQNELTTWGLSFENKLLSLNGRVLPSERIIQGGRAFEYNPWTA 473
Query: 493 RWN--LNNKKVVRPAKIEHWAVVNFSARCDV-RGLVRDLIKCARLKGIPIDEPYEEIFEE 549
W+ + ++ +++W + DV + L++ L K + GI + +E+
Sbjct: 474 DWSKEMRGLPLISCMSLDNWLMFYTRRNADVAQSLLQTLNKVSGPMGIRMQRAVMIEYED 533
Query: 550 NGQFRRAPPLVRVEKMFERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQ 609
R E + +Q+ + ++ +LP + D Y KK + +Q
Sbjct: 534 -----------RQESLLRALQQNVARETQMVVVILPTNRK-DKYDCVKKYLCVDCPTPSQ 581
Query: 610 CI-SPTRVNDQYL----TNVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSH 664
C+ S T Q L T + +++N K+GG + VE IP+ +I+G+D H
Sbjct: 582 CVVSRTISKPQALMTVATKIALQMNCKMGG--ELWSVE----IPLRQ---LMIVGIDCYH 632
Query: 665 GSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRELL 724
+ SI A+V+S + + CV Q+ E+ID L + +G ++ L
Sbjct: 633 DTAAGKR--SIGAMVASLNQGMSRWFSKCV-LQNRGQEIIDALKGSL-----QGALKAYL 684
Query: 725 IDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQK 784
Y++S P II++RDGV + V++ E+ QI+++ K + + + PK V+V +K
Sbjct: 685 K--YNNS---LPSRIIVYRDGVGDGMLQSVVDYEVPQIMQSIKTMGQDYEPKLSVVVVKK 739
Query: 785 NHHTKFFQ--PGSPDNVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIG 842
++FF G N PPGTVID ++ P DF++ + A G PTHY+V+ D+ G
Sbjct: 740 RISSRFFARIDGKIANPPPGTVIDTEVTRPEWYDFFIVSQAVRFGCVAPTHYNVVFDNSG 799
Query: 843 FSPDELQELVHSLSYVYQRSTTAISVVAPICYAHLAATQIGQ 884
PD +Q L + L ++Y + V AP YAH A +GQ
Sbjct: 800 LKPDHMQRLTYKLCHMYYNWQGIVRVPAPCQYAHKLAFLVGQ 841
>PIWI_DROME (Q9VKM1) Piwi protein
Length = 843
Score = 135 bits (340), Expect = 5e-31
Identities = 153/687 (22%), Positives = 303/687 (43%), Gaps = 79/687 (11%)
Query: 221 IRVLDIILRQHAAKQGCLLVRQSFFHNDPKNYADVGGGVLGC-RGFHSSFRTTQSGLSLN 279
++VL++ILR+ LV ++ F DP+ ++ + G+ +S R + + L
Sbjct: 194 LQVLNLILRRSMKGLNLELVGRNLF--DPRAKIEIREFKMELWPGYETSIRQHEKDILLG 251
Query: 280 IDVSTTMIIQPGPVVDFLIANQNVRDPFSLDWAKAKRTLKNLRIKASPSNQEYKITGLSE 339
++ T +++ + D I + +P + + + + +L + +N+ Y+I +
Sbjct: 252 TEI-THKVMRTETIYD--IMRRCSHNP-ARHQDEVRVNVLDLIVLTDYNNRTYRINDVDF 307
Query: 340 LPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGKPKR--- 396
+ TF+ K + +I+ EY++ I +R I+ + K
Sbjct: 308 GQTPKSTFSCKGR------------DISFVEYYLTKYNIRIRDHNQPLLISKNRDKALKT 355
Query: 397 -----PTYVPVELCSLVSLQRYTKALTTLQR--SSLVEKSRQKPLERMNVLNQALKTSNY 449
+P ELC + L ++ L R SS + ++ +R+ N L+ +
Sbjct: 356 NASELVVLIP-ELCRVTGLNAEMRSNFQLMRAMSSYTRMNPKQRTDRLRAFNHRLQNTPE 414
Query: 450 GNEPMLKNCGITIASGFTQVEGRVLQAPRLKFGNGEDFNPRNGRWNLN---NKKVVRPAK 506
+ +L++ + + T+V+GR++ + F NG+ N W + + + P+
Sbjct: 415 SVK-VLRDWNMELDKNVTEVQGRIIGQQNIVFHNGKVPAGENADWQRHFRDQRMLTTPSD 473
Query: 507 -IEHWAVVNFSARC-DVRGLVRDLIKCARLKGIPIDEPYEEIF--EENGQFRRAPPLVRV 562
++ WAV+ ++R L+ L + A G+ I P E I + G + RA
Sbjct: 474 GLDRWAVIAPQRNSHELRTLLDSLYRAASGMGLRIRSPQEFIIYDDRTGTYVRA------ 527
Query: 563 EKMFERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCISPTRVNDQYLT 622
M + ++ + P +LCL+P N++ Y KK+ + + TQ ++ ++ L
Sbjct: 528 --MDDCVRSD----PKLILCLVPN-DNAERYSSIKKRGYVDRAVPTQVVTLKTTKNRSLM 580
Query: 623 NVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPT---IILGMDVSHGSPGQSDIPSIAAVV 679
++ KI +L N LG P + ++P + +G D++ + + + A++
Sbjct: 581 SIATKIAIQL---NCKLGYT-----PWMIELPLSGLMTIGFDIAKSTRDRKR--AYGALI 630
Query: 680 SSREWPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRELLIDFYSSSGKRKPDNI 739
+S + S Y + V S + + L+ +++ +R+ Y ++ P I
Sbjct: 631 ASMDLQQNSTYFSTVTECSAFDVLANTLWPMIAKA-----LRQ-----YQHEHRKLPSRI 680
Query: 740 IIFRDGVSESQFNQVLNIELNQIIEACKF---LDETWNPKFLVIVAQKNHHTKFFQPGSP 796
+ +RDGVS Q+ E+ IIE K + P+ IV ++ +T+FF G
Sbjct: 681 VFYRDGVSSGSLKQLFEFEVKDIIEKLKTEYARVQLSPPQLAYIVVTRSMNTRFFLNGQ- 739
Query: 797 DNVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLS 856
N PPGT++D+ I P DFY+ + GT PT Y+VL +G SP+++Q+L + +
Sbjct: 740 -NPPPGTIVDDVITLPERYDFYLVSQQVRQGTVSPTSYNVLYSSMGLSPEKMQKLTYKMC 798
Query: 857 YVYQRSTTAISVVAPICYAHLAATQIG 883
++Y + V A YA AT +G
Sbjct: 799 HLYYNWSGTTRVPAVCQYAKKLATLVG 825
>GCC7_CAEEL (Q21770) Germ cell expressed protein R06C7.1
Length = 945
Score = 97.1 bits (240), Expect = 2e-19
Identities = 181/804 (22%), Positives = 310/804 (38%), Gaps = 108/804 (13%)
Query: 125 NGKDFAYDGEKTLFTIGSLARNKLEFTVVLEDVISNRNNGNCSPDGASTNDSDKKRMRRP 184
+G YDG+ TL+T +L ++L+ V +GA T + D K + P
Sbjct: 148 DGNQIVYDGQSTLYTTVNLF-SELDANGTKSKVFQI--------NGADTGNDDLKTL--P 196
Query: 185 YHSKTFKVEISFAAKIPLQAIVNALRGQESE-NYQEAIRVLDIILRQHAAKQ----GCLL 239
S + + + + Q E N +E + L++ L QH ++ GC
Sbjct: 197 CISLEIYAPRDNSITLSSENLGKRTADQNIEVNNREYTQFLELALNQHCVRETNRFGCFE 256
Query: 240 VRQSFFHN------DPKNYADVGGGVLGCRGFHSSFRTT-------QSGLSLNIDVSTTM 286
+ +F N D ++ DVG G G + + Q+ SL ID
Sbjct: 257 HGKVYFLNATEEGFDQRDCVDVGDGKQLYPGLKKTIQFIEGPYGRGQNNPSLVIDGMKAA 316
Query: 287 IIQPGPVVD--FLIANQNVRDPFS-LDWAKAKRTLKNLRIKASPSNQE--YKITGLSELP 341
+ V+ F I Q+ + + + KA +K L ++ +N++ +I G+
Sbjct: 317 FHKEQTVIQKLFDITGQDPSNGLNNMTREKAAAVIKGLDCYSTYTNRKRHLRIEGIFHES 376
Query: 342 CKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRY-SADLP-CINVGKPKRPTY 399
+ F + + ++ EY+ + KI L+Y +A+L C + G Y
Sbjct: 377 ATKTRFELPDG-----------KTCSIAEYYADKYKISLQYPNANLVVCKDRGNNN---Y 422
Query: 400 VPVELCSLVSLQRYTKALTTLQRSSLVEKS---RQKPLERMNVLNQALKTSNYGNEPMLK 456
P EL ++ QR T T +S K +RM + + NE +L
Sbjct: 423 FPAELMTVSRNQRVTIPQQTGNQSQKTTKECAVLPDVRQRMIITGKNAVNITLENE-LLV 481
Query: 457 NCGITIASGFTQVEGRVLQAPRLKFGNGEDFNPRNGRWNLNNKKVVRPAKIEH----WAV 512
GI + S V+ R L L + + G+W V+PA + +AV
Sbjct: 482 ALGIKVYSEPLMVQARELDGKELVYQRSVMSDM--GKWRAPPGWFVKPATVPDLWAAYAV 539
Query: 513 VNFSARC---DVRGLVRDLIKCARLKGIPIDEPYEEIFEENGQFRRAPPLVRVEKMFERI 569
N R DV LV I + KG+ I P E L EK+ ++
Sbjct: 540 GNPGCRFSIGDVNQLVGMFIDSCKKKGMVIKPPCET------------GLYSTEKIMTQL 587
Query: 570 QKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCISPTRVNDQY-------LT 622
+K ++L ++ + L+ +K IV Q + ++ N L
Sbjct: 588 EKVAASKCKYVL-MITDDAIVHLHKQYKALEQRTMMIV-QDMKISKANAVVKDGKRLTLE 645
Query: 623 NVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSR 682
N++ K N KLGGLN + +I+G+ VS +P P+ +
Sbjct: 646 NIINKTNVKLGGLNYTVSDAKKSMTD-----EQLIIGVGVS--AP-----PAGTKYMMDN 693
Query: 683 EWPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRELL---IDFYSSSGKRKPDNI 739
+ L + E + + S ++ I ++L ID + + K P I
Sbjct: 694 KGHLNPQIIGFASNAVANHEFVGDFVLAPSGQDTMASIEDVLQNSIDLFEKNRKALPKRI 753
Query: 740 IIFRDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFFQP------ 793
II+R G SE +L E+ + + K + IV K H +FF+
Sbjct: 754 IIYRSGASEGSHASILAYEIP--LARAIIHGYSKEIKLIFIVVTKEHSYRFFRDQLRSGG 811
Query: 794 -GSPDNVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELV 852
+ N+PPG V+DN + +P F++ H + GT++ Y VL DD D L+EL
Sbjct: 812 KATEMNIPPGIVLDNAVTNPACKQFFLNGHTTLQGTAKTPLYTVLADDCKAPMDRLEELT 871
Query: 853 HSLSYVYQRSTTAISVVAPICYAH 876
+L + +Q + + S+ P+ A+
Sbjct: 872 FTLCHHHQIVSLSTSIPTPLYVAN 895
>YQ53_CAEEL (Q09249) Hypothetical protein C16C10.3 in chromosome III
Length = 1032
Score = 94.0 bits (232), Expect = 2e-18
Identities = 133/582 (22%), Positives = 238/582 (40%), Gaps = 83/582 (14%)
Query: 333 KITGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVG 392
+++ ++E + +F MK +D E E+TV EYF+ I L+Y LP +
Sbjct: 414 RVSSIAENNAENTSFMMK--------DDKGEREVTVAEYFLLQYNIKLKYPR-LPLVVSK 464
Query: 393 KPKRPTYVPVELCSLVSLQRY--TKALTTLQRSSLVEKSRQKPLERMNVLNQALKTS-NY 449
+ K ++ P+EL + QR K T+Q S++ ++ P + ++ L+ +
Sbjct: 465 RFKHESFFPMELLRIAPGQRIKVNKMSPTVQ-SAMTGRNASMPQHHVKLVQDILRDNLKL 523
Query: 450 GNEPMLKNCGITIASGFT-QVEGRVLQAPRLKFGNGEDFNPRNGRWNLNNK-KVVRPAKI 507
+ GI + S Q+ ++L ++KF G+ + P R + K V PA+I
Sbjct: 524 EQNKYMDAFGIKLMSTEPIQMTAKLLPPAQIKF-KGQTYMPDMSRPAFRTQDKFVEPARI 582
Query: 508 EHWAVVNFSARCDVR---GLVRDLIKCARLKGIPIDEPYEE--IFEENGQFRRAPPLVRV 562
+V F +R L R GI +++ + I E N A +
Sbjct: 583 RKIGIVVFDNCIQMRQAEDFCDKLSNFCRDNGITVEKDSRDWSIRELNSSDSVAIQNLMK 642
Query: 563 EKMFERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCISPTRVND---- 618
+ + +R+ +L + K D++ K G+ T + V+
Sbjct: 643 KWLDDRVD---------ILVGIAREKKPDVHDILKYFE-ESIGLQTIQLCQQTVDKMMGG 692
Query: 619 ----QYLTNVLMKINAKLGGLNSVLGVEMNPSI---PIVSKVPTI---------ILGMDV 662
Q + NV+ K N K GG N VE+ ++ + S T+ +G ++
Sbjct: 693 QGGRQTIDNVMRKFNLKCGGTNFF--VEIPNAVRGKAVCSNNETLRKKLLEHVQFIGFEI 750
Query: 663 SHG--------SPGQSD-IPSIAAVVSSREWPLISKYRACVRTQSP-KVEMIDNLFKQVS 712
SHG S Q D PS+ V S ++TQ K++ +D F +
Sbjct: 751 SHGASRTLFDRSRSQMDGEPSVVGVSYSLTNSTQLGGFTYLQTQKEYKLQKLDEFFPKC- 809
Query: 713 EKEDEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNIELNQIIEACKFLDET 772
+ Y K P I+I+R G E FN+V E+ ++ +
Sbjct: 810 ------------VRSYKEHSKTLPTRIVIYRVGAGEGNFNRVKE-EVEEMRRTFDKIQPG 856
Query: 773 WNPKFLVIVAQKNHHTKFFQP------GSPDNVPPGTVIDNKICHPRNNDFYMCAHAGMI 826
+ P +VI+AQ+ H + F + N+P GT ++N + ++F + + +I
Sbjct: 857 YRPHLVVIIAQRASHARVFPSCISGNRATDQNIPSGTCVENVLTSYGYDEFILSSQTPLI 916
Query: 827 GTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAISV 868
GT RP Y +L++D +S +EL L + ++ +Q S SV
Sbjct: 917 GTVRPCKYTILVNDAKWSKNELMHLTYFRAFGHQVSYQPPSV 958
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 116,757,868
Number of Sequences: 164201
Number of extensions: 5436309
Number of successful extensions: 20616
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 42
Number of HSP's successfully gapped in prelim test: 39
Number of HSP's that attempted gapping in prelim test: 19835
Number of HSP's gapped (non-prelim): 290
length of query: 927
length of database: 59,974,054
effective HSP length: 120
effective length of query: 807
effective length of database: 40,269,934
effective search space: 32497836738
effective search space used: 32497836738
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0031.13


